Abstract
YidC depletion affects membrane protein insertion and leads to a defect in the growth of the Escherichia coli cell. We analyzed global changes in gene expression upon YidC depletion to determine the importance of YidC for cellular functions using a gene chip method to compare the transcriptomes of JS71 (control) and JS7131 (yidC depletion strain). Of the more than 4,300 genes identified, 163 were upregulated and 99 were downregulated upon YidC depletion, including genes which are responsible for DNA/RNA repair; energy metabolism; various transporters, proteases and chaperones; stress response; and translation and transcription functions. Real-time PCR was performed on selected genes to confirm the results. Specifically, we found upregulation of the genes encoding the energy transduction proteins F1Fo ATP synthase and cytochrome bo3 oxidase due to perturbation in assembly when YidC was depleted. We also determined that the high-level induction of the PspA stress protein under YidC depletion conditions is roughly 10-fold higher than the activation due to the addition of protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP), which dissipates the proton motive force. In addition, the gene chip data reveal the Cpx stress pathway is activated upon YidC depletion. The data show the broad physiological contribution of YidC to the bacterial cell and the considerable ramification to the cell when it is depleted.
According to genomic and proteomic analyses, approximately 20% of proteins encoded by the Escherichia coli genome are membrane proteins (4, 14), and these membrane proteins play important roles in energy metabolism, membrane transport of molecules, stress response, and other biological processes. All membrane proteins must be inserted and correctly folded in the membrane in order to function properly. Bacterial cells utilize specialized insertion machineries to facilitate these processes. In E. coli, membrane proteins are primarily inserted by the SecYEG translocase and YidC insertase (43).
The YidC insertase has been shown to be critical for the insertion of several Sec-independent proteins, such as the M13 procoat protein and F1Fo ATP synthase subunit c (2, 28, 29, 37, 38, 45). In addition to working independently, YidC was found to cooperate with the SecYEG translocase in catalyzing the membrane integration of F1Fo ATP synthase subunit a and cytochrome bo oxidase subunit II (CyoA) (1, 8, 36, 44). These results explain why depletion of YidC has a marked effect on the assembly of the cytochrome bo oxidase and F1Fo ATP synthase (39). YidC may also function as a chaperone for the folding of polytopic membrane proteins such as LacY (17) and MalF (41) and is required for the assembly of the maltose transport complex (41).
Recently, it was reported that YidC is present within a large-molecular-weight complex that includes the membrane protease, FtsH, a protease capable of degrading misassembled membrane proteins (35). The hypothesis is that this complex may function in the quality control of membrane proteins, with YidC functioning as a chaperone to fold membrane proteins and FtsH-degrading proteins that have not obtained their proper membrane conformation.
To better understand the importance of YidC to cell physiology, high-throughput methods such as gene chip and proteomic experiments can be performed. Such tools can help scientists understand the protein function and the cellular role of the protein by examining the global changes that occur in the transcriptome and the protein level in a cell when the protein under study is eliminated or reduced.
In this paper, we present a comprehensive analysis of the effects of YidC depletion in E. coli using the well-established JS71 and JS7131 strains. JS71 was derived from MC1060 and has 2 copies of yidC: the endogenous gene and one copy under the control of the arabinose-inducible pBAD promoter (28). JS7131 was constructed from JS71 where the original yidC gene on the chromosome was disrupted by a deletion and a complementary copy of yidC under the control of an araBAD promoter was incorporated into the chromosome. Thereby, the expression of YidC in JS7131 can be induced with arabinose and repressed with glucose (28). Samples were collected 5 h after the addition of glucose, when the growth defect was evident, to examine primary effects from the absence of YidC and secondary effects from the absence of YidC's substrates.
A gene chip and two-dimensional (2D) gel electrophoresis method was utilized to analyze the change of the gene expression pattern and protein pattern, respectively, when YidC was depleted from the cell. Real-time PCR and Western blot analysis were used to confirm the microarray and proteomic analysis studies, respectively. The growth, phenotype, and motility of the cells were also analyzed. This study provides important information on how the bacterial cell responds to a defect in membrane protein biogenesis. It is also the first study that describes the effect of eliminating YidC function on the level of the transcriptome and proteome and on cell physiology.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli JS71 and JS7131 strains (28) are from our laboratory collection. JS71 was used as the control strain and JS7131 was used as the experimental strain in this study. Starter cultures for both strains were grown overnight in LB medium from single colonies at 37°C, and 0.2% arabinose was added to JS7131. Before back dilution, the JS7131 culture was washed with LB medium three times to remove all traces of arabinose. Overnight cultures were then diluted 1 to 50 into LB media plus 0.2% glucose. After dilution, both JS71 and JS7131 were vigorously shaken and grown for 5 h before cells were harvested.
Growth curve and microscopy.
Cells were grown as described above with 3 ml of cultures of each strain to obtain the growth curve. The growth of the cells was monitored by measuring the optical density at 600 nm (OD600) at 1 h after back dilution and then each hour thereafter for 12 h. Cell cultures were grown in LB-glucose medium for 5 h for both JS71 (YidC expression condition) and JS7131 (YidC depletion condition) strains and stained using 0.3% methylene blue for better visualization of the cells under the microscope. Pictures were taken using a Nikon FDX-35 digital camera attached to the Nikon Eclipse E800 microscope at a ×100 magnification.
Cell motility assay.
LB motility agar plates were used to compare the motility of JS71 expressing normal YidC levels and JS7131 after YidC depletion. The LB motility agar was made with 2.5% LB broth Miller (Novagen) and 0.3% agar. Glucose (0.2%) and arabinose (0.002%) were added to the plates to ensure that JS7131 survived the subsequent long-time incubation, while the expression of YidC protein was kept at a low level. After 5 h of YidC depletion in liquid culture, equal amounts of JS71 and JS7131 cells (normalized using a microscope and counting chamber) were added to the LB motility agar plates and the plates were incubated at 37°C for 24 h. The diameters of the swimming zones formed by the motile bacteria were measured to determine their motility.
RNA and cDNA preparation for gene chip experiments.
Cells were grown in 10-ml cultures of each strain for RNA preparation. Cells were mixed with 2 volumes of RNAprotect bacterial reagent (Qiagen) to stabilize mRNA. An RNeasy minikit was used to purify mRNA followed by on column DNase digestion. The quality of the purified total RNA was checked by measuring OD260 and OD280. Samples were either stored at −20°C or used immediately for cDNA preparation.
cDNA was synthesized using high-capacity cDNA reverse transcription kits (Applied Biosystems). RNase inhibitor was added to prevent the degradation of RNA during PCR cycles. The reaction was carried out in an MJ Research Minicycler. The conditions were 25°C for 10 min, 37°C for 120 min, and 85°C for 5 s for 30 cycles. cDNA was then purified from the reaction mix by using the QIAquick PCR purification kit. The concentration of cDNA was measured at OD260 using an extinction coefficient of 35 ng/μl per OD unit.
Gene chip experiments and data analysis.
The gene chip experiments and raw data analysis were performed in the Biomedical Genomics core at the Columbus Children's Hospital. Total RNA was checked with a capillary electrophoresis Bioanalyzer 2100 (Agilent) for integrity and measured with a Nanodrop 1000 spectrophotometer (Nanodrop, Wilmington, DE) for quantification. Biotin-labeled cDNA was then synthesized from total RNA using the Ovation biotin RNA amplification and labeling system (NuGen Technologies, Inc., San Carlos, CA) following the manufacturer's protocol. Twenty-five nanograms of total RNA was used for synthesizing the first-strand cDNA, followed by double-stranded cDNA amplification. The amplified cDNA was purified using a NucleoSpin extract kit (BD Clontech). Fragmentation and biotin labeling were achieved using NuGEN FL-Ovation cDNA biotin module V2 (NuGen Technologies, Inc., San Carlos, CA). The quality and quantity of the fragmented/biotinylated cDNA were analyzed using the Nanodrop spectrophotometer and Bioanalyzer 2100, respectively.
E. coli Genome 2.0 GeneChips (Affymetrix, Inc.) were used for this experiment, with approximately 10,000 probe sets for all 20,366 genes that are present in four strains of E. coli. For each gene chip, 2 μg of cDNA was applied. The hybridization process was carried out at 45°C for 16 h. The gene chip was then washed and stained in a Fluidics Station 450 machine (Affymetrix, Inc.). A GeneChip Scanner 3000 (Affymetrix, Inc.) was used to scan the gene chip, and the data files were obtained using GeneChip Operating Software (GCOS) (Affymetrix, Inc.). Signals for all probe sets were generated by software ArrayAssist 3.4 (Stratagene) using the RMA algorithm. Three biological replicates where RNA was prepared from three independent cultures were performed for each experimental group (JS7131) and control group (JS71). The P value was used to assess the statistical significance of that estimate. The value ranges from 0 to 1, with a smaller P value providing more confidence in the regulation pattern. For this study, data with P values of <0.05 are generally considered confident and reliable. DAVID tools (http://david.abcc.ncifcrf.gov/home.jsp) were used for the functional group analysis. Only genes with a >2-fold change in expression levels were submitted for this functional group analysis.
Real-time PCR.
To verify the gene chip results, real-time PCR was employed by analyzing selected genes. The primers used for each selected gene were designed using Primer Express software v3.0 (Applied Biosystems) and synthesized by Integrated DNA Technologies. Reactions were set up as follows: 10 μl cDNA at 20 ng/ml, 12.5 μl Power SYBR green PCR master mix (Applied Biosystems), and 1.25 μl each of forward and reverse primers, each at 0.2 mM. PCR was carried out at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Samples were collected at each 60°C step using a 7300 real-time PCR machine from Applied Biosystems and analyzed by 7300 system SDS software using gene glgA as a reference, because it showed no change in gene chip data. For the real-time PCR data, we include the error bars (representing the standard deviation) that were calculated from triplicates for each gene.
Protein sample preparation for 2D gel experiments.
Cells were grown as described above as 6-liter cultures of each strain for the protein sample preparation. The protocol for preparing the E. coli inner membrane fraction was adopted from reference 9, with some modifications. The inner membrane protein leader peptidase and the cytosolic protein GroEL were used in a Western blot study to confirm that the soluble and inner membrane fractions were purified. After 5 h of growth in LB medium containing glucose, JS7131 cells became fragile, so all steps for making spheroplasts were omitted. Cells were resuspended in ice-cold DE buffer (1 mM dithiothreitol [DTT] and 20 mM EDTA, pH 7.2) and French pressed. The French-pressed cells were spun down at 6,000 × g for 10 min to remove the cell debris, and then the supernatant was centrifuged for 1 h at 4°C at 250,000 × g to pellet the membranes. Supernatant was saved for the soluble fraction protein sample. The membranes were subjected to sucrose gradient centrifugation to isolate the inner membrane vesicles (IMVs) (8). The isolated IMVs were pelleted and were resuspended in 50 mM Tris-HCl, 1 mM MgOAc, 1 mM DTT, and 2% dodecyl maltoside (DDM). Protein concentration was determined using the bicinchoninic acid (BCA) kit from Pierce. All samples used in the 2D gel experiments had a protein concentration of 10 mg/ml.
