Abstract
Salmonella enterica serovar Enteritidis has emerged as a major health problem worldwide in the last few decades. DNA loci unique to S. Enteritidis can provide markers for detection of this pathogen and may reveal pathogenic mechanisms restricted to this serovar. An in silico comparison of 16 Salmonella genomic sequences revealed the presence of an ∼12.5-kb genomic island (GEI) specific to the sequenced S. Enteritidis strain NCTC13349. The GEI is inserted at the 5′ end of gene ydaO (SEN1377), is flanked by 308-bp imperfect direct repeats (attL and attR), and includes 21 open reading frames (SEN1378 to SEN1398), encoding primarily phage-related proteins. Accordingly, this GEI has been annotated as the defective prophage SE14 in the genome of strain NCTC13349. The genetic structure and location of φSE14 are conserved in 99 of 103 wild-type strains of S. Enteritidis studied here, including reference strains NCTC13349 and LK5. Notably, an extrachromosomal circular form of φSE14 was detected in every strain carrying this island. The presence of attP sites in the circular forms detected in NCTC13349 and LK5 was confirmed. In addition, we observed spontaneous loss of a tetRA-tagged version of φSE14, leaving an empty attB site in the genome of strain NCTC13349. Collectively, these results demonstrate that φSE14 is an unstable genetic element that undergoes spontaneous excision under standard growth conditions. An internal fragment of φSE14 designated Sdf I has been used as a serovar-specific genetic marker in PCR-based detection systems and as a tool to determine S. Enteritidis levels in experimental infections. The instability of this region may require a reassessment of its suitability for such applications.
The genus Salmonella comprises a heterogeneous group of Gram-negative bacteria, differentiable by biochemical and serological properties. More than 2,500 Salmonella serovars have been identified according to the serospecificities of the somatic and flagellar antigens. Some serovars, exemplified by Salmonella enterica serovar Typhimurium and S. Enteritidis, can infect a broad range of hosts. However, a subset of serovars, such as S. Typhi, a human-specific pathogen, show a high degree of adaptation to a specific host.
In the last few decades, S. Enteritidis has emerged as a major health problem worldwide (31). This pathogen colonizes the reproductive organs of infected birds without causing discernible illness and survives host defenses during the formation of the egg (25, 27). The production of a capsule-like O antigen structure by certain wild-type strains of S. Enteritidis (30, 46) has been associated with reproductive tract tropism and improved survival within eggs (26, 27, 45). Egg contamination can originate before oviposition by direct contamination of the yolk, albumen, or eggshell membranes with bacteria from the infected reproductive organs of the birds or after or during oviposition by penetration of bacteria from contaminated feces through the eggshell (8, 14, 25). Transmission of the bacterium to humans occurs mainly through the consumption of contaminated eggs or egg products (8, 14, 25). Upon infection of a human host, S. Enteritidis causes self-limiting gastroenteritis similar to that caused by other nontyphoidal Salmonella serovars.
According to information gathered from 84 countries responding to a global survey conducted by the World Health Organization (WHO), S. Enteritidis and S. Typhimurium accounted for ∼70% of all human and nonhuman isolates of Salmonella reported worldwide between 1995 and 2008. In fact, S. Enteritidis alone accounted for 61.4% of the ∼1.5 million human isolates of Salmonella reported during this period, according to the WHO Global Foodborne Infections Network Country Databank (http://www.who.int/salmsurv). Remarkably, S. Enteritidis is the second most prevalent cause of Salmonella infection in humans, after S. Typhimurium, in the United States (10).
The high global prevalence of S. Enteritidis makes the development of a rapid, sensitive, and highly specific detection system critical to collect accurate epidemiologic data. The identification of loci that serve as specific markers for DNA-based identification of this pathogen may also provide insights into pathogenic mechanisms restricted to this serovar. Genomic regions that are unique to given serovars are especially suitable for such epidemiologic detection (3). For instance, Agron and colleagues identified an S. Enteritidis-specific genomic region of ∼4,060 bp adjacent to the ydaO gene, carrying six open reading frames (ORFs) that they designated lygA to lygF (1). A PCR-based assay successfully detected the presence of an internal fragment of this serovar-specific region in most strains in a diverse collection of clinical and environmental S. Enteritidis isolates and not in 73 non-Enteritidis isolates of S. enterica representing 34 different serovars (1). Since then, this region has been widely used as an S. Enteritidis-specific molecular marker in the development of several PCR-based assays for detection and epidemiological typing of Salmonella serovars in clinical and environmental samples (2, 11, 32, 37, 44, 53). Recently, an S. Enteritidis-specific real-time quantitative PCR (qPCR) assay based on the detection of this region was developed (15). This qPCR assay has been used in a series of studies of the distribution and replication kinetics of S. Enteritidis in experimentally infected animals (16-21).
We performed a bioinformatic study to identify genomic regions specific to S. Enteritidis and found a genomic island (GEI) that includes the S. Enteritidis-specific locus lyg (1). This island has been annotated as the defective prophage SE14 in the genome of S. Enteritidis strain NCTC13349 (52). Although we demonstrate that the location in the genome and the overall genetic structure of the island are conserved in wild-type isolates of S. Enteritidis from different origins, we detected strains that do not carry the island in their genomes. Finally, we demonstrate here that the island corresponds to an unstable element that undergoes spontaneous excision from the genome of S. Enteritidis under standard growth conditions.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in the present study are listed in Table S1 in the supplemental material. Our analysis included 50 clinical strains of S. Enteritidis isolated in Santiago, Chile, and obtained from our laboratory collection and 33 strains of S. Enteritidis isolated from poultry and related products and obtained from Servicio Agrícola y Ganadero (SAG), Chile. These strains were confirmed to be S. enterica serovar Enteritidis by standard microbiological, biochemical, and serological methods. Bacteria were routinely grown in Luria-Bertani (LB) medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) at 37°C with aeration. LB medium was supplemented with ampicillin (Amp; 100 mg/liter), chloramphenicol (Cam; 20 mg/liter), kanamycin (Kan; 50 mg/liter), tetracycline (Tet; 10 mg/liter), trimethoprim (Tmp; 100 mg/liter), or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 50 mg/liter) as appropriate. N minimal medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.1 mM Tris-HCl (pH 7.4), 10 mM MgCl2] was supplemented with 0.2% (vol/vol) glycerol and 0.1% (wt/vol) Casamino Acids. Media were solidified by the addition of agar (15 g/liter).
Standard DNA techniques.