The Proteome Works system from Bio-Rad was used to perform the 2D gel experiment. For isoelectric focusing (IEF), a mixture of 8 M urea, 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 2 mM tributyl phosphine (TBP) buffer was used for the soluble fraction and a mixture of 7 M urea, 2 M thiourea, 2% CHAPS, 2% sulfabetaines (B3 to -10), and 2 mM TBP buffer was used for inner membrane protein fraction. Two different conditions were used to analyze membrane proteins: typically, pH 3 to 10 IEF strips were used for the inner membrane fraction, but for better resolution, pH 4 to 7 IEF strips were employed. Gels were stained with SYPRO ruby to achieve high sensitivity. Gel figures were obtained by the VersaDoc system and compared using PDquest Version 7.40 from Bio-Rad. Selected spots were cut out by Protean 2D Spotcutter and sent for in-gel digestion and mass spectrometry (MS).
In-gel digestion.
The in-gel protease digestion and mass spectrometry were performed at the Campus Chemical Instrument Center, Ohio State University. In-gel digestion was performed using the Montage in-gel digestion kit (Millipore, Bedford, MA) following the manufacturer's protocol. Protein spots were cut out and incubated in 50% methanol-5% acetic acid for 1 h twice and dehydrated in acetonitrile. Rehydration was done in DTT solution (5 mg/ml in 100 mM ammonium bicarbonate) for 30 min. Iodoacetamide (15 mg/ml) in 100 mM ammonium bicarbonate solution was then added to the rehydrated gel spots, and the mixture was incubated for 30 min in the dark. After that, gel spots were washed using acetonitrile and ammonium bicarbonate (100 mM) and dried in a SpeedVac. To digest the proteins, first, 50 μl of sequencing-grade trypsin (Promega Madison, WI) or chymotrypsin (Roche Indianapolis, IN) at 20 μg/ml in 50 mM ammonium bicarbonate was added to the dried gel pieces for 10 min. Then, 20 μl of 50 mM ammonium bicarbonate was added to the soaked gel and the digestion was carried out overnight at room temperature. The resulting peptides were extracted from the polyacrylamide gel with 50% acetonitrile and 5% formic acid and concentrated to a volume of about 25 μl.
Mass spectrometry.
Peptide detection and sequencing were performed using a capillary-liquid chromatography-nanospray tandem mass spectrometer (nano-LC/MS/MS). Samples were injected onto the trapping column (LC-Packings; Dionex Co., Sunnyvale, CA) and washed with 50 mM acetic acid. A ProteoPep II C18 column with a dimension of 5 cm and 75-μm inside diameter (New Objective, Inc., Woburn, MA) was packed directly in the nanospray tip and used for chromatographic separations. Peptides were eluted off the column into the LTQ system (Thermo Finnigan) using a gradient of 2 to 80% acetonitrile over 50 min, with a flow rate of 300 nl/min. The MS/MS was performed under standard conditions. Briefly, a nanospray source was operated in positive-ion mode with a 3-kV spray voltage and with a 200°C capillary temperature. A full scan was recorded first between 350 and 2,000 Da, followed by another MS/MS scan to generate product ion spectra to determine the amino acid sequence of the 10 most abundant peaks in the spectrum. The collision-induced dissociation (CID) fragmentation energy was set to 35%. Dynamic exclusion was enabled with a repeat count of 30 s, exclusion duration of 350 s, a low-mass width of 0.5 Da, and a high-mass width of 1.50 Da.
The raw data from the MS/MS experiment containing sequence information were analyzed by Mascot Daemon by Matrix Science version 2.2.1 (Boston, MA) and matched against SwissProt database version 54.1. During analysis, methionine oxidation and carbamidomethyl cysteine modifications were considered; two missed cleavages from the enzyme digestion were permitted. Only proteins with a Mascot score of 50 or higher, a P value of <0.05, and at least two unique matched peptides were considered as positive hits. The identified proteins were then individually checked manually by matching their calculated molecular mass, pI, and predicted cellular localization data with the 2D gel data to give the final assignment.
PspA induction and accumulation of pro-OmpA in carbonyl cyanide m-chlorophenylhydrazone (CCCP)-treated cells.
To determine the concentration of CCCP required for pro-OmpA to accumulate in the cytoplasm of the cell, the cultures were treated with different concentrations of CCCP. Specifically, JS7131 cells were grown in LB medium plus 0.2% arabinose for 5 h and then transferred into M9 medium containing 0.5% fructose for 15 min and treated with various concentrations of CCCP (dissolved in DMSO) for 15 min prior to labeling with [35S] methionine for 1 min. For the control group, equal amounts of DMSO were added to the LB medium before radiolabeling the cells. Also, JS7131 was grown in LB medium with glucose to deplete YidC and analyzed the same way, except no CCCP was added. Samples were analyzed by immunoprecipitation with OmpA antibody (from our lab collection). To measure the effects of CCCP on the growth of JS7131 in LB medium plus 0.2% arabinose, JS7131 cells were grown in LB medium for 2 h to the mid-log phase. CCCP was then added to the culture at different concentrations, and OD600 was measured for an additional 3 h. We also grew JS7131 in LB medium containing 0.2% glucose or 0.2% arabinose with no CCCP treatment. For the PspA induction study, JS7131 cells were treated with CCCP the same way as for the growth curve analysis. SDS-PAGE was performed using a 12% polyacrylamide gel. Where indicated, the gel was stained with Coomassie brilliant blue G-250 or analyzed by Western blotting. To reduce the amount of background bands on the Coomassie-stained gel, the soluble proteins in the samples were extracted from the E. coli cells using the B-PER bacterial protein extraction reagent (Pierce).
Western blotting.
The proteins YidC, PspA, maltose binding protein (MBP), DnaJ, and GroEL were analyzed to confirm the protein levels as determined by 2D gel analysis. They were detected by immunoblot analysis using the SuperSignal West Pico chemiluminescent substrate (Pierce). The antibodies used were raised against YidC (from our lab), PspA (Jan Tommassen at Utrecht University), MBP (New England Biolabs), DnaJ (Stressgen Biotechnologies), and GroEL (Sigma). Anti-DnaJ, anti-GroEL, and anti-YidC are polyclonal antibodies generated in rabbits against purified proteins or peptides. Anti-MBP is a monoclonal antibody (murine). As the control for the inner membrane proteome, Foa, Fob, Foc, and MalF were also analyzed by Western blotting under the YidC depletion condition. Antibodies against the Foa, Fob, and Foc subunits of F1Fo ATPase were kindly provided by Robert H. Fillingame at the University of Wisconsin—Madison. MalF antibody was provided by Beth Traxler at the University of Washington. Inner membrane proteins were purified using sucrose gradients and ultracentrifugation following a previously described protocol (9).
RESULTS
YidC depletion affects growth and cell morphology.
The growth of JS71 and JS7131 was measured at 37°C after 0.2% glucose was added to the medium. Since YidC synthesis was repressed in JS7131, its growth rate slowed down significantly compared to that of JS71 (Fig. 1), and after 7 h, JS7131 stopped growing. We confirmed by Western blotting that YidC is depleted in JS7131 to >90% after 5 h of growth in LB glucose medium (see Fig. S1 in the supplemental material). It should be noted that JS71 is entering the stationary phase at the 4-h time point (for the semi-log plot, see Fig. S2 in the supplemental material).
FIG. 1.
YidC depletion affects cell growth in liquid medium. Growth of JS71 (control) and JS7131 (YidC depletion) was measured by OD600. Absorbances were measured from 3 independent cultures of both JS71 and JS7131. A 0.2% concentration of glucose was added at time point 0. Error bars represent standard deviation from the 3 independent culture measurements.
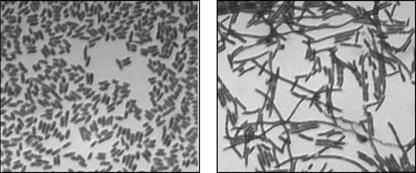
The cell morphology was examined using a microscope under YidC expression and YidC depletion conditions. Figure 2 reveals the YidC-depleted JS7131 cells grown for 5 h with glucose were significantly longer than JS71 cells (which showed normal E. coli morphology) and were nonmotile. The results indicate that the cells did not divide normally when YidC was depleted, possibly due to a disruption of the membrane structure.
FIG. 2.
Cell morphology is affected by YidC depletion. Images (×100 magnification) were obtained after the cells were grown for 5 h under the YidC depletion condition for JS7131 (right panel) or the normal condition for JS71 (left panel).
Transcriptome analysis during the YidC depletion condition. (i) Overall regulation pattern.
To better understand the changes in gene expression during YidC depletion, a high-throughput experiment was carried out under glucose conditions using JS71 grown as a control sample and JS7131 as a test sample. Gene chip techniques were employed to comprehensively analyze the change of the transcriptome pattern during the YidC depletion condition compared to the normal condition. According to the gene chip results (see the Excel file showing the entire data set in the supplemental material), among the total of 10,208 probes (which represent 4,355 genes and 1,427 intergenic regions) on the gene chip, 449 probes showed >2-fold upregulation, while 317 probes showed <0.5-fold downregulation, both with P values of <0.05. The upregulated probes include 163 genes, 129 hypothetical genes or undefined transcripts, and 157 intergenic regions. The downregulated probes include 99 genes, 173 hypothetical genes or undefined transcripts, and 45 intergenic regions. Table 1 lists the results with >3-fold changes, and Table 2 lists those for <0.3-fold changes, each with a P value of <0.05. Some genes which showed significant change in their expression levels with relatively high copy numbers were selected for real-time PCR experiments to confirm the gene chip data (Fig. 3). Among others, this included the strongly upregulated cadA and pspA genes and the strongly downregulated flu gene. We also examined the expression of these genes at 3 h as well. Real-time PCR confirmed that the mgtA, cadA, tig, and pspA genes were strongly induced at 5 h of YidC depletion as well as at the 3-h YidC depletion time. It also confirmed that the expression of YidC which is controlled by the araBAD promoter was repressed by growth of JS7131 in LB-glucose medium. As expected, real-time PCR showed expression of flu was repressed.
TABLE 1.
Significantly upregulated genes in this studya
| Probe identification | Fold change in expression in JS7131/JS71 | P value | Gene name | Functional annotationb |
|---|---|---|---|---|
| DNA/RNA related | ||||
| 1768419_s_at | 3.22 | 5E−03 | exo | 5′→3′ exonuclease |
| 1766724_s_at | 3.49 | 1E−02 | recD | DNA helicase, ATP-dependent dsDNA/ssDNA exonuclease V subunit, ssDNA endonuclease |
| 1766069_s_at | 3.25 | 5E−04 | deaD | Inducible ATP-independent RNA helicase |
| 1768185_at | 3.97 | 4E−02 | ybfD | Putative DNA ligase |
| 1761660_s_at | 3.59 | 1E−02 | rne | RNase E |
| 1760225_s_at | 3.16 | 5E−02 | rpoB | RNA polymerase, β-subunit |
| 1767680_at | 3.16 | 7E−03 | Regulatory RNA | |
| Energy | ||||
| 1759374_s_at | 3.34 | 3E−02 | cyoB | Cytochrome o ubiquinol oxidase subunit I |
| 1769018_s_at | 6.70 | 1E−03 | cyoA | Cytochrome o ubiquinol oxidase subunit II |
| 1764404_s_at | 4.31 | 3E−02 | cyoD | Cytochrome o ubiquinol oxidase subunit IV |
| 1761759_s_at | 3.79 | 5E−02 | nuoB | NADH dehydrogenase I chain B |
| 1763447_s_at | 4.87 | 2E−02 | nuoM | NADH dehydrogenase I chain M |
| 1761548_s_at | 3.15 | 3E−02 | bax | Putative ATP-binding protein |
| 1767503_s_at | 3.50 | 2E−02 | yihK | Putative GTP-binding factor |
| 1768459_s_at | 3.38 | 2E−02 | yhjL | Putative oxidoreductase subunit |
| 1764381_s_at | 3.30 | 5E−02 | atpF | Membrane-bound ATP synthase, Fo sector, subunit b |
| Fimbrin | ||||
| 1767098_s_at | 4.05 | 1E−02 | fimA | Major type 1 subunit fimbrin (pilin) |
| 1763102_s_at | 3.16 | 3E−03 | fimI | Fimbrin-like protein FimI precursor |
| Metabolism | ||||
| 1769070_s_at | 3.49 | 2E−04 | dxs | 1-Deoxyxylulose-5-phosphate synthase; flavoprotein |
| 1767819_s_at | 3.02 | 9E−03 | accB | Acetyl-CoA carboxylase, BCCP subunit; carrier of biotin |
| 1762548_s_at | 5.46 | 4E−05 | apt | Adenine phosphoribosyltransferase |
| 1761486_s_at | 3.55 | 9E−04 | add | Adenosine deaminase |
| 1762363_at | 4.84 | 3E−02 | malS | α-Amylase |
| 1759122_s_at | 3.03 | 2E−05 | thrA | Aspartokinase I, homoserine dehydrogenase I |
| 1761305_s_at | 3.30 | 2E−03 | mrdA | Cell elongation, e phase; peptidoglycan synthetase; penicillin-binding protein 2 |
| 1762501_s_at | 3.56 | 5E−02 | fdoH | Formate dehydrogenase O, iron-sulfur subunit |
| 1762677_s_at | 3.18 | 9E−03 | gatA | Galactitol-specific enzyme IIA of PTS system |
| 1762781_at | 3.59 | 5E−02 | aceK | Isocitrate dehydrogenase kinase/phosphatase |
| 1764617_s_at | 32.86 | 1E−03 | cadA | Lysine decarboxylase 1 |
| 1769223_s_at | 3.07 | 6E−03 | argA | N-Acetylglutamate synthase; amino acid acetyltransferase |
| 1764499_s_at | 3.17 | 1E−02 | pldA | Outer membrane phospholipase A |
| 1760363_s_at | 5.12 | 2E−03 | ppsA | Phosphoenolpyruvate synthase |
| 1769056_s_at | 3.80 | 1E−02 | hemK | Possible protoporphyrinogen oxidase |
| 1766494_s_at | 4.31 | 4E−07 | yiaE | Putative dehydrogenase |
| 1764702_s_at | 5.73 | 4E−02 | yhdG | Putative dehydrogenase |
| 1763775_at | 6.69 | 5E−03 | Putative glycan biosynthesis enzyme | |
| 1767386_at | 8.77 | 3E−02 | Putative ligase | |
| 1762947_s_at | 11.68 | 1E−02 | gatZ | Putative tagatose 6-phosphate kinase 1 |
| 1768388_s_at | 5.07 | 4E−03 | glpD | sn-Glycerol-3-phosphate dehydrogenase (aerobic) |
| 1763273_s_at | 6.15 | 9E−04 | sodA | Superoxide dismutase, manganese |
| 1767672_s_at | 8.40 | 7E−03 | gatY | Tagatose-bisphosphate aldolase 1 |
| 1763484_s_at | 6.92 | 5E−02 | thiC | Thiamine biosynthesis, pyrimidine moiety |
| 1764403_s_at | 4.31 | 4E−03 | thiE | Thiamine biosynthesis, thiazole moiety |
| 1767599_s_at | 3.71 | 9E−03 | thyA | Thymidylate synthetase |
| 1764322_at | 3.81 | 3E−02 | avtA | Valine-pyruvate aminotransferase |
| 1767715_s_at | 3.62 | 2E−04 | Putative polymerase proteinase | |
| 1763494_s_at | 3.74 | 1E−03 | Putative amidase | |
| 1763111_s_at | 3.41 | 4E−02 | Acetolactate synthase II, valine insensitive, large subunit, silent in K-12 | |
| 1766804_s_at | 3.81 | 1E−02 | glpK | Glycerol kinase |
| Transporter | ||||
| 1765773_s_at | 3.13 | 5E−03 | oppC | Homolog of Salmonella oligopeptide transport permease protein |
| 1768775_s_at | 5.57 | 5E−03 | rbsB | d-Ribose periplasmic binding protein |
| 1760971_s_at | 5.32 | 1E−03 | rbsA | ATP-binding component of d-ribose high-affinity transport system |
| 1760798_s_at | 3.20 | 2E−03 | proV | ATP-binding component of transport system for glycine, betaine, and proline |
| 1760041_s_at | 3.26 | 1E−03 | malK | ATP-binding component of transport system for maltose |
| 1759395_s_at | 3.88 | 9E−03 | dppA | Dipeptide transport protein |
| 1763403_s_at | 4.78 | 1E−04 | rbsC | d-Ribose high-affinity transport system |
| 1763733_s_at | 7.82 | 5E−02 | rbsD | d-Ribose high-affinity transport system, membrane-associated protein |
| 1764804_s_at | 3.77 | 3E−03 | pstS | High-affinity phosphate-specific transport system, periplasmic phosphate-binding protein |
| 1764279_s_at | 13.33 | 1E−02 | mgtA | Mg2+ transport ATPase, P-type 1 |
| 1766260_s_at | 3.67 | 2E−03 | tsx | Nucleoside channel, receptor of phage T6 and colicin K |
| 1768919_s_at | 3.95 | 7E−04 | sdaC | Probable serine transporter |
| 1768684_s_at | 3.78 | 1E−02 | manX | PTS enzyme IIAB, mannose specific |
| 1763069_s_at | 3.74 | 3E−02 | manY | PTS enzyme IIC, mannose specific |
| 1762917_s_at | 7.26 | 2E−05 | ptsG | PTS system, glucose-specific IICB component |
| 1764801_at | 3.28 | 5E−02 | yieG | Putative membrane/transport protein |
| 1765980_s_at | 6.93 | 5E−03 | yjdL | Putative peptide transporter |
| 1764271_s_at | 3.04 | 6E−03 | yhiV | Putative transport system permease protein |
| 1760764_s_at | 6.31 | 4E−05 | Putative transporter protein | |
| 1759972_s_at | 20.69 | 1E−02 | cadB | Transport of lysine/cadaverine |
| Protease and chaperone | ||||
| 1763873_s_at | 3.48 | 8E−06 | ppiA | Peptidyl-prolyl cis-trans isomerase A (rotamase A) |
| 1762546_s_at | 4.49 | 2E−02 | fimC | Periplasmic chaperone, required for type 1 fimbriae |
| 1764696_s_at | 3.74 | 4E−05 | ptr | Protease III |
| 1767122_s_at | 4.85 | 4E−03 | tig | Trigger factor, a molecular chaperone involved in cell division |
| Stress response | ||||
| 1761114_s_at | 20.03 | 3E−04 | cspA | Cold shock protein 7.4, transcriptional activator of hns |
| 1762932_s_at | 4.56 | 6E−03 | cspB | Cold shock protein; may affect transcription |
| 1765796_s_at | 8.00 | 2E−05 | pspD | Phage shock protein |
| 1768435_s_at | 8.30 | 2E−04 | pspB | Phage shock protein |
| 1760618_s_at | 3.38 | 5E−04 | pspE | Phage shock protein |
| 1769101_s_at | 13.60 | 5E−04 | pspA | Phage shock protein, inner membrane protein |
| 1762530_s_at | 6.78 | 8E−03 | pspC | Phage shock protein, activates phage shock protein expression |
| 1769169_s_at | 3.12 | 2E−02 | yjiY | Putative carbon starvation protein |
| Translation and transcription | ||||
| 1762512_s_at | 4.10 | 2E−03 | rpsA | 30S ribosomal subunit protein S1 |
| 1767291_s_at | 3.32 | 4E−02 | rpsJ | 30S ribosomal subunit protein S10 |
| 1765175_s_at | 3.48 | 4E−03 | rpsP | 30S ribosomal subunit protein S16 |
| 1768873_s_at | 3.19 | 5E−02 | rpsB | 30S ribosomal subunit protein S2 |
| 1762255_s_at | 3.08 | 2E−02 | rplR | 50S ribosomal subunit protein L18 |
| 1767377_s_at | 3.62 | 2E−02 | rplS | 50S ribosomal subunit protein L19 |
| 1760752_s_at | 3.44 | 5E−02 | rplB | 50S ribosomal subunit protein L2 |
| 1761244_s_at | 3.25 | 3E−02 | rplV | 50S ribosomal subunit protein L22 |
| 1761605_s_at | 3.47 | 2E−03 | glyQ | Glycine tRNA synthetase, α-subunit |
| 1765772_s_at | 3.54 | 5E−03 | glyS | Glycine tRNA synthetase, β-subunit |
| 1761999_s_at | 4.08 | 4E−02 | fusA | GTP-binding protein chain elongation factor EF-G |
| 1767618_s_at | 4.06 | 9E−04 | prfA | Peptide chain release factor RF-1 |
| 1766499_s_at | 3.09 | 1E−03 | pheS | Phenylalanine tRNA synthetase, α-subunit |
| 1764306_s_at | 3.32 | 4E−05 | rnpA | RNase P, protein component; protein C5; processes tRNA, 4.5S RNA |
| 1765454_s_at | 3.24 | 7E−04 | rho | Transcription termination factor Rho, polarity suppressor |
| 1764493_s_at | 3.46 | 4E−03 | trmD | tRNA methyltransferase; tRNA (guanine-7-)-methyltransferase |
| 1759109_s_at | 3.01 | 1E−02 | Putative elongation factor | |
| Other | ||||
| 1762757_s_at | 3.18 | 4E−02 | pal | Peptidoglycan-associated lipoprotein |
| 1767981_s_at | 3.66 | 4E−03 | ompF | Outer membrane protein 1a (Ia, b, F) |
| 1766425_s_at | 4.11 | 4E−02 | priB | Primosomal replication protein N |
| 1761338_s_at | 5.59 | 1E−03 | Putative Bor protein of prophage CP-933X | |
| 1765995_s_at | 3.65 | 6E−03 | Putative membrane protein | |
| Hypothetical | ||||
| 1760914_s_at | 7.69 | 3E−04 | Hypothetical protein | |
| 1766415_s_at | 6.88 | 5E−04 | Hypothetical protein | |
| 1764179_s_at | 3.64 | 8E−04 | Hypothetical protein | |
| 1761529_s_at | 3.41 | 2E−03 | Hypothetical protein | |
| 1759951_s_at | 3.55 | 2E−02 | Hypothetical protein | |
| 1766884_s_at | 3.60 | 4E−02 | Hypothetical protein | |
| 1762024_s_at | 3.31 | 1E−02 | Hypothetical protein | |
| 1767540_s_at | 5.41 | 1E−02 | yeaJ | ORF, unknown function |
| 1769192_s_at | 5.41 | 5E−06 | yeaF | ORF, hypothetical protein |
| 1763019_s_at | 4.73 | 4E−04 | yafK | ORF, hypothetical protein |
| 1767568_s_at | 3.25 | 8E−04 | yhjU | ORF, hypothetical protein |
| 1762187_s_at | 3.30 | 3E−03 | ORF, hypothetical protein | |
| 1760906_s_at | 4.87 | 5E−03 | ORF, hypothetical protein | |
| 1766652_s_at | 4.14 | 5E−03 | ORF, hypothetical protein | |
| 1762056_s_at | 3.15 | 6E−03 | ynaE | ORF, hypothetical protein |
| 1760762_s_at | 4.06 | 7E−03 | ORF, hypothetical protein | |
| 1766638_s_at | 3.84 | 9E−03 | ybeA | ORF, hypothetical protein |
| 1763486_s_at | 3.14 | 1E−02 | yceD | ORF, hypothetical protein |
| 1759527_s_at | 3.01 | 1E−02 | yhjW | ORF, hypothetical protein |
| 1761324_at | 4.36 | 2E−02 | ORF, hypothetical protein | |
| 1764771_s_at | 4.36 | 2E−02 | ycbK | ORF, hypothetical protein |
| 1766279_at | 8.09 | 3E−02 | ycgW | ORF, hypothetical protein |
| 1767883_s_at | 4.82 | 3E−02 | yfjA | ORF, hypothetical protein |
| 1762710_s_at | 3.43 | 4E−02 | ybgF | ORF, hypothetical protein |
| 1765691_s_at | 3.13 | 5E−02 | ORF, hypothetical protein |
The most significantly upregulated genes from the gene chip data are shown. Only those showing a >3-fold change with reliable P values (P < 0.05) are listed.