Total genomic DNA was obtained from fresh overnight cultures using the GenElute bacterial genomic DNA kit according to the instructions of the manufacturer (Sigma) and eluted with 1 mM Tris-HCl, pH 8.5, buffer. Plasmid DNA was obtained from fresh overnight cultures using the QIAprep spin miniprep kit (Qiagen) and eluted with the above-mentioned buffer. PCR products for cloning or sequencing were purified using the QIAquick PCR purification kit as recommended by the manufacturer (Qiagen). Restriction digestion using endonuclease HincII (Fermentas) and ligation using T4 DNA ligase (NEB) were conducted as recommended by the manufacturers. DNA samples were routinely analyzed by electrophoresis in 1% agarose gels (with 0.5× Tris-acetate-EDTA buffer) and visualized under UV light after ethidium bromide staining.
Bioinformatic identification of GEIs in S. Enteritidis.
The highest-percentage hit for any 100-bp window within each ORF has been used previously as a convenient metric to compare Salmonella genomes to one another and to those of members of the nearest genus, Escherichia (41, 49). This tool was applied to each annotated ORF in the genome sequence of S. Enteritidis NCTC13349, which was compared to genomic sequences of 15 other Salmonella strains (including S. enterica subsp. I serovar Typhimurium isolates LT2, SL1344, DT104, and 14028, serovar Typhi isolates Ty2 and CT18, serovar Paratyphi A SARB42, serovar Paratyphi B SPB7, serovar Paratyphi C RKS4594, serovar Gallinarum 281/91, serovar Dublin CT02021853, and serovar Choleraesuis SC-B67; S. enterica subsp. arizonae [subsp. IIIa] CDC346-86; S. enterica subsp. diarizonae [subsp. IIIb] CDC01-0005; and Salmonella bongori isolate 12419). An ORF was considered to be present in the analyzed genome if the best recorded hit for any internal 100-bp window showed 95 to 100% identity. This threshold was based on the observation that over 95% of syntenic reciprocal best hits have over 95% identity in their best 100-bp window of identity (41) (data not shown). ORFs were considered to be absent from the analyzed genome if the best recorded hit displayed less than 85% identity. This threshold was chosen based on the observation that syntenic reciprocal best hits in comparisons between Salmonella and the nearest other genus, Escherichia, average more than 85% identity in their best 100-base window of identity (41) (data not shown). S. Enteritidis NCTC13349 ORFs that were absent from any of the 15 compared genomes were designated S. Enteritidis specific.
PCR amplification.
Primers for PCR amplification were designed based on the reported sequence of S. Enteritidis PT4 strain NCTC13349 (52) (see Table S2 in the supplemental material) by using Vector NTI Advance 9.0 software. PCR amplifications were performed using a MultiGene TC9600-G thermal cycler (Labnet) and GoTaq Flexi DNA polymerase (Promega) in a standard volume of 20 μl. Reaction mixes contained 1× buffer, 2 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates, 200 nM (each) primers, 0.5 μl of template DNA (50 to 100 ng), and 0.5 U of DNA polymerase. Standard conditions for amplification were 3 min at 94°C, 30 cycles of incubation at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, and a final extension step at 72°C for 5 min. Conditions for tiling-PCR amplification were 3 min at 94°C, 30 cycles of incubation at 94°C for 30 s, 58°C for 30 s, and 72°C for 4 min, and a final extension step at 72°C for 7 min. Conditions for inverse and nested PCR amplification were 3 min at 94°C, 30 cycles of incubation at 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, and a final extension step at 72°C for 5 min. One microliter of a 1/100 dilution of the inverse PCR amplification mixture was used as template for the nested PCR amplification.
Mutant construction and analyses.
Mutant strains with specific deletions and concomitant insertions of a Kan, Cam, or Tet resistance cassette were constructed using the lambda Red recombination method (13). PCR primers 60 bases long were synthesized with 40 nucleotides (nt) on the 5′ ends corresponding to the ends of the desired deletions (see Table S2 in the supplemental material). In the case of mutants with deletions of gene SEN1378, the φSE14 attR site, or most of the island (genes SEN1379 to SEN1396A), the 3′ 20 nt of each primer was annealed to the 5′ or 3′ end of an antibiotic resistance cassette flanked by the FLP recombination target (FRT) sites present in plasmids pCLF2 and pCLF3 (Cam resistance), and pCLF4 (Kan resistance). In the case of mutants with deletions of genes SEN1385 to SEN1387 (i.e., φSE14::tetRA mutants), the 3′ 20 nt of each primer was annealed to the 5′ or 3′ end of a tetRA cassette from a T-POP insertion in the chromosome of S. Typhi SAS0137 (see Table S1 in the supplemental material). PCR amplifications were carried out under standard conditions using primers described in Table S2 in the supplemental material. S. Enteritidis strain NCTC13349 containing the plasmid pKD46 was grown to an optical density at 600 nm (OD600) of 0.5 at 30°C in LB medium containing Amp and l-arabinose (10 mM). Bacteria were made electrocompetent by sequential washes with ice-cold sterile 10% glycerol (42) and transformed with approximately 500 ng of each purified PCR product. Transformants were selected on LB agar plates containing Kan, Cam, or Tet at 37°C. The presence of each mutation was confirmed by PCR amplification using primers flanking the sites of substitution.
To generate a nonpolar in-frame deletion of gene SEN1378, leaving an intact copy of the φSE14 attL site, the ΔSEN1378::kan mutant was transformed with the temperature-sensitive plasmid pCP20, which synthesizes the FLP recombinase (12, 13). Transformants were selected at 30°C on LB agar plates containing Amp. A few colonies were streaked two times onto LB agar plates at 37°C and tested for the loss of the antibiotic resistance cassettes by being patched onto LB agar plates containing Kan and Amp. The presence of the ΔSEN1378::FRT mutant allele was confirmed by PCR amplification using primers flanking the sites of substitution. The same overall procedure was followed to generate a Δ(SEN1379-SEN1396A)::FRT derivative of the Δ(SEN1379-SEN1396A)::kan mutant.
To generate recA derivatives of NCTC13349, the mutant allele recA1 was cotransduced with the srl-203::Tn10dCam (Camr)- or srl-202::Tn10 (Tetr)-linked allele from S. Typhimurium donor strain MST1531 or MST1801 (see Table S1 in the supplemental material) by using the high-frequency transducing phage P22 HT105/1 int-201 as described previously (39). Transductants were selected at 37°C on LB agar plates containing Cam or Tet and screened for sensitivity to UV light.
Cloning of genes SEN1396A and SEN1398 and complementation of ΔattR mutants.