dsDNA, double-stranded DNA; ssDNA, single-stranded DNA; acetyl-CoA, acetyl coenzyme A; BCCP, biotin carboxyl carrier protein; PTS, phosphotransferase; ORF, open reading frame.
TABLE 2.
Significantly downregulated genes in this studya
| Protein type and probe identification | Fold change in expression in JS7131/JS71 | P value | Gene name | Functional annotationb |
|---|---|---|---|---|
| Energy | ||||
| 1767636_s_at | 0.22 | 1E−03 | frdA | Fumarate reductase, anaerobic, flavoprotein subunit |
| 1767792_s_at | 0.24 | 3E−03 | appC | Probable third cytochrome oxidase, subunit I |
| 1767833_s_at | 0.23 | 2E−04 | appB | Probable third cytochrome oxidase, subunit II |
| 1760548_s_at | 0.27 | 2E−04 | yhaH | Putative cytochrome |
| 1759127_s_at | 0.12 | 9E−06 | yghA | Putative oxidoreductase |
| 1764339_s_at | 0.24 | 1E−04 | ybdR | Putative oxidoreductase |
| Metabolism | ||||
| 1767109_s_at | 0.28 | 3E−04 | hdhA | 7-α-Hydroxysteroid dehydrogenase |
| 1768184_s_at | 0.23 | 7E−06 | hyaB | Hydrogenase-1, large subunit |
| 1765453_s_at | 0.19 | 8E−04 | hyaA | Hydrogenase-1, small subunit |
| 1768953_s_at | 0.26 | 1E−04 | appA | Phosphoanhydride phosphorylase; pH 2.5 acid phosphatase; periplasmic |
| 1768393_s_at | 0.23 | 9E−05 | purE | Phosphoribosylaminoimidazole (AIR) carboxylase, catalytic subunit |
| 1762205_s_at | 0.20 | 2E−04 | purC | Phosphoribosylaminoimidazole-succinocarboxamide (SAICAR) synthetase |
| Transporter | ||||
| 1768513_x_at | 0.19 | 3E−02 | KdpF protein of high-affinity potassium transport system | |
| 1760130_s_at | 0.25 | 2E−05 | yohK | Putative seritonin transporter |
| Stress response | ||||
| 1763125_s_at | 0.09 | 7E−05 | glgS | Glycogen biosynthesis, rpoS dependent |
| 1761076_s_at | 0.25 | 1E−04 | psiF | Induced by phosphate starvation |
| 1759179_s_at | 0.14 | 2E−04 | osmB | Osmotically inducible lipoprotein |
| Translation and transcription | ||||
| 1762381_s_at | 0.27 | 3E−03 | Spot 42 RNA | |
| 1763089_s_at | 0.29 | 8E−04 | 6S RNA | |
| 1763985_s_at | 0.25 | 6E−03 | 4.5S RNA, component of ribonucleoprotein particle | |
| 1765371_s_at | 0.18 | 1E−05 | osmE | Activator of ntrL gene |
| Other | ||||
| 1760228_s_at | 0.23 | 2E−05 | Putative filament protein | |
| 1761941_s_at | 0.30 | 4E−05 | Putative polyprotein | |
| 1761735_s_at | 0.30 | 2E−05 | Amino-terminal fragment of WrbA | |
| 1766745_s_at | 0.28 | 1E−04 | Entericidin B | |
| 1760040_at | 0.08 | 7E−04 | flu | Outer membrane fluffing protein, similar to adhesin |
| 1763857_s_at | 0.17 | 4E−04 | relF | Polypeptide destructive to membrane potential |
| 1763864_s_at | 0.20 | 2E−04 | Putative homeobox protein | |
| 1767642_s_at | 0.11 | 3E−02 | yibP | Putative membrane protein |
| 1762401_s_at | 0.22 | 2E−04 | yciF | Putative structural proteins |
| 1768436_s_at | 0.07 | 4E−03 | ||
| 1760679_at | 0.17 | 1E−03 | ||
| Hypothetical | ||||
| 1761754_s_at | 0.08 | 1E−04 | Hypothetical protein | |
| 1759995_s_at | 0.25 | 1E−02 | Hypothetical protein | |
| 1761327_s_at | 0.27 | 2E−03 | Hypothetical protein | |
| 1767107_s_at | 0.28 | 3E−02 | Hypothetical protein | |
| 1763445_s_at | 0.07 | 5E−04 | Hypothetical protein | |
| 1760150_s_at | 0.30 | 4E−02 | Hypothetical protein | |
| 1760953_at | 0.18 | 2E−02 | ORF, unknown function | |
| 1765333_s_at | 0.21 | 2E−02 | ORF, unknown function | |
| 1760940_s_at | 0.29 | 6E−04 | ORF, unknown function | |
| 1766731_s_at | 0.09 | 5E−05 | yohJ | ORF, hypothetical protein |
| 1761873_s_at | 0.09 | 8E−05 | yciG | ORF, hypothetical protein |
| 1759494_s_at | 0.13 | 9E−05 | ycfR | ORF, hypothetical protein |
| 1768326_s_at | 0.14 | 2E−04 | ydeI | ORF, hypothetical protein |
| 1766187_s_at | 0.15 | 6E−06 | ORF, hypothetical protein | |
| 1763516_s_at | 0.19 | 3E−04 | yceP | ORF, hypothetical protein |
| 1760727_s_at | 0.19 | 3E−05 | ygaM | ORF, hypothetical protein |
| 1766489_s_at | 0.19 | 2E−06 | ybfA | ORF, hypothetical protein |
| 1762657_s_at | 0.20 | 1E−03 | ORF, hypothetical protein | |
| 1764792_s_at | 0.21 | 4E−04 | yhbO | ORF, hypothetical protein |
| 1761638_s_at | 0.22 | 3E−03 | ymgE | ORF, hypothetical protein |
| 1761022_s_at | 0.23 | 3E−03 | yecH | ORF, hypothetical protein |
| 1764073_s_at | 0.23 | 2E−03 | yhcO | ORF, hypothetical protein |
| 1762735_s_at | 0.24 | 9E−04 | yhhA | ORF, hypothetical protein |
| 1768189_s_at | 0.24 | 8E−04 | yphA | ORF, hypothetical protein |
| 1768649_at | 0.25 | 1E−05 | ycdF | ORF, hypothetical protein |
| 1768587_s_at | 0.26 | 4E−04 | yciE | ORF, hypothetical protein |
| 1761690_s_at | 0.26 | 1E−03 | ORF, hypothetical protein | |
| 1767977_s_at | 0.26 | 4E−04 | yjfY | ORF, hypothetical protein |
| 1766480_s_at | 0.27 | 1E−03 | ORF, hypothetical protein | |
| 1768363_s_at | 0.27 | 3E−04 | ybdQ | ORF, hypothetical protein |
| 1766893_s_at | 0.28 | 6E−05 | ORF, hypothetical protein | |
| 1764881_s_at | 0.28 | 2E−04 | ybeH | ORF, hypothetical protein |
| 1765823_s_at | 0.29 | 2E−04 | yeaQ | ORF, hypothetical protein |
| 1765253_s_at | 0.29 | 7E−05 | yjbJ | ORF, hypothetical protein |
| 1767891_s_at | 0.29 | 2E−04 | ynhG | ORF, hypothetical protein |
| 1767800_s_at | 0.30 | 2E−04 | yqjI | ORF, hypothetical protein |
| 1768822_s_at | 0.30 | 8E−04 | ybiM | ORF, hypothetical protein |
| 1759909_s_at | 0.30 | 1E−05 | ORF, hypothetical protein | |
| 1768573_s_at | 0.30 | 8E−04 | ORF, hypothetical protein | |
| 1761089_at | 0.30 | 3E−04 | relE | ORF, hypothetical protein |
| 1768030_s_at | 0.30 | 3E−05 | ytfK | ORF, hypothetical protein |
| 1766739_s_at | 0.30 | 8E−04 | ychH | ORF, hypothetical protein |
The most significantly downregulated genes from the gene chip data are shown. Only those with <0.3-fold changes with reliable P values (P <0.05) are listed.
ORF, open reading frame.
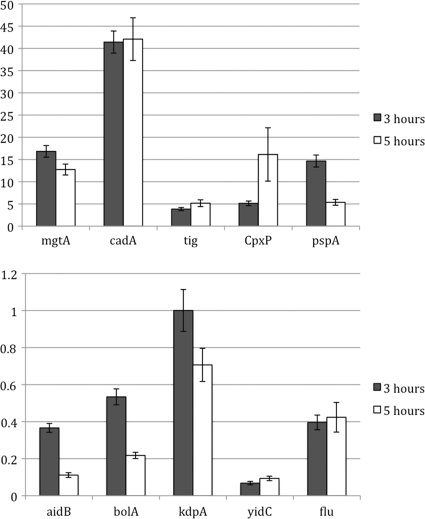
FIG. 3.
Selected genes are tested by real-time PCR to confirm the microarray data. The y axis represents the relative expression level of each gene under YidC depletion conditions compared to the normal condition. A value of 1 would represent no change in expression upon YidC depletion. The mRNA levels at both the 3- and 5-h time points were tested. Error bars are calculated from triplicates for each experiment.
Next, we used DAVID tools to group the significantly affected genes (larger than 2-fold and less than 0.5-fold) by their functions. To make the analysis easier, only genes with a known function were submitted to the program. The results from this grouping analysis are summarized in Fig. 4.
FIG. 4.
Gene category summary. All genes showing a different expression pattern (>2 fold difference and P value of <0.05) were grouped according to their functions. The x axis indicates the number of genes within each functional group. Black bars represent those groups of genes which were upregulated by microarray analysis under YidC depletion conditions for 5 h. Gray bars represent those groups of genes which were downregulated by microarray analysis under YidC depletion conditions for 5 h.
(ii) Stress response.
Since YidC is an inner membrane insertase and chaperone, depletion of YidC will affect the insertion and folding of its substrates, thus leaving these proteins either in the incorrect cellular location or in the incorrect folding state. At the same time, some of YidC's substrates, such as F1Fo ATP synthase subunits a and c and CyoA (subunit II of cytochrome bo oxidase) have important physiological functions in energy transduction for the cell when assembled into their respiratory complex (39). Therefore, it is conceivable that depletion of YidC would trigger a stress response system, allowing the cell to handle potential problems, which include both primary effects from the absence of YidC and secondary effects from the absence of YidC's substrates. Indeed, we found that the phage shock response, cold shock response, and the Cpx pathway were switched on following YidC depletion (Table 1).
Both the phage shock protein family and the cold shock protein family genes were highly upregulated (Table 1). pspA, -B, -C, -D, and -E showed 13.6-, 8.3-, 6.8-, 8.0-, and 3.4-fold upregulation, respectively. For the cold shock protein family, 5 out of 9 cold shock proteins were increased at the mRNA level, including cspA, -B, -C, -G, and -I (Table 1; see the Excel file in the supplemental material). By microarray analysis, cspA is the gene with the highest upregulation, at 20-fold. Previous studies showed that cold shock proteins are generally mRNA-binding proteins which handle the secondary structure of mRNA, working together with some RNA helicases. Interestingly, two RNA helicase genes, deaD and rhlE (see the Excel file in the supplemental material) were also upregulated in our YidC depletion experiments.
Previous reports have shown that Cpx/sigma E pathways were switched on during the YidC depletion condition (32). In our data, 3 isomerase genes from the Cpx pathway, fkpA (1.55-fold) (coding for an FK506 binding protein [FKBP]-type peptidyl-prolyl cis-trans isomerase), dsbA (1.59-fold) (a periplasmic protein disulfide isomerase), and ppiA (3.48-fold) (coding for peptidyl-prolyl cis-trans isomerase A) were upregulated (see the Excel file in the supplemental material). Unfortunately, the cpxP probe is missing from our gene chip probe array. Therefore, we set up an additional real-time PCR experiment to check the cpx transcription profile. The result showed a 15-fold upregulation of cpxP at 5 h YidC depletion (Fig. 3). In contrast, rpoH, which was used as the reporter for activation of the sigma E pathway (32), showed no significant change (see the Excel file in the supplemental material). To further investigate the activation of both pathways, 5 genes from the Cpx pathway and 6 genes from the sigma E pathway were selected and their expression profiles under the YidC depletion condition were examined by real-time PCR (Fig. 5). For the Cpx pathway, all 3 genes (dsbA, ompC, and ppiA) that are expected to be upregulated if the pathway is switched on were upregulated at both 3 and 5 h, while motB and flgM, which are expected to be downregulated if the pathway is switched on, were downregulated at both time points (Fig. 5). In contrast, at the 3-h time point, only half of the selected genes (ropE, mdoG, and ompA) in the sigma E pathway matched their expected regulation patterns if the pathway was switched on. Others were regulated in the opposite manner (cutC and ompX) (Fig. 5). At the 5-h time point, none of the sigma E genes showed significant changes in expression when YidC was depleted. degP, which typically is upregulated when either the sigma E or Cpx stress pathway is activated, was not significantly changed (Fig. 5; see the Excel file in the supplemental material).
FIG. 5.
Regulation of specific Cpx- and sigma E-controlled genes by real-time PCR under YidC depletion conditions. The expected regulation pattern of genes is indicated for each pathway listed in the chart. The y axis represents the relative expression level by real-time PCR of each gene under YidC depletion conditions compared to the normal condition. (A) Relative expression level of selected genes for Cpx pathway at both 3 and 5 h of YidC depletion. (B) Relative expression level of selected genes for sigma E pathway at both 3 and 5 h of YidC depletion.
We conclude that the Cpx pathway is switched on based on the fact that CpxP is induced and the fact that a number of genes in this pathway (see Table S1 in the supplemental material) are switched on upon YidC depletion. Increased induction of CpxP has been shown to be a good indicator that the Cpx pathway is activated (5); i.e., CpxP can only be switched on by activation by this stress pathway (T. Silhavy, personal communication). We suspect that the Cpx pathway-controlled genes that were not induced in our microarray study (see Table S1 in the supplemental material) were influenced by other factors that occurred during YidC depletion. Many of these genes have other promoters in the regulon and show complex regulation (7). The evidence that the sigma E pathway is activated in our microarray study is not strong (see Discussion) because there are a number of genes that are regulated in the opposite direction, and some genes that are known to be highly expressed by activation of the sigma E pathway were not upregulated (see Table S2 in the supplemental material).
(iii) Proteases and chaperones.
Proteases and chaperones are expected to play important roles in handling malfolded membrane proteins during the YidC depletion condition. The genes coding for the two chaperones, fimC (periplasmic chaperone) and tig (trigger factor, a chaperone involved in protein folding), were highly induced at 4.5- and 4.9-fold, respectively (Table 1). Surprisingly, other chaperone genes, namely, dnaK, dnaJ, groES, and groEL, were not upregulated in our microarray analysis (see the Excel file in the supplemental material), in contrast to a recent microarray study of Streptococcus mutans when the signal recognition particle (SRP) component Ffh was knocked out (10).
Several protease genes were shown to be significantly upregulated upon YidC depletion (greater than 2-fold change) (see the Excel data set in the supplemental material), including ompT, ptr (protease III), hflC, and hflK, which form a membrane complex with the membrane protease hflB (FtsH) and negatively regulate the protease activity of FtsH. Some other protease genes such as iap (aminopeptidase in alkaline phosphatase), yaeL, lspA, htpX, glpG, and hflB were also induced (140% to ∼192% compared to the control).
(iv) The role of phage shock response in YidC-depleted cells.
The phage shock response is thought to help maintain the proton motive force (PMF) under membrane stress conditions such as infection by phage or treatment with a protonophore (13). Since YidC depletion results in a decrease in the PMF due to a defect in the assembly and activity of cytochrome bo3 oxidase and F1Fo ATP synthase (39), we expected that the phage shock response is induced after YidC depletion. Therefore, we compared PspA expression after YidC depletion with the effects of addition of CCCP, a protonophore to collapse the PMF, to E. coli cells (22).
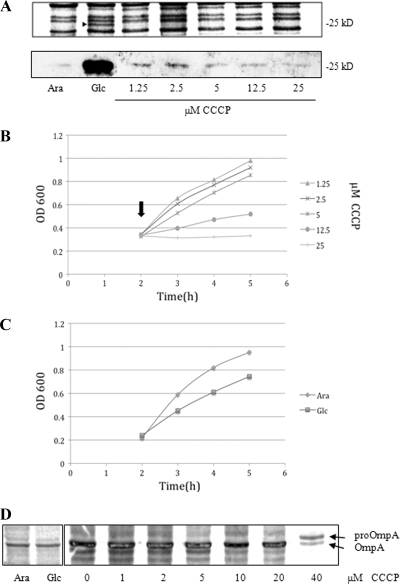
Surprisingly, our microarray data derived from YidC-depleted cells are very different from the data from another gene chip study in which the effects of CCCP were analyzed (22). That study did not see an activation of the Psp operon when CCCP was added, although transcription of a number of other genes was affected (see Table S3). We found significant PspA induction upon YidC depletion (Fig. 6A). Equal amounts (with cells normalized to the same OD600) of JS7131 cells grown in glucose (YidC-depleted conditions), arabinose (YidC expression conditions), and arabinose containing various concentrations of CCCP were collected and subjected to SDS-PAGE and Western blot analysis (Fig. 6A). When YidC was depleted, PspA was induced (see arrowhead, Glc, upper panel), while no PspA was detectable on the Coomassie-stained gel after CCCP treatment (Fig. 6A, upper panel). Similarly, Western blot analysis using PspA antibody showed that there is a strongly induced PspA band in YidC-depleted cells and only a weak PspA band in CCCP-treated cells.
FIG. 6.
YidC depletion induces a higher level of phage shock response than CCCP treatment. (A) JS7131 was grown in glucose (0.2%) or arabinose (0.2%), treated with various concentrations of CCCP, and then analyzed by Coomassie G-250 staining (top panel) or Western blotting using PspA antibody (lower panel). Soluble proteins were removed before running the gel for Coomassie stain (see Materials and Methods). (B) The growth curve was measured for JS7131 in LB medium containing arabinose (0.2%) with various concentrations of CCCP. (C) The growth curve was determined for JS7131 in LB medium with arabinose (0.2%) or glucose (0.2%). (D) The addition of 40 μM CCCP to the JS71 culture resulted in the accumulation of 35S-labeled pro-OmpA in the cell. The amounts of precursor and mature OmpA in a 1-min pulse-label were determined as described in Materials and Methods.
The effects of the different concentrations of CCCP on the growth of the cell were analyzed. We found that the addition of CCCP (see arrow) at concentrations above 2.5 μM slowed growth of JS7131 (in LB medium supplemented with 0.2% arabinose). The growth curves of the cultures treated with 5 and 12.5 μM CCCP were comparable to that of YidC-depleted JS7131 cells (Fig. 6B and C). After the addition of 25 μM CCCP, the cells stopped growing. The effects of CCCP on pro-OmpA export were examined. Pro-OmpA only accumulated when CCCP was added at 40 μM (Fig. 6D). Our effects of CCCP on pro-OmpA accumulation are similar to what Possot et al. (23) had reported: i.e., 50 μM CCCP caused an almost complete block in pullulanase secretion. They showed that already 5 μM CCCP dissipated the Δψ below the sensitivity of the assay using tetraphenyl phosphonium ion (TPP+) uptake, suggesting the PMF was almost completely collapsed. In conclusion, our PspA study here shows that the PspA induction that is observed upon YidC depletion appears independent of the PMF.
(v) Other possible effects.
Another reason why the expression of some genes show changes in our study may be due to JS71, but not JS7131, entering stationary phase after 5 h of growth (Fig. 1; see Fig. S1 in the supplemental material). For example, stationary-phase-induced genes would be switched on in JS71 because it is entering stationary phase at the 4-h point. Since JS7131 is not entering stationary phase, this could lead to a decrease in the expression of those genes in JS7131 compared to JS71.