Primers ISE-Y13(F) and ISE-Y14(R) were used to amplify a 476-bp fragment of the NCTC13349 genome including genes SEN1396A and SEN1398 and the 3′ end of gene SEN1399. The PCR product was purified and ligated to plasmid vector pGEM-T Easy (Promega). Escherichia coli DH5α was transformed with the ligation mix, and transformants were selected on LB agar plates containing Amp and X-Gal. Plasmid DNA from several independent transformants was prepared, and the presence of the insert in each plasmid was confirmed by PCR amplification using primers T7 and SP6, flanking the cloning site in pGEM-T Easy. The orientation of the inserts was determined by PCR amplification using primer combinations ISE-Y13(F) plus SP6 and ISE-Y14(R) plus SP6. Plasmids having the insert cloned in each orientation into pGEM-T Easy were used to transform ΔattR derivatives of strain NCTC13349 in complementation assays for φSE14 excision.
Selection for spontaneous mutants missing φSE14.
Three independent NCTC13349 φSE14::tetRA clones were grown overnight in LB medium at 37°C with aeration. Each culture was serially diluted in sterile phosphate-buffered saline (PBS), and aliquots (5 to 10 μl) of the dilutions were plated onto LB agar plates and incubated overnight at 37°C to determine the number of CFU per milliliter of culture. From cultures suitably diluted in PBS, aliquots of 100 μl having ∼106 CFU were plated onto freshly made Bochner-Maloy medium (5 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl, 10 g/liter NaH2PO4·H2O, 50 mg/liter chlortetracycline hydrochloride, 12 mg/liter fusaric acid, 0.1 mM ZnCl2, 15 g/liter agar) (40). After 48 h of incubation at 37°C, colonies from the Bochner-Maloy plates were patched onto LB agar, replica plated onto LB agar containing tetracycline, and incubated overnight at 37°C. Each confirmed Tets derivative was tested for the presence of φSE14 by PCR-based detection of (i) each of four defined fragments of the element, (ii) the extrachromosomal circular form (CF), and (iii) the empty attB site left in the chromosome after the excision. The frequency of spontaneous excision and loss of φSE14 for each of the three independent NCTC13349 φSE14::tetRA clones was calculated as the total number of Tets CFU confirmed to have lost the island after selection on Bochner-Maloy plates, divided by the total number of CFU plated.
DNA sequence analyses of attB and attP sites postexcision.
PCR amplification products obtained from strains NCTC13349, LK5, MZ743, and MZ745 and three spontaneous mutants lacking φSE14 by using primers ISE-Y1(F) and ISE-Y8(R) and from strains NCTC13349 and LK5 by using primers ISE-Y11(F) and ISE-Y12(R) were purified and ligated to plasmid vector pGEM-T Easy. E. coli DH5α was transformed with the ligation mixes by electroporation, and transformants were selected on LB agar plates supplemented with Amp and X-Gal. Plasmid DNA was isolated from two independent clones from each transformation, and the double-stranded sequence of the cloned insert in each plasmid was determined by Macrogen(Korea) using primers T7 and SP6, flanking the cloning site in pGEM-T Easy.
Animal studies with BALB/c mice.
An Ampr derivative of mutant strain NCTC13349 Δ(SEN1379-SEN1396A)::FRT was tested for virulence in mixed infections with a Tmpr derivative of wild-type strain NCTC13349 to determine the competitive index (CI) in BALB/c mice after intraperitoneal infection. Strains used for infection were grown overnight at 37°C with aeration, mixed in a 1:1 ratio, and serially diluted in sterile PBS to the appropriate concentration for inoculation. Aliquots of the dilutions were plated onto LB medium-Tmp agar plates (for wild-type CFU) and LB medium-Amp agar plates (for mutant CFU) to determine the exact titers and ratios of strains inoculated. Groups of five BALB/c mice (8- to 10-week-old females) were inoculated intraperitoneally with ∼106 CFU from the mutant and wild-type strain mixture in 100 μl of PBS. Mice were humanely euthanized at 48 h postinfection, and the spleens and livers were removed and homogenized in 3 ml of ice-cold sterile PBS. Each organ homogenate was serially diluted, and aliquots of the dilutions were plated onto LB medium-Tmp and LB medium-Amp agar plates to determine the exact ratio of strains recovered. CIs were calculated as the ratio of mutant to wild-type CFU inoculated, divided by the ratio of mutant to wild-type CFU recovered from each organ.
RESULTS
Bioinformatic identification of an S. Enteritidis-specific GEI.
In order to identify genes and GEIs specific to S. Enteritidis, we performed an in silico comparison of the genome sequences available in public databases for 16 strains of Salmonella. Our study included data from sequenced strains of S. bongori, S. enterica subsp. enterica serovars Typhimurium, Typhi, Paratyphi A, Paratyphi B, Paratyphi C, Gallinarum, Enteritidis, Dublin, and Choleraesuis, S. enterica subsp. diarizonae, and S. enterica subsp. arizonae.
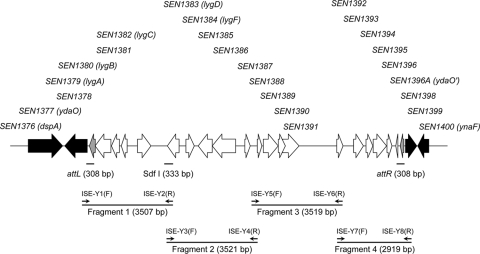
Our analysis revealed the presence of an ∼12.5-kb segment in the genome of the sequenced S. Enteritidis PT4 strain NCTC13349 that is absent in 14 of the other genomes studied. The only exception was the genome of S. Typhi strain CT18, which carried an internal fragment of this island presenting extensive sequence identity to prophage ST18 (Table 1). This GEI is inserted at the 5′ end of gene SEN1377 (also named ydaO or ttcA) in the genome of NCTC13349 and includes 21 ORFs (SEN1378 to SEN1398) (Fig. 1), most of which encode proteins of phage origin (Table 1). Notably, this GEI is flanked by relatively long, imperfect direct repeats (DR) of 308 bp (referred to as attL and attR) presenting six mismatches (see Fig. S2 in the supplemental material). Two ORFs in the GEI (SEN1378 and SEN1398) encode small putative proteins identical to the C-terminal end of the integrase encoded by the Rac prophage and related prophages in E. coli strains. Although SEN1378 and SEN1398 are likely to be pseudogenes, the presence of long attL and attR sites suggested that this GEI may be excised from the chromosome under certain conditions.
TABLE 1.