To evaluate whether this is influencing our microarray data, we performed real-time PCR at 3 and 5 h on two genes that are known to be induced at stationary phase. The stationary-phase-induced genes aidB and bolA were selected because transcription of these genes is markedly increased upon entering stationary phase (34) (see Table S4 in the supplemental material). The aidB gene encodes a dehydrogenase that has a role in DNA metabolism (40); the bolA-encoded protein is a regulator of cell wall biosynthetic enzymes (30). We compared levels of gene expression of aidB and bolA in JS7131 (compared to JS71) after 3 and 5 h of growth in LB medium plus glucose. We found that levels of gene expression of aidB and bolA were decreased at 5 h relative to the level at 3 h, consistent with JS71 entering stationary phase and its influence on the gene expression data (Fig. 3). A second approach to check if the microarray data are being influenced by the different growth phases of the JS71 and JS7131 strains is to look at other genes under rpoS control. RpoS is essential for induction of various stationary-phase genes, including aidB. If these RpoS-controlled stationary-phase genes are being switched on in JS71 as it begins to enter stationary phase, it will result in the genes being downregulated in JS7131 compared to JS71. We see this for the osmotically regulated gene osmB (0.14-fold) for trehalose breakdown, the glycogen synthesis gene glgS (0.09-fold), the acid phosphatase gene appA (0.26-fold), and the bolA gene (0.40-fold) (see the Excel file in the supplemental material) involved in cell morphology (Table 2). Finally, comparison of our YidC depletion microarray data with data from a microarray study that identified 53 stationary-phase-induced genes (34) showed that one-third of the stationary-phase-induced genes were affected in our YidC depletion study (leading to a decrease in the expression of the genes in JS7131/JS71) (see Table S4 in the supplemental material). Nevertheless, we think that the different growth phases of JS7131 and JS71 are marginally influencing the expression pattern of the sigma E pathway and Cpx pathway genes compared to the influence of YidC depletion on the Cpx pathway genes (Fig. 5).
(vi) Other significantly up/downregulated genes.
Table 1 (and see the Excel file in the supplemental material) shows that the ribosome and aminoacyl-tRNA biosynthesis pathways were affected under YidC depletion conditions. Nineteen genes for ribosomal proteins of the 30S subunit, 20 genes for ribosomal proteins of the 50S subunit, 8 tRNA synthetases, 2 elongation factors (EFs), and 1 initiation factor (IF) were highly induced. The translocase subunit secY also showed a 2.3-fold increase at mRNA level, while neither secE nor secG was affected. Also, a large number of genes involved in ubiquinone biosynthesis, glycine, serine, and threonine metabolism, and the ABC transporter system were upregulated (Table 1; see the Excel file in the supplemental material). Interestingly, many of these transporter systems were upregulated in their ATP-binding components, such as rbsA (ATP-binding component of a d-ribose transport system), pstB (ATP-binding component of a phosphate transport system), potA (ATP-binding component of spermidine/putrescine transport), proV (ATP-binding component of transport system for glycine, betaine, and proline), and malK (ATP-binding component of maltose transport system) (see the Excel file in the supplemental material for potA; see Table 1 for the other genes). The oxidative phosphorylation pathway was also affected, with NADH dehydrogenase I (1.7- to ∼4.9-fold upregulation; see the Excel file in the supplemental material for the full list) and cytochrome bo3 oxidase (2.5- to ∼6.8-fold upregulation; see the Excel file in the supplemental material) showing upregulation for most of their components at the mRNA level, while the succinate dehydrogenase/fumarate reductase and the cytochrome bd complex were downregulated for most of their components at the mRNA level. The most noticeable downregulated groups of genes belong to the chemotaxis family, including cheA (0.46-fold), cheB (0.47-fold), cheY (0.44-fold), and cheW (0.35-fold) and the potassium transport system, including kdpA (0.40-fold), kdpB (0.41-fold), kdpC (0.47-fold), kdpD (0.33-fold), and kdpE (0.48-fold). For the genes in the kdp operon, we suspect that the changing pattern was actually due to the upregulation of these genes in strain JS71 because the culture density was higher than that of JS7131. Consistent with this is that the downregulation of kdpA (JS7131/JS71) is more pronounced at 5 h compared to the 3 h of YidC depletion, as determined by real-time PCR; JS71 is entering stationary phase at the 5-h growth time (Fig. 3). The higher density of JS71 may lower the available potassium ions in the medium, which in turn could induce the expression of the high-affinity potassium transporter genes.
cadA and cadB were the two most highly upregulated genes in our microarray study. cadA was upregulated 32.8-fold, while cadB was upregulated 20.7-fold (Table 1). The cad operon is generally induced under acidic external pH or anoxic conditions. For example, cadA was shown to be upregulated under acidic conditions (15). Since YidC depletion significantly lowers the PMF due to a perturbation in the assembly of the respiratory complexes F1Fo ATP synthase and cytochrome bo oxidase and affects several important genes within the proton transport pathway, the pH balance is likely to be out of control, and certain genes were activated to cope with this. We also used real-time PCR to confirm that cadA was highly expressed after 5 h of depletion (Fig. 3).
YidC depletion affects cell motility.
Two flagellar biosynthesis genes, fliS and fliC, were downregulated under YidC depletion conditions. To test if flagellar function was indeed affected, cell motility assays were carried out. Equal amounts of JS7131 and JS71 cells (normalized to the same number of cells) were applied to motility agar containing 0.2% glucose and incubated at 37°C for 24 h (Fig. 7). After 24 h of incubation, the diameter of the JS71 swimming zone was approximately 5 times larger than that with JS7131. JS7131 grows much slower than JS71 under the YidC depletion condition, and eventually YidC depletion leads to cell death, so we added a low concentration of arabinose (final concentration, 0.002%) to the plate to ensure JS7131 was kept alive during the 24-h test period. The results confirmed that YidC depletion had a negative effect on flagellar function.
FIG. 7.
YidC depletion affects cell motility. After growth for 5 h in LB medium supplemented with 0.2% glucose, equal numbers (as determined using a microscope and counting chamber) of JS71 (YidC expressed) and JS7131 (YidC depleted) cells were applied to a motility agar plate containing 0.2% glucose and 0.002% arabinose. This arabinose concentration was tested to be sufficient to keep JS7131 cells alive but did not allow growth within 24 h. Plates were incubated at 37°C for 24 h. The size of the JS7131 colony was 0.6 ± 0.1 cm. The size of the JS71 colony was 3.2 ± 0.3 cm. Sizes were measured independently using 5 plates for each of the strains. Only one representative plate is shown here.
Proteome analysis during the YidC depletion condition. (i) Overall regulation pattern.
To understand how the cell responds to YidC depletion, we analyzed the global change of the proteome by 2D gel electrophoresis experiments. To improve the resolution, we separated the inner membrane fraction from the soluble fraction. Equal amounts of protein from each fraction from both JS71 and JS7131 were applied to immobilized pH gradient (IPG) strips. The 2D gels were stained with SYPRO ruby gel stain. Figure 8 shows 2 representative gels for both YidC expression and YidC depletion conditions for the soluble fraction. The membrane fraction was not used for a quantitative analysis since the separating resolution on the 2D gels was not sufficient. The protein spots showing different intensities under YidC depletion were selected, as indicated by arrows on the figure. Protein spots were cut out and analyzed by mass spectrometry. The results are listed in Table 3. Some proteins are listed as the precursor form. This is because the database has no separate entries for preprotein and the mature proteins, and the program used to analyze the mass spectrometry data could not differentiate between a preprotein and the mature protein.
FIG. 8.
Comparison of the protein patterns in the soluble fractions from JS71 (control) and JS7131 (YidC depletion) cells. Two representative 2D gels are shown (JS71 soluble and JS7131 soluble). Gels were stained with SYPRO ruby. The arrow indicates the spots that were cut out for MS analysis.
TABLE 3.
List of significantly affected proteins from 2D gel experimentsa
| Protein type and spot no. | Protein | MW | pI | JS7131/JS71 change in protein level | Fold change in protein level | Gene expression (microarray) |
|---|---|---|---|---|---|---|
| Metabolism | ||||||
| 2 | Alkyl hydroperoxide reductase subunit C | 20,862 | 5.03 | Up | 2.00 | 1.14 (P = 0.4) |
| 5 | l-Serine dehydratase 1 | 49,388 | 5.18 | Up | 2.51 | 1.33 (P = 0.2) |
| 7 | Alkyl hydroperoxide reductase, subunit F | 56,487 | 5.47 | Up | 2.41 | 1.42 (P = 0.2) |
| 10 | Enhancing lycopene biosynthesis protein 2 | 23,252 | 4.68 | Down | 0.45 | 0.85 (P = 0.01) |
| 12 | Pyruvate kinase I | 51,039 | 5.77 | Down | 0.50 | 0.72 (P = 0.9) |
| 13 | Enolase | 45,683 | 5.32 | Down | 0.35 | 0.84 (P = 0.3) |
| 15 | 7-α-Hydroxysteroid dehydrogenase | 26,990 | 5.22 | Down | 0.43 | 0.28 (P = 0.0003) |
| 16 | KHG/KDPG aldolaseb | 22,441 | 5.57 | Down | 0.23 | No exact match |
| 17 | Flavohemoprotein | 43,954 | 5.48 | Down | 0.27 | No exact match |
| Protease and chaperones | ||||||
| 8 | Chaperone protein ClpB | 95,697 | 5.37 | Up | 3.26 | 0.56 (P = 0.007) |
| 9 | 10-kDa chaperonin GroES | 10,381 | 5.15 | Up | 2.77 | 0.71 (P = 0.02) |
| 3 | 60-kDa chaperonin GroEL | 57,464 | 4.85 | Up | 3.90 | 0.91 (P = 0.7) |
| 11 | DnaK | 69,130 | 4.83 | Down | 0.34 | 0.81 (P = 0.1) |
| Stress response | ||||||
| 4 | Universal stress protein A | 16,113 | 5.11 | Up | 2.73 | 1.23 (P = 0.03) |
| Transporter | ||||||
| 6 | Maltose-binding periplasmic protein precursor | 43,360 | 5.53 | Up | 9.04 | 2.53 (P = 0.0008) |
| 14 | Cystine-binding periplasmic protein precursor | 29,021 | 6.21 | Down | 0.42 | 0.43 (P = 0.0005) |
| Other | ||||||
| 1 | Protein YgiW precursor | 14,059 | 5.08 | Up | 3.57 | 0.54 (P = 0.0007) |
Significantly affected means a >2-fold increase or >2-fold decrease.
KHG, 4-hydroxy-2-oxoglutarate; KDPG, phospho-2-dehydro-3-deoxygluconate.
We succeeded in identifying 17 proteins in which the amounts of the proteins in the soluble fraction were changed by YidC depletion either greater than a 2-fold increase or greater than a 2-fold decrease. The predicted molecular mass and pI reasonably match the measured molecular mass and pI for each protein. Nine proteins were found in larger amounts and 8 in smaller amounts in the YidC-depleted cells. Notably, we detected only a fraction of the known proteins on the 2D gels. This proteomic study was carried out to identify relatively abundant proteins with altered protein levels in the fractions when YidC is depleted. Our intent was not to identify exhaustively all of the proteins affected by YidC depletion. We confirmed the altered levels of several proteins by Western blotting (Fig. 9). In addition, we showed the level of Foc is strongly reduced in the membrane fraction when YidC is depleted (39, 45) and Foa is reduced (45). MalF was also found to be decreased when YidC is depleted, as previously reported (41).
FIG. 9.