Genes and gene products of φSE14 in S. enterica serovar Enteritidis NCTC13349
| Gene | Alternative name(s) | Coordinatesa | Product size (aa)d | Predicted functional identification and/or description of product | Homolog encoded in S. Typhi strain CT18 (% identity to gene product) |
|---|---|---|---|---|---|
| SEN1377b | ydaO, ttcA | 1468428-1469363 (complement) | 311 | C32 tRNA thiolase (enzyme for thiolation of cytidine in position 32 of tRNA)e | |
| SEN1378c | int | 1469415-1469696 (complement) | 93 | Putative integrase (identical to portion of Rac prophage integrase); truncated protein | |
| SEN1379c | lygA | 1469680-1470303 (complement) | 207 | Putative phage-encoded exodeoxyribonuclease VIII; truncated version of RecE | |
| SEN1380 | lygB | 1470325-1470642 (complement) | 105 | Predicted phage protein with unknown function | |
| SEN1381 | kil | 1470727-1470948 (complement) | 73 | Putative FtsZ inhibitor protein Kil (analogous to Rac prophage protein); cell division inhibitor | |
| SEN1382 | lygC | 1471371-1471907 | 178 | Putative membrane protein related to phage superinfection exclusion proteins | |
| SEN1383 | lygD | 1472555-1473022 (complement) | 155 | Putative DNA binding protein belonging to the xenobiotic response element (XRE) family of transcriptional regulators | |
| SEN1384 | lygF | 1473295-1473624 | 109 | Putative phage-encoded DNA binding protein | |
| SEN1385 | 1473786-1474340 (complement) | 184 | Putative phage-encoded membrane protein with unknown function | Putative protein STY2058 (99) | |
| SEN1386 | 1474337-1475269 (complement) | 310 | Predicted phage protein with unknown function | Putative protein STY2057 (95) | |
| SEN1387 | hok, mok | 1475639-1475851 | 70 | Putative small toxic protein in the Hok/Gef family of prophage maintenance proteins | Putative protein STY2054 (92) |
| SEN1388 | 1476127-1476312 | 61 | Predicted phage protein with unknown function | ||
| SEN1389 | 1476375-1476974 | 199 | Hypothetical protein with unknown function | Putative protein STY2052 (87) | |
| SEN1390 | 1476974-1477264 | 96 | Putative phage protein with unknown function | Putative protein STY2051 (85) | |
| SEN1391 | 1477141-1477797 | 218 | Putative phage antitermination protein | Putative protein STY2050 (80) | |
| SEN1392 | 1479296-1479544 | 82 | Predicted phage protein with unknown function | ||
| SEN1393 | 1479968-1480363 | 131 | Putative phage protein homologous to plasmid and bacterial tellurite and colicin resistance determinants TerB and its homologs | ||
| SEN1394 | 1480457-1480744 | 95 | Putative prophage holin protein | ||
| SEN1395 | 1480741-1481286 | 181 | Predicted lysozyme protein | ||
| SEN1396 | 1481283-1481492 | 69 | Putative phage-encoded exported protein with unknown function | ||
| SEN1396Ac | ydaO′, ttcA′ | 1481723-1481776 (complement) | 17 | C32 tRNA thiolase (enzyme for thiolation of cytidine in position 32 of tRNA); truncated protein | |
| SEN1398c | int | 1481828-1481968 (complement) | 46 | Putative integrase (identical to portion of Rac prophage integrase); truncated protein |
Coordinates are those of the S. Enteritidis NCTC13349 sequence (EMBL accession number AM933172).
φSE14 carries 21 annotated genes (SEN1378 to SEN1398) in the genome of S. Enteritidis NCTC13349. The features of the neighboring gene SEN1377 (also called ydaO or ttcA) are included as a reference.
Predicted pseudogene.
aa, amino acids.
Data are from reference 36.
FIG. 1.
Schematic representation of φSE14 from S. Enteritidis strain NCTC13349. White arrows represent genes carried in the island, and black arrows represent neighboring genes in the chromosome of NCTC13349. Gray arrows indicate integrase genes. The positions of the DR (attL and attR) flanking the island and of the internal fragment Sdf I are indicated. The locations of primers designed to amplify four fragments of the island by tiling-PCR are shown.
In agreement with our in silico analysis, the 5′ end of the GEI we identified corresponds to the S. Enteritidis-specific locus lyg identified by Agron and colleagues (1). Recently, this GEI has been described as the prophage-like element φSE14 in the genome of S. Enteritidis strain NCTC13349 (coordinates 1469312 to 1482032), and this element was reported to be absent from the genomes of S. Typhimurium strain LT2 and S. Gallinarum strain 287/91 (52).
Genetic organization of φSE14 in wild-type strains of S. Enteritidis.
A set of eight primers based on the reported sequence of strain NCTC13349 was designed to study the genetic structure of φSE14 in S. Enteritidis strains by tiling-PCR. The primers were designed to amplify four fragments (2.9 to 3.5 kb) of the island with overlapping sequences of ∼100 to 150 bp (Fig. 1; see also Table S2 in the supplemental material). Two of the primers used, ISE-Y1(F) and ISE-Y8(R), are external to φSE14 (flanking attR and attL, respectively) so that we could confirm the locations of the island in the genomes of the isolates studied. Generation of an ∼700-bp PCR product using these external primers indicates the absence of φSE14 at the location corresponding to the position of this genetic element in strain NCTC13349.
Initially, PCR amplifications were performed to detect the defined fragments of φSE14 in the genomes of S. Enteritidis (NCTC13349 and LK5), S. Typhimurium (LT2, SL1344, and ATCC 14028), and S. Typhi (Ty2, CT18, SARB63, and SARB64) reference strains. Each fragment was detected in both S. Enteritidis strains, and none of them were detected in strains of S. Typhimurium and S. Typhi (data not shown). However, a strong PCR product of ∼700 bp from the three S. Typhimurium strains tested was obtained using the external primers ISE-Y1(F) and ISE-Y8(R), confirming the absence of φSE14 in these S. Typhimurium genomes. Furthermore, a specific product of ∼3 kb was obtained from the four S. Typhi strains we tested, reflecting the presence of an insert of ∼2.4 kb upstream of gene ydaO in these S. Typhi genomes, which confirms observations in previous reports of the sequences of CT18 and Ty2 strains (22, 47). In addition, a faint PCR product of ∼700 bp was obtained from both reference strains of S. Enteritidis (see below).
The four fragments of φSE14 were detected in 97 of 101 isolates in a collection of wild-type S. Enteritidis strains obtained from humans, rodents, birds, and poultry-related products (Fig. S2 in the supplemental material shows representative amplification data from a limited number of strains). Notably, the faint amplification product of ∼700 bp was also obtained from these 97 strains when PCR analyses were performed using primers external to φSE14. No fragment of the island was detected in the remaining four strains, all of which generated a strong PCR product of ∼700 bp with the external primers, confirming the absence of φSE14 in these genomes (see Fig. S2 in the supplemental material). In fact, in a previous study of differences in gene content among S. Enteritidis isolates using comparative genomic hybridizations (50), these strains (MZ743, MZ745, SARB17, and SARB18) showed no hybridization signals with any of the probes representing genes carried by φSE14.
The PCR products obtained for the four fragments of φSE14 from the 99 strains (including NCTC13349 and LK5) carrying the island were subjected to restriction fragment length polymorphism (RFLP) analysis using the restriction endonuclease HincII. Theoretically, the fragments presented three to four HincII sites in their sequences. After digestion, each fragment generated a unique RFLP pattern, and each set of corresponding fragments from the 99 strains tested had identical patterns (data not shown). These results indicate that there are no detectable differences in the genetic structure and the genome location of φSE14 in S. Enteritidis strains. Thus, the entire prophage-like element is either present or absent in the genomes of the S. Enteritidis strains studied, regardless of the source of isolation.
Detection of circular excision products of φSE14 and an empty chromosomal attB site in strains of S. Enteritidis.
The PCR products obtained from strains NCTC13349, LK5, MZ743, and MZ745 using external primers ISE-Y1(F) and ISE-Y8(R) were cloned, and the sequences of two independent clones from each strain were determined. The analysis of the sequence data revealed the presence of an empty attB site in the genomes of the strains analyzed. In all cases, 308-bp-long sites containing variable moieties of attL and attR at the 5′ and 3′ ends of attB, respectively, were detected (see Fig. S2A in the supplemental material). The attB sequences obtained from both clones of MZ743 were identical. The same was true for the attB sequences from both clones of MZ745, reflecting the unique recombination event that led to the excision of φSE14 in each strain. On the other hand, the attB sequences obtained from each pair of clones from NCTC13349 and LK5 were different (see Fig. S2A in the supplemental material), reflecting different events of excision by crossover occurring at different locations during the recombination between attL and attR sites in the bacterial populations. Collectively, these results strongly suggest that φSE14 undergoes spontaneous excision from the genome by recombination between the flanking attL and attR sites. If this is true, the recombination event must lead to the formation of an extrachromosomal CF of the island.
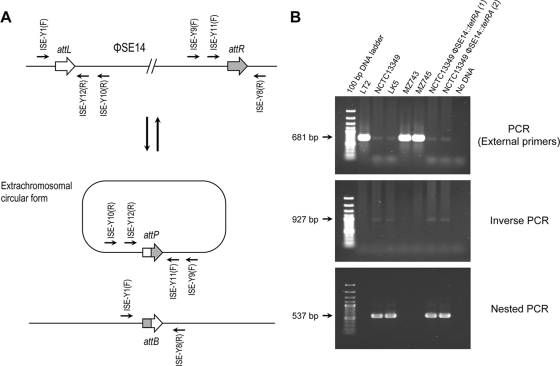
To test this hypothesis, an inverse PCR amplification approach was implemented. Primers ISE-Y9(F) and ISE-Y10(R) (see Table S2 in the supplemental material) were designed to generate a 927-bp PCR product only if a CF was formed after the excision of φSE14 (Fig. 2A). A second primer pair was designed to confirm the presence of the CF by nested PCR (see Table S2 in the supplemental material). Thus, primers ISE-Y11(F) and ISE-Y12(R) amplify a 581-bp fragment internal to the product of the first inverse PCR assay. As shown in Fig. 2B, both the inverse and nested PCR amplification assays gave products of the expected sizes from strains NCTC13349 and LK5, indicating the presence of a CF of φSE14 in both genetic backgrounds. This finding was observed for every S. Enteritidis strain carrying the island that we tested (data not shown). No amplification products were obtained from S. Typhimurium LT2 and S. Enteritidis strains MZ743 and MZ745, none of which carry φSE14 in their genomes.
FIG. 2.
The excision of φSE14 generates an extrachromosomal CF of the island. (A) Schematic representation of the excision event showing the locations of primers designed for the detection of the extrachromosomal element. (B) Detection of an extrachromosomal CF of φSE14 in S. Enteritidis strains. The PCR assays were performed as described in Materials and Methods by using primers ISE-Y1(F) and ISE-Y8(R) for external PCR, ISE-Y9(F) and ISE-Y10(R) for inverse PCR, and ISE-Y11(F) and ISE-Y12(R) for nested PCR. The sequence of each primer can be found in Table S2 in the supplemental material. Two independent clones harboring a tetRA-tagged version of φSE14 in the NCTC13349 background (φSE14::tetRA) were included in the analysis.
PCR products obtained from strains NCTC13349 and LK5 by using inverse primers ISE-Y11(F) and ISE-Y12(R) were cloned, and the sequences of two independent clones from each strain were determined. In each case, the analysis of the sequence data confirmed the presence of 308-bp-long attP sites containing variable moieties of attR and attL (see Fig. S2B in the supplemental material). The sequences obtained from each pair of clones from NCTC13349 and LK5 were different, revealing different excision events occurring in the bacterial populations sampled by the inverse PCR. These results strongly suggest that φSE14 excision leads to the generation of a circular intermediate. This intermediate was also detected when DNA from samples of NCTC13349 cultures in LB or N minimal medium collected throughout the growth phase was used as a template in the nested PCR assay (data not shown), indicating that the excision event occurs throughout the growth curve under different culture conditions.
Spontaneous excision of φSE14 in NCTC13349.
The Tn10-carried tetRA genes that encode resistance to tetracycline have been shown previously to confer sensitivity to fusaric acid, and this property can be used to select for the loss of the Tn10 marker (5). A modified Bochner medium (40) was used to select for mutant strains that had lost a tetRA-tagged version of φSE14 from their genomes. Derivatives of strain NCTC13349 with targeted substitutions of a tetRA cassette for genes SEN1385 to SEN1387 were constructed. The substitution was designed to keep the attL and attR sites of φSE14 at the same distance at which they exist in the NCTC13349 genome. As shown in Fig. 2B, the CF of φSE14 was detected in this genetic background using both the inverse and the nested PCR assays, indicating that the replacement of genes SEN1385 to SEN1387 by the tetRA cassette does not affect the excision of φSE14.
By using the modified Bochner medium, 24 Tets clones from NCTC13349 φSE14::tetRA were selected. The absence of the island in only three of these clones was confirmed by PCR. Thus, spontaneous excision and loss of the tetRA-tagged version of φSE14 in NCTC13349 occurred at a frequency of ∼10−6. In addition, a strong amplification product of ∼700 bp obtained from each of these three clones by using primers external to φSE14 was cloned and sequenced. The analysis of the sequence data confirmed the presence of an empty attB site in the genomes of the three strains analyzed (see Fig. S2C in the supplemental material). The attB sequences obtained from the clones were different, indicating that unique recombination events led to the excision of φSE14 in each strain. These results further confirmed that φSE14 excision occurs spontaneously in S. Enteritidis during growth under standard conditions.