Western blot analysis of selected proteins. To verify the 2D gel results, protein samples were collected from cell lysates derived from JS7131 and JS71 after 5 h of growth in LB medium plus 0.2% glucose and subjected to Western blot analysis using the indicated antibodies. YidC, phage shock protein PspA, MBP, and the chaperones DnaJ and GroEL were analyzed. In the case of MalF and the Fo ATPase subunits, it was necessary to enrich the proteins by isolating the membranes. Inner membrane vesicles from JS7131 and JS71 under the 5-h YidC depletion condition were purified and analyzed by Western blotting using antibodies against Foa, Fob, Foc, and MalF.
(ii) Stress response.
Universal stress protein UspA, which had shown a 23% upregulation on the mRNA level (see the Excel file in the supplemental material), showed a 2.7-fold increase on the 2D gel. UspA functions to protect the cell from DNA damage (6). This gene is upregulated when E. coli cells are subjected to growth arrest conditions (19, 20), regardless of the reason for the cell stasis. Thus, we believe the level of UspA protein is elevated because the growth of the cells is arrested after YidC depletion in JS7131.
The phage shock protein A was well resolved on the 2D gel of the membrane fraction (data not shown) and showed a 13-fold increase consistent with its mRNA showing a 13-fold increase on the gene chip. This agrees with several previous studies which showed that phage shock protein response was switched on during the YidC depletion condition (12).
(iii) Proteases and chaperones.
The chaperones that were enhanced after YidC depletion on the 2D gels were GroEL (4-fold), GroES (3-fold), and ClpB (3-fold) (Fig. 8; Table 3). Since the YidC depletion has been shown to result in the misfolding of certain polytopic membrane proteins (17), the increase in the levels of chaperones was somewhat expected. However, none of them showed significant upregulation on the mRNA level (see the Excel file in the supplemental material), which could be a problem due to a poor probe design. To verify the mRNA level, groEL and groES were subjected to real-time PCR experiments using different probes designed in our lab. In real-time PCR experiments, while groEL showed no significant change, groES showed an 8.1-fold upregulation (P = 0.0006). Why gene expression of GroEL is not significantly changed but the protein level increases is not clear; it may be that under YidC depletion conditions, the GroEL protein itself is stabilized.
(iv) Metabolism and transporters.
YidC depletion also leads to changes at the protein levels of proteins and enzymes involved in cellular metabolism (Table 3). Likewise, the maltose binding protein is increased 9-fold in the soluble fraction (which includes periplasmic and cytoplasmic proteins) when YidC is depleted for 5 h. Exactly why this change occurs is unclear. On the other hand, the periplasmic cysteine-binding protein FliY is found in reduced levels in the soluble fraction on the 2D gel. This may be due to JS71 entering the early stationary phase, causing higher expression of this well-known stationary-phase-induced gene (see Fig. S2 in the supplemental material) and leading to a relative decrease for the FliY protein in the JS7131 strain, which is not in stationary phase. This agrees with 0.4-fold downregulation of its mRNA on the gene chip. FliY acts as a cell density sensor and transfers the signal to alternative sigma factor sigma F. Since the JS71 culture is denser than JS7131 after 5 h of growth, FliY is more likely to be induced.
The proteomic and gene chip studies reported here show that YidC depletion results in multiple changes in the cell at the protein and mRNA level. These changes presumably allow the cell in part to cope with the defects in membrane protein insertion and folding of proteins.
DISCUSSION
The primary reason for undertaking this study was to determine if there is a quality control system that is induced to handle the stress when membrane protein assembly and folding are impaired by YidC depletion. A clear role of the Cpx envelop stress response was found. We determined also in this study that the PspA induction that occurs during YidC depletion is largely independent of the PMF. We performed a microarray analysis at 5 h so that we could evaluate the broad physiological contribution of YidC to the bacterial cell and the considerable consequences to the cell when it is depleted. Of course, this includes primary, secondary, and tertiary effects. As a primary effect, YidC-dependent substrates such as subunits a and c of the F1Fo ATP synthase and CyoA start to be inhibited for membrane insertion after 1 h of depletion and are strongly inhibited in membrane insertion after 3 h of depletion (J. Yuan and R. E. Dalbey, unpublished data).
In the microarray analysis, we confirmed the induction of the phage shock response during YidC depletion (12). We also found that the universal stress response protein A was upregulated in JS7131 under the YidC depletion condition. This is most likely because of an induced growth arrest, previously shown to switch on the expression of this protein (18). Another family of stress response proteins shown to be highly upregulated is the cold shock protein Csp family, which generally functions as RNA chaperones. The heat shock proteins were generally not affected. Of the 17 heat shock proteins that can be analyzed by the gene chip method, only 2 of them, HtrC and Ddg, showed significant upregulation (see the Excel file in the supplemental material). However, the heat shock proteins GroEL and GroES were found in higher abundance in the soluble fraction when YidC was depleted. Since YidC depletion affects the PMF of the cell (39), we compared our microarray study with YidC-depleted cells with a published microarray study from another group where E. coli cells were treated with CCCP to dissipate the proton gradient across the inner membrane (22). The two sets of micro-gene chip data were drastically different, showing that YidC is important for physiological processes other than to assemble the respiratory complexes needed to generate a PMF.
YidC depletion affects gene expression of ATP synthase and electron transport chain complexes.
Our data indicate that the three integral membrane protein subunits of the Fo sector of the ATP synthase are significantly upregulated at the mRNA level upon YidC-depletion. Specifically, the genes encoding subunits a, b, and c from the Fo sector of ATP synthase were upregulated 2- to 3-fold (see the Excel file in the supplemental material). Likewise, the expression of the genes encoding the F1 sector proteins was upregulated 1.5- to 3-fold (see the Excel file in the supplemental material).
In the YidC-depleted cells, the expression of the genes encoding CyoABCDE (cytochrome bo3 ubiquinol oxidase) was upregulated (ranging from 2.5- to 6.8-fold) (Table 1; see the Excel file in the supplemental material). cyoB and cyoD were selected and checked by real-time PCR, which showed 2.27- and 2.29-fold upregulation, respectively. This upregulation is reasonable as a response to assemble more cytochrome bo3 oxidase and F1FO ATP synthase complexes and to compensate for the reduced amount of these complexes in the cell inner membrane. The identity of the signal that switches on transcription of the ATP synthase and cytochrome bo3 oxidase operons warrants further investigation. We suspect that YidC-depleted cells are unable to maintain the cytoplasmic pH due to the impaired proton pumps. This explains the upregulated cadA and cadB genes, which were previously shown to be induced under the low internal cellular pH condition (16).
Along with the genes encoding F1Fo ATP synthase and cytochrome bo3 oxidase, other electron transport chain complexes also showed an interesting regulation pattern when YidC is depleted. Genes encoding the subunits for the NADH dehydrogenase I complex in the electron transport chain were upregulated: nuoABCEFGHIJKLMN showed 1.7 to 4.9-fold upregulation (see the Excel file in the supplemental material). For real-time PCR analysis, nuoB and nuoM were selected, and we found 3.19- and 2.96-fold upregulation, respectively. We suspect the NADH dehydrogenase II complex that normally operates under aerobic conditions is perturbed by the YidC depletion and complex I is induced instead. At the same time, genes encoding the subunits for the fumarate reductase complex were downregulated. Specifically, genes frdABCD of fumarate reductase were downregulated 0.2- to ∼0.4-fold (see below). Interestingly, it has recently been shown that FrdA is strongly affected by YidC depletion of anaerobically growing E. coli (24).
Our hypothesis is that the inhibition in the assembly of electron transport chain complexes likely affects the redox balance inside YidC-depleted cells, causing quinols and oxygen to accumulate in the cell. To verify this hypothesis, we first checked the fumarate nitrate reductase (Fnr) pathway, which is one of the major regulatory pathways used to handle redox balance problems. Fnr is an O2-sensing transcriptional regulator, which can activate or inactivate transcription of certain genes when low oxygen levels are present. Under these conditions, Fnr can form dimers and thereby bind their promoter regions. For the frdABCD, aspA, fumB, and pfl genes, Fnr acts as an activator at low oxygen levels (11, 31, 33). Thus, in the presence of oxygen, the transcription level is low in all of these genes. In our data set, frdABCD genes were downregulated and also pfl was slightly downregulated. In the presence of oxygen, an increase in the transcription level of cyoABCD and sdhABCD genes had been reported (3, 21). Our results show that the expression of the cyoABCD genes was upregulated and the expression of sdhABCD genes was slightly upregulated when YidC is depleted, indicating that sufficient oxygen was available and Fnr was not functioning as a repressor. Our data agree well with data reported in a previous study which used a gene chip method to study the effects of oxygen availability and Fnr pathway. For a total of 36 genes analyzed in that paper which were claimed to use Fnr dimer as a repressor, the expression level is expected to be elevated when oxygen is present (see Table S5 in the supplemental material) (27). Indeed, 18 of these genes were upregulated according to our gene chip data, while others either were not found in our analysis or were not affected (see the Excel file in the supplemental material). For a total of 58 genes which were claimed to use Fnr dimer as an activator (see Table S6 in the supplemental material) (27), the expression level should be maintained at a normal level in our experiment, where more oxygen is present. We found that 47 of these genes showed no significant change, 2 of them were not found, 7 were upregulated, and 2 were downregulated (see the Excel file in the supplemental material). We conclude that oxygen is not limiting under YidC depletion conditions.
The high-level induction of the psp operon under the YidC-depleted condition is primarily not due to the disturbance of the PMF.
PspA is a highly expressed protein when YidC is depleted in the cell (12, 39). The induction of pspA in YidC-depleted cells was thought to be the result of an impaired PMF because YidC depletion leads to a decrease in the PMF (39) and pspA was also induced after E. coli K561 cells were exposed to 40 or 60 μM CCCP (42). It is well established that CCCP, a protonophore, collapses the proton gradient across the inner membrane. Kleerebezem et al. had shown that the PspA protein helps to maintain the PMF when a mutant PhoE protein is overexpressed and makes the membrane leaky (13). However, our data show that CCCP treatment only leads to a 2- to 4-fold induction of pspA, compared to 30-fold pspA induction when YidC is depleted (Fig. 6). Also, we noticed that increasing the CCCP concentrations from 1.25 to 25 μM did not lead to an increase of PspA. Additionally, we found that the high concentration of CCCP (40 μM) that Weiner and Model used for their study had a much stronger effect on protein export and growth of the cell than YidC depletion (Fig. 6) (42).
Our data are supported by another gene chip study in which the authors showed that CCCP (40 μM) did not activate by 2-fold the phage shock response operon (22; Volker Wendisch, University of Muenster, personal communication). In this previous gene chip study, CCCP was added for 3 h of growth at a final CCCP concentration of 40 μM. While we realize one has to be careful comparing different microarray studies (where different growth media and genetic backgrounds of the strain are used), we find it intriguing that the overall pattern of regulated genes was completely different from what we observed for the YidC-depleted cells (22; V. Wendisch, personal communication). For example, among 27 genes which were highly upregulated (>5-fold) in the CCCP-treated cells, only one of them was found to be upregulated in our YidC depletion study (see Table S4 in the supplemental material). For the 10 genes which were highly downregulated (<30%) in CCCP-treated cells, none was found to be downregulated in the YidC-depleted cells in our study. Strikingly, the phage shock operon was not measurably induced upon CCCP treatment (22; V. Wendisch, personal communication), while YidC depletion caused significant induction of phage shock proteins. Therefore, we conclude that PspA is strongly induced when membrane protein insertion is perturbed to compensate for the problems that result when the membrane is damaged, aside from a drop in the PMF. The most likely scenario is that multiple factors activate the psp operon once YidC is depleted, and it will be interesting to determine what factors are involved in this activation.