Roles of SEN1378, SEN1398, attR, and RecA in φSE14 excision.
Derivatives of wild-type strain NCTC13349 were constructed with specific deletions of the SEN1378 gene, leaving an intact copy of the attL site in the genome. The CF of φSE14 was readily detected in these ΔSEN1378 mutants by using both the inverse and the nested PCR assays (see Fig. S3 in the supplemental material), indicating that SEN1378 is not required for φSE14 excision. Targeted deletions of a 283-bp internal fragment of attR (containing genes SEN1396A and SEN1398) were then constructed in the NCTC13349 genetic backgrounds. Both the inverse and the nested PCR assays failed to detect the CF in these ΔattR derivatives (see Fig. S4 in the supplemental material). This pattern was also observed when one of the ΔattR mutants was complemented in trans with a high-copy-number plasmid carrying genes SEN1396A and SEN1398 (data not shown). These results indicate that the integrity of the attR site is essential for the excision of φSE14 and that genes SEN1396A and SEN1398 are dispensable for the excision process. The fact that SEN1378 and SEN1398 are dispensable for the excision event is consistent with their description as pseudogenes (Table 1). Finally, to determine if RecA plays any role in the excision of φSE14, recA1 mutant derivatives of strain NCTC13349 were constructed and the presence of the CF was determined. The parental strain and the recA1 mutant derivatives produced a CF that was readily detected by our inverse and nested PCR assays (see Fig. S5 in the supplemental material), indicating that RecA recombinase is not required for the excision of φSE14.
Impact of φSE14 absence on the virulence of S. Enteritidis in mice.
To evaluate the role played by φSE14 in the pathobiology of S. Enteritidis, a derivative of strain NCTC13349 with an internal deletion of most of the island (genes SEN1379 to SEN1397) was generated. This mutant was tested for its ability to compete with the parental wild-type strain for organ colonization in Salmonella-susceptible BALB/c mice at 2 days postinfection. Notably, the mutant was capable of colonizing the liver (CI ± standard error of the mean [SEM] = 1.251 ± 0.1628) and spleen (CI ± SEM = 1.311 ± 0.1171) as efficiently as strain NCTC13349. Accordingly, data from a global screening of S. Enteritidis mutants under negative selection in vivo indicated that no gene carried in φSE14 is required for short-term colonization of BALB/c mice infected by the intraperitoneal route with a complex mixture of transposon insertion mutants in the NCTC13349 genetic background (C. A. Silva and C. A. Santiviago, unpublished data). Overall, these results indicate that the presence of φSE14 is dispensable for systemic colonization in our murine model of infection.
DISCUSSION
In this work, a serovar-specific GEI in the genome of S. Enteritidis was identified. Bioinformatic analyses revealed that this GEI carries mostly genes showing extensive homology to phage-related genes (Table 1). This genetic element has been annotated as the prophage SE14 in the genome of S. Enteritidis strain NCTC13349 (52), despite the fact that it carries no phage structural gene. Although we prefer to define this element as a GEI so as not to presuppose its function, we retained the φSE14 designation that has been given to this region previously (52).
We demonstrate here that φSE14 is an unstable genetic element that undergoes spontaneous excision from the genome under standard growth conditions. The excision of φSE14 was detected in a subpopulation of every strain carrying this island. Interestingly, the presence of S. Enteritidis subpopulations has been reported to play important roles in the adaptation of the bacterium to different niches, including animal hosts (26-29, 35). Thus, it is possible that somatic instability of a subset of the population is a feature that is exploited to improve the survival of the wild-type bacterium.
Excision of φSE14 occurs through recombination between the 308-bp-long attL and attR DR flanking the island. Evidence of different excision events occurring in the populations of individual tested strains revealed that recombination takes place at different locations in these imperfect DR. The presence of unusually long attL and attR sites argues against an illegitimate recombination event and supports homologous or specific recombination as the mechanism responsible for excision of this element. Homologous recombination between flanking large repeats in the chromosome occurs normally by the action of RecA and a set of alternative pathways. Although RecA is not required for φSE14 excision, we cannot eliminate the possible participation of alternative homologous recombination pathways that are known to participate in the excision of other genetic elements (33, 38). In addition, φSE14 harbors two genes (SEN1378 and SEN1398) that encode proteins identical to the C-terminal end of the integrase encoded by the Rac prophage. Our results show that SEN1378 and SEN1398 are dispensable for the excision of φSE14, probably because they are pseudogenes. Since cross talk between integrases encoded by different GEIs has been reported to occur (34), the participation of foreign integrases in the excision of φSE14 remains a possibility.
Excision of GEIs from the bacterial chromosome often leads to the formation of a detectable extrachromosomal CF (6, 9, 23, 34, 43). Although inverse PCR and sequence analyses indicate that spontaneous excision of φSE14 most likely results in the formation of a CF, we do not yet have direct physical evidence to prove the existence of such an excision product, as in the case of other unstable GEIs (4, 6). Furthermore, in addition to the possibility of CF formation, we cannot yet exclude the possibility that φSE14 can also exist as tandem repeats in the bacterial chromosome, as our inverse PCR products are also consistent with this possibility. Such tandem amplification has been described previously for several GEIs in different organisms (4, 24, 48, 51).
The location and overall genetic structure of φSE14 are remarkably conserved in S. Enteritidis strains, regardless of the geographic origins and sources of isolation of the strains. Of a collection of more than 100 wild-type S. Enteritidis strains isolated from humans, rodents, birds, and poultry-related products, only four strains do not carry this GEI. Two of these strains (MZ743 and MZ745) were isolated in the late 1940s (50), and the other two strains (SARB17 and SARB18) are part of the SARB reference collection assembled during the early 1980s (7). Since φSE14 is unstable and the excision of this element occurs spontaneously during growth under standard in vitro conditions, it is possible that these four strains may have lost φSE14 after extensive passage in the laboratory over decades. However, the presence of φSE14 in every recent isolate of S. Enteritidis and in the majority of the older strains analyzed suggests that strong selection occurs in vivo to maintain this island. Our data indicate that no gene carried in φSE14 is required for short-term colonization of systemic organs of BALB/c mice infected by the intraperitoneal route. Even though these observations argue against a major role for φSE14 in the virulence of S. Enteritidis in mice, we cannot rule out the possibility that some of these genes are required for the colonization of other niches in the host, including the intestinal tract and the oviduct, where the presence of this element may be under positive selection.
Finally, an internal fragment of φSE14, called Sdf I (Fig. 1), has been used widely as an S. Enteritidis-specific molecular marker in the development of several PCR-based detection and typing assays for Salmonella serovars in clinical and environmental samples (1, 2, 11, 32, 37, 44, 53). By using these assays, Sdf I has been detected only in S. Enteritidis strains, supporting our finding that φSE14 is an S. Enteritidis-specific locus. An S. Enteritidis-specific qPCR assay based on the detection of Sdf I was also recently developed and used to study the regular distribution patterns for S. Enteritidis in the gastrointestinal tracts and internal organs from orally infected mice (15, 17). This qPCR assay has been used extensively to study the replication kinetics of S. Enteritidis in internal organs and the distribution of the pathogen in the gastrointestinal tracts of experimentally infected ducks (16, 18-21). Although our findings support the specificity of φSE14 as a molecular marker for S. Enteritidis, the instability of this genetic element requires a reevaluation of its usefulness for quantitative applications until the in vivo stability of the island in different animal hosts can be determined.
Supplementary Material
Acknowledgments
We are indebted to Stanley Maloy (SDSU, San Diego, CA), Guido Mora (UNAB, Santiago, Chile), Juan Carlos Hormazábal (ISP, Chile), María Esther Saldías (SAG, Chile), and Francisco Silva (Hospital Clínico, Universidad de Chile) for generous gifts of bacterial strains. We thank Fred Long (VRISD) for invaluable help in bioinformatic analyses. We also thank Claudia Durán and Sabrita Chandía (ICBM) for assistance in microbiological techniques and Christine Shields (TAMU) for assistance in animal work.
This work was supported by grant ADI-08/2006 to I.C. from CONICYT (Chile) and the World Bank. C.J.B. and C.A.S. were supported by fellowships from CONICYT (Chile). M.M. and S.P. were supported partly by NIH grants R01AI073971 and R01AI052237. H.L.A.-P. was supported by NIH grants R21AI083964, R01AI083646, and R56AI077645.
Footnotes
Published ahead of print on 19 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agron, P. G., R. L. Walker, H. Kinde, S. J. Sawyer, D. C. Hayes, J. Wollard, and G. L. Andersen. 2001. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 67:4984-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, J., M. Sota, A. B. Vivanco, I. Perales, R. Cisterna, A. Rementeria, and J. Garaizar. 2004. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J. Clin. Microbiol. 42:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrach, N., S. Porwollik, P. Cheng, A. Cho, F. Long, S. H. Choi, and M. McClelland. 2008. Salmonella serovar identification using PCR-based detection of gene presence and absence. J. Clin. Microbiol. 46:2581-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barten, R., and T. F. Meyer. 2001. DNA circle formation in Neisseria gonorrhoeae: a possible intermediate in diverse genomic recombination processes. Mol. Gen. Genet. 264:691-701. [DOI] [PubMed] [Google Scholar]
- 5.Bochner, B. R., H. C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourhy, P., L. Salaun, A. Lajus, C. Medigue, C. Boursaux-Eude, and M. Picardeau. 2007. A genomic island of the pathogen Leptospira interrogans serovar Lai can excise from its chromosome. Infect. Immun. 75:677-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139(Pt. 6):1125-1132. [DOI] [PubMed] [Google Scholar]
- 8.Braden, C. R. 2006. Salmonella enterica serotype Enteritidis and eggs: a national epidemic in the United States. Clin. Infect. Dis. 43:512-517. [DOI] [PubMed] [Google Scholar]
- 9.Bueno, S. M., C. A. Santiviago, A. A. Murillo, J. A. Fuentes, A. N. Trombert, P. I. Rodas, P. Youderian, and G. C. Mora. 2004. Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi. J. Bacteriol. 186:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CDC. 2008. Salmonella surveillance: annual summary, 2006. U.S. Department of Health and Human Services, CDC, Atlanta, GA. http://www.cdc.gov/ncidod/dbmd/phlisdata/salmonella.htm.
- 11.Charlton, B. R., R. L. Walker, B. H. Kinde, C. R. Bauer, S. E. Channing-Santiago, and T. B. Farver. 2005. Comparison of a Salmonella Enteritidis-specific polymerase chain reaction assay to delayed secondary enrichment culture for the detection of Salmonella Enteritidis in environmental drag swab samples. Avian Dis. 49:418-422. [DOI] [PubMed] [Google Scholar]
- 12.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Buck, J., F. Van Immerseel, F. Haesebrouck, and R. Ducatelle. 2004. Colonization of the chicken reproductive tract and egg contamination by Salmonella. J. Appl. Microbiol. 97:233-245. [DOI] [PubMed] [Google Scholar]
- 15.Deng, S. X., A. C. Cheng, M. S. Wang, and P. Cao. 2007. Gastrointestinal tract distribution of Salmonella enteritidis in orally infected mice with a species-specific fluorescent quantitative polymerase chain reaction. World J. Gastroenterol. 13:6568-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng, S. X., A. C. Cheng, M. S. Wang, and P. Cao. 2008. Serovar-specific real-time quantitative detection of Salmonella Enteritidis in the gastrointestinal tract of ducks after oral challenge. Avian Dis. 52:88-93. [DOI] [PubMed] [Google Scholar]
- 17.Deng, S. X., A. C. Cheng, M. S. Wang, P. Cao, B. Yan, N. C. Yin, S. Y. Cao, and Z. H. Zhang. 2008. Quantitative studies of the regular distribution pattern for Salmonella enteritidis in the internal organs of mice after oral challenge by a specific real-time polymerase chain reaction. World J. Gastroenterol. 14:782-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng, S. X., A. C. Cheng, M. S. Wang, X. R. Li, and B. Yan. 2009. Replication kinetics of Salmonella enteritidis in internal organs of ducklings after oral challenge: a quantitative time-course study using real-time PCR. Vet. Res. Commun. 33:273-280. [DOI] [PubMed] [Google Scholar]
- 19.Deng, S. X., A. C. Cheng, M. S. Wang, B. Yan, N. C. Yin, S. Y. Cao, Z. H. Zhang, and P. Cao. 2008. The pathogenesis of Salmonella Enteritidis in experimentally infected ducks: a quantitative time-course study using TaqMan polymerase chain reaction. Poult. Sci. 87:1768-1772. [DOI] [PubMed] [Google Scholar]
- 20.Deng, S. X., A. C. Cheng, M. S. Wang, B. Yan, N. C. Yin, S. Y. Cao, Z. H. Zhang, and P. Cao. 2008. A study of the distribution patterns and levels of Salmonella Enteritidis in the immune organs of ducklings after oral challenge by serovar-specific real-time PCR. Avian Dis. 52:507-512. [DOI] [PubMed] [Google Scholar]
- 21.Deng, S. X., A. C. Cheng, M. S. Wang, and L. G. Ye. 2009. Quantitative analysis of Salmonella Enteritidis loads in ducklings after nasal inoculation. Poult. Sci. 88:1888-1892. [DOI] [PubMed] [Google Scholar]