Quality control system for bacterial membrane protein.
We found in our microarray study that the expression of several protease and chaperone genes was upregulated, possibly to cope with the defects in membrane protein insertion, folding, and assembly when the YidC level is reduced (see the Excel file in the supplemental material). This includes the genes encoding peptidyl prolyl cis-trans isomerase A, protease III (a carboxypeptidase), DnaJ, FimC, and trigger factor. In addition the levels of ClpB, GroEs, and GroEL proteins in the soluble fraction were increased when YidC was depleted. Surprisingly, expression of a number of inner membrane proteases such as FtsH, GlpG, HtpX, YaeL, and YfbL (see the Excel file in the supplemental material for the complete list) were upregulated to only a small extent (140% to 192% compared to control). Since there is no obvious inner membrane protease specifically involved in handling protein stress due to misfolding of membrane proteins, the quality control of inner membrane proteins is most likely handled by several proteases.
Our results support the hypothesis that the Cpx pathway plays a role in dealing with improper membrane protein biogenesis, in addition to its well-established functions in handling altered lipid composition, overproduction of the outer membrane lipoprotein, and high pH (25). We observed upregulation of dsbA and ppiA genes from our gene chip study (see the Excel file in the supplemental material) and the cpxP gene from real-time PCR data. We compared our gene chip data with data from another study which identified 26 genes that are regulated by the Cpx pathway (see Table S1 in the supplemental material) (7). Eleven of the genes showed a similar regulation pattern in our study, and 2 of them were not found, while others were either not significantly affected or did not give reliable data. While not all of the Cpx-regulated genes are activated in the YidC depletion study, there is still good agreement. Importantly, current studies support the notion that CpxP (which is upregulated 15-fold) can only be induced by the Cpx pathway (T. Silhavy, personal communication). Therefore, we conclude that the Cpx stress pathway is activated by YidC depletion, and this supports a previous study that proposed a Cpx activation after YidC is depleted based on cpxP and rpoHP3 promoter LacZ fusions (32).
Our data clearly do not support the activation of the sigma E pathway, as previously proposed (32). For sigma E pathway-controlled genes, among 31 transcription units, which were shown to be regulated by the sigma E factor with the conserved promoter motif upstream of the genes (see Table S2 in the supplemental material) (26), 14 of them showed a sigma E-controlled regulation pattern, 4 of them showed the opposite regulation pattern, and 13 of them did not show a significant change. Also, rpoE mRNA is usually a good indicator of sigma E activity, and there was no change in our microarray data. Therefore, no significant correlation was found in this analysis.
Conclusion.
In this study, the global pattern of gene expression at the mRNA and protein level was obtained and analyzed under YidC depletion conditions in E. coli. We demonstrated that the motility of E. coli cells is impaired and the cell morphology is altered when YidC is depleted. Our data show that YidC depletion affects various bacterial electron transport chain complexes, and their genes are upregulated to handle these problems. When analyzing the stress response caused by YidC depletion, we found that the Cpx pathway is induced upon depletion of YidC. Surprisingly, our CCCP experiments suggested that the phage shock response that is induced by YidC depletion is not directly the result of the partially collapsed PMF.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM63862 to R.E.D. and by DFG grant Ku749/6-1.
Footnotes
Published ahead of print on 8 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Celebi, N., L. Yi, S. J. Facey, A. Kuhn, and R. E. Dalbey. 2006. Membrane biogenesis of subunit II of cytochrome bo oxidase: contrasting requirements for insertion of N-terminal and C-terminal domains. J. Mol. Biol. 357:1428-1436. [DOI] [PubMed] [Google Scholar]
- 2.Chen, M., K. Xie, J. Yuan, L. Yi, S. J. Facey, N. Pradel, L. F. Wu, A. Kuhn, and R. E. Dalbey. 2005. Involvement of SecDF and YidC in the membrane insertion of M13 procoat mutants. Biochemistry 44:10741-10749. [DOI] [PubMed] [Google Scholar]
- 3.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. [DOI] [PubMed] [Google Scholar]
- 5.Danese, P. N., and T. J. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diez, A., N. Gustavsson, and T. Nystrom. 2000. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK-dependent regulatory pathway. Mol. Microbiol. 36:1494-1503. [DOI] [PubMed] [Google Scholar]
- 7.Dorel, C., P. Lejeune, and A. Rodrigue. 2006. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res. Microbiol. 157:306-314. [DOI] [PubMed] [Google Scholar]
- 8.du Plessis, D. J., N. Nouwen, and A. J. Driessen. 2006. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281:12248-12252. [DOI] [PubMed] [Google Scholar]
- 9.Geller, B. L., N. R. Movva, and W. Wickner. 1986. Both ATP and the electrochemical potential are required for optimal assembly of pro-OmpA into Escherichia coli inner membrane vesicles. Proc. Natl. Acad. Sci. U. S. A. 83:4219-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasona, A., K. Zuobi-Hasona, P. J. Crowley, J. Abranches, M. A. Ruelf, A. S. Bleiweis, and L. J. Brady. 2007. Membrane composition changes and physiological adaptation by Streptococcus mutans signal recognition particle pathway mutants. J. Bacteriol. 189:1219-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, H. M., and R. P. Gunsalus. 1987. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 169:3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, S. E., L. J. Lloyd, K. K. Tan, and M. Buck. 2003. Secretion defects that activate the phage shock response of Escherichia coli. J. Bacteriol. 185:6707-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleerebezem, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15:162-171. [PMC free article] [PubMed] [Google Scholar]
- 14.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 15.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng, S. Y., and G. N. Bennett. 1992. Regulation of the Escherichia coli cad operon: location of a site required for acid induction. J. Bacteriol. 174:2670-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagamori, S., I. N. Smirnova, and H. R. Kaback. 2004. Role of YidC in folding of polytopic membrane proteins. J. Cell Biol. 165:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nystrom, T., and F. C. Neidhardt. 1992. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 6:3187-3198. [DOI] [PubMed] [Google Scholar]
- 19.Nystrom, T., and F. C. Neidhardt. 1996. Effects of overproducing the universal stress protein, UspA, in Escherichia coli K-12. J. Bacteriol. 178:927-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nystrom, T., and F. C. Neidhardt. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11:537-544. [DOI] [PubMed] [Google Scholar]
- 21.Park, S. J., C. P. Tseng, and R. P. Gunsalus. 1995. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol. Microbiol. 15:473-482. [DOI] [PubMed] [Google Scholar]
- 22.Polen, T., D. Rittmann, V. F. Wendisch, and H. Sahm. 2003. DNA microarray analyses of the long-term adaptive response of Escherichia coli to acetate and propionate. Appl. Environ. Microbiol. 69:1759-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Possot, O. M., L. Letellier, and A. P. Pugsley. 1997. Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol. Microbiol. 24:457-464. [DOI] [PubMed] [Google Scholar]
- 24.Price, C. E., and A. J. Driessen. 2008. YidC is involved in the biogenesis of anaerobic respiratory complexes in the inner membrane of Escherichia coli. J. Biol. Chem. 283:26921-26927. [DOI] [PubMed] [Google Scholar]
- 25.Raivio, T. L. 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56:1119-1128. [DOI] [PubMed] [Google Scholar]
- 26.Rhodius, V. A., W. C. Suh, G. Nonaka, J. West, and C. A. Gross. 2006. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salmon, K., S. P. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 278:29837-29855. [DOI] [PubMed] [Google Scholar]
- 28.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 29.Samuelson, J. C., F. Jiang, L. Yi, M. Chen, J. W. de Gier, A. Kuhn, and R. E. Dalbey. 2001. Function of YidC for the insertion of M13 procoat protein in E. coli: translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J. Biol. Chem. 276:34847-34852. [DOI] [PubMed] [Google Scholar]
- 30.Santos, J. M., M. Lobo, A. P. Matos, M. A. De Pedro, and C. M. Arraiano. 2002. The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol. Microbiol. 45:1729-1740. [DOI] [PubMed] [Google Scholar]
- 31.Sawers, G. 1993. Specific transcriptional requirements for positive regulation of the anaerobically inducible pfl operon by ArcA and FNR. Mol. Microbiol. 10:737-747. [DOI] [PubMed] [Google Scholar]
- 32.Shimohata, N., S. Nagamori, Y. Akiyama, H. R. Kaback, and K. Ito. 2007. SecY alterations that impair membrane protein folding and generate a membrane stress. J. Cell Biol. 176:307-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiro, S., and J. R. Guest. 1987. Activation of the lac operon of Escherichia coli by a mutant FNR protein. Mol. Microbiol. 1:53-58. [DOI] [PubMed] [Google Scholar]
- 34.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Bloois, E., H. L. Dekker, L. Froderberg, E. N. Houben, M. L. Urbanus, C. G. de Koster, J. W. de Gier, and J. Luirink. 2008. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 582:1419-1424. [DOI] [PubMed] [Google Scholar]
- 36.van Bloois, E., G. J. Haan, J. W. de Gier, B. Oudega, and J. Luirink. 2006. Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J. Biol. Chem. 281:10002-10009. [DOI] [PubMed] [Google Scholar]
- 37.van Bloois, E., G. Jan Haan, J. W. de Gier, B. Oudega, and J. Luirink. 2004. F(1)F(0) ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Lett. 576:97-100. [DOI] [PubMed] [Google Scholar]
- 38.Van Der Laan, M., P. Bechtluft, S. Kol, N. Nouwen, and A. J. Driessen. 2004. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Der Laan, M., M. L. Urbanus, C. M. Ten Hagen-Jongman, N. Nouwen, B. Oudega, N. Harms, A. J. Driessen, and J. Luirink. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. U. S. A. 100:5801-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkert, M. R., L. I. Hajec, Z. Matijasevic, F. C. Fang, and R. Prince. 1994. Induction of the Escherichia coli aidB gene under oxygen-limiting conditions requires a functional rpoS (katF) gene. J. Bacteriol. 176:7638-7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner, S., O. Pop, G. J. Haan, L. Baars, G. Koningstein, M. M. Klepsch, P. Genevaux, J. Luirink, and J. W. de Gier. 2008. Biogenesis of MalF and the MalFGK(2) maltose transport complex in Escherichia coli requires YidC. J. Biol. Chem. 283:17881-17890. [DOI] [PubMed] [Google Scholar]
- 42.Weiner, L., and P. Model. 1994. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. U. S. A. 91:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie, K., and R. E. Dalbey. 2008. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat. Rev. Microbiol. 6:234-244. [DOI] [PubMed] [Google Scholar]
- 44.Yi, L., N. Celebi, M. Chen, and R. E. Dalbey. 2004. Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 279:39260-39267. [DOI] [PubMed] [Google Scholar]
- 45.Yi, L., F. Jiang, M. Chen, B. Cain, A. Bolhuis, and R. E. Dalbey. 2003. YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry 42:10537-10544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.