- 22.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 24.Doublet, B., G. R. Golding, M. R. Mulvey, and A. Cloeckaert. 2008. Secondary chromosomal attachment site and tandem integration of the mobilizable Salmonella genomic island 1. PLoS One 3:e2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gantois, I., R. Ducatelle, F. Pasmans, F. Haesebrouck, R. Gast, T. J. Humphrey, and F. Van Immerseel. 2009. Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol. Rev. 33:718-738. [DOI] [PubMed] [Google Scholar]
- 26.Guard-Bouldin, J., R. K. Gast, T. J. Humphrey, D. J. Henzler, C. Morales, and K. Coles. 2004. Subpopulation characteristics of egg-contaminating Salmonella enterica serovar Enteritidis as defined by the lipopolysaccharide O chain. Appl. Environ. Microbiol. 70:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guard-Petter, J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421-430. [DOI] [PubMed] [Google Scholar]
- 28.Guard-Petter, J. 1998. Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl. Environ. Microbiol. 64:2166-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guard-Petter, J., L. H. Keller, M. M. Rahman, R. W. Carlson, and S. Silvers. 1996. A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol. Infect. 117:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guard-Petter, J., C. T. Parker, K. Asokan, and R. W. Carlson. 1999. Clinical and veterinary isolates of Salmonella enterica serovar Enteritidis defective in lipopolysaccharide O-chain polymerization. Appl. Environ. Microbiol. 65:2195-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herikstad, H., Y. Motarjemi, and R. V. Tauxe. 2002. Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 129:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera-Leon, S., J. R. McQuiston, M. A. Usera, P. I. Fields, J. Garaizar, and M. A. Echeita. 2004. Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. J. Clin. Microbiol. 42:2581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill, S. A., T. Woodward, A. Reger, R. Baker, and T. Dinse. 2007. Role for the RecBCD recombination pathway for pilE gene variation in repair-proficient Neisseria gonorrhoeae. J. Bacteriol. 189:7983-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochhut, B., C. Wilde, G. Balling, B. Middendorf, U. Dobrindt, E. Brzuszkiewicz, G. Gottschalk, E. Carniel, and J. Hacker. 2006. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol. Microbiol. 61:584-595. [DOI] [PubMed] [Google Scholar]
- 35.Humphrey, T. J., A. Williams, K. McAlpine, M. S. Lever, J. Guard-Petter, and J. M. Cox. 1996. Isolates of Salmonella enterica Enteritidis PT4 with enhanced heat and acid tolerance are more virulent in mice and more invasive in chickens. Epidemiol. Infect. 117:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jäger, G., R. Leipuviene, M. G. Pollard, Q. Qian, and G. R. Bjork. 2004. The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:750-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, S., J. G. Frye, J. Hu, P. J. Fedorka-Cray, R. Gautom, and D. S. Boyle. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 44:3608-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundblad, V., A. F. Taylor, G. R. Smith, and N. Kleckner. 1984. Unusual alleles of recB and recC stimulate excision of inverted repeat transposons Tn10 and Tn5. Proc. Natl. Acad. Sci. U. S. A. 81:824-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maloy, S. R. 1990. Experimental techniques in bacterial genetics. Jones and Bartlett Publishers, Boston, MA.
- 40.Maloy, S. R., and W. D. Nunn. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145:1110-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, S. Porwollik, A. Sabo, R. Meyer, T. Bieri, P. Ozersky, M. McLellan, C. R. Harkins, C. Wang, C. Nguyen, A. Berghoff, G. Elliott, S. Kohlberg, C. Strong, F. Du, J. Carter, C. Kremizki, D. Layman, S. Leonard, H. Sun, L. Fulton, W. Nash, T. Miner, P. Minx, K. Delehaunty, C. Fronick, V. Magrini, M. Nhan, W. Warren, L. Florea, J. Spieth, and R. K. Wilson. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. F. 1994. Bacterial transformation by electroporation. Methods Enzymol. 235:375-385. [DOI] [PubMed] [Google Scholar]
- 43.Murphy, R. A., and E. F. Boyd. 2008. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J. Bacteriol. 190:636-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Regan, E., E. McCabe, C. Burgess, S. McGuinness, T. Barry, G. Duffy, P. Whyte, and S. Fanning. 2008. Development of a real-time multiplex PCR assay for the detection of multiple Salmonella serotypes in chicken samples. BMC Microbiol. 8:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker, C. T., B. Harmon, and J. Guard-Petter. 2002. Mitigation of avian reproductive tract function by Salmonella enteritidis producing high-molecular-mass lipopolysaccharide. Environ. Microbiol. 4:538-545. [DOI] [PubMed] [Google Scholar]
- 46.Parker, C. T., E. Liebana, D. J. Henzler, and J. Guard-Petter. 2001. Lipopolysaccharide O-chain microheterogeneity of Salmonella serotypes Enteritidis and Typhimurium. Environ. Microbiol. 3:332-342. [DOI] [PubMed] [Google Scholar]
- 47.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 48.Pavlovic, G., V. Burrus, B. Gintz, B. Decaris, and G. Guedon. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759-774. [DOI] [PubMed] [Google Scholar]
- 49.Porwollik, S., J. Frye, L. D. Florea, F. Blackmer, and M. McClelland. 2003. A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 31:1869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porwollik, S., C. A. Santiviago, P. Cheng, L. Florea, and M. McClelland. 2005. Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J. Bacteriol. 187:6545-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravatn, R., S. Studer, D. Springael, A. J. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomson, N. R., D. J. Clayton, D. Windhorst, G. Vernikos, S. Davidson, C. Churcher, M. A. Quail, M. Stevens, M. A. Jones, M. Watson, A. Barron, A. Layton, D. Pickard, R. A. Kingsley, A. Bignell, L. Clark, B. Harris, D. Ormond, Z. Abdellah, K. Brooks, I. Cherevach, T. Chillingworth, J. Woodward, H. Norberczak, A. Lord, C. Arrowsmith, K. Jagels, S. Moule, K. Mungall, M. Sanders, S. Whitehead, J. A. Chabalgoity, D. Maskell, T. Humphrey, M. Roberts, P. A. Barrow, G. Dougan, and J. Parkhill. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trafny, E. A., K. Kozlowska, and M. Szpakowska. 2006. A novel multiplex PCR assay for the detection of Salmonella enterica serovar Enteritidis in human faeces. Lett. Appl. Microbiol. 43:673-679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.