Abstract
Antimicrobial peptides are critical for innate antibacterial defense. Both Gram-negative and Gram-positive microbes have mechanisms to alter their surfaces and resist killing by antimicrobial peptides. In Vibrio cholerae, two natural epidemic biotypes, classical and El Tor, exhibit distinct phenotypes with respect to sensitivity to the peptide antibiotic polymyxin B: classical strains are sensitive and El Tor strains are relatively resistant. We carried out mutant screens of both biotypes, aiming to identify classical V. cholerae mutants resistant to polymyxin B and El Tor V. cholerae mutants sensitive to polymyxin B. Insertions in a gene annotated msbB (encoding a predicted lipid A secondary acyltransferase) answered both screens, implicating its activity in antimicrobial peptide resistance of V. cholerae. Analysis of a defined mutation in the El Tor biotype demonstrated that msbB is required for resistance to all antimicrobial peptides tested. Mutation of msbB in a classical strain resulted in reduced resistance to several antimicrobial peptides but in no significant change in resistance to polymyxin B. msbB mutants of both biotypes showed decreased colonization of infant mice, with a more pronounced defect observed for the El Tor mutant. Mass spectrometry analysis showed that lipid A of the msbB mutant for both biotypes was underacylated compared to lipid A of the wild-type isolates, confirming that MsbB is a functional acyltransferase in V. cholerae.
Pathogenic bacteria that colonize the digestive tract must overcome a variety of stresses imposed upon them by the host. Epithelial cells in the crypts of the intestinal lumen (Paneth cells and enterocytes) produce large amounts of antimicrobial peptides called defensins (16). Defensins, like most antimicrobial peptides, are thought to act by associating with the lipopolysaccharide (LPS) on the bacterial surface (through electrostatic interactions) and then permeabilizing the membranes, leading to cell death (37, 48). Gram-negative bacteria have developed a wide range of strategies to overcome the antimicrobial activity of these peptides, including production of proteases that degrade the peptides (41), production of secretory proteins that bind the peptides and prevent them from accessing their target (21), production of efflux systems that actively pump antimicrobial peptides back into the environment if they access the bacterial cytoplasm (36), and incorporation of positively charged groups into lipid A, which reduces the net anionic charge of the bacterial surface and decreases the affinity of the peptides for the membrane (10, 13, 14).
LPS of Gram-negative bacteria is composed of three main parts: (i) the O-antigen polysaccharide (O-PS); (ii) the relatively conserved core polysaccharide (core-PS); and (iii) lipid A, the hydrophobic lipid component responsible for biological activities within the host (9, 25). The lipid A region of the LPS is anchored in the bacterial outer membrane, and the hydrophilic core-PS and O-PS project outward into the environment. LPS comprises 70% of the bacterial outer membrane and is the main surface-associated antigen recognized by the innate immune system. Toll-like receptors in the host recognize the lipid A portion of the LPS in association with MD2 and CD14 and stimulate inflammation to attract immune cells and clear bacterial infections (5, 27). The strong immune response to lipid A is the reason that LPS has historically been referred to as “endotoxin” (20). Some pathogens regulate the structure of their lipid A and its acylation patterns in order to adapt to the host environment, thereby contributing to greater fitness within the host (12, 31).
Vibrio cholerae causes cholera, an epidemic diarrheal disease. Disease occurs when contaminated food or water is ingested, resulting in a voluminous secretory diarrhea that can lead to dehydration and death if left untreated. The V. cholerae species is not homogeneous, with distinctions made on the basis of serogroup, serotype, biotype, production of cholera toxin, and potential for epidemic spread. While more than 200 serogroups have been identified, only two of these, O1 and O139, are associated with epidemic cholera (33). V. cholerae O1 strains can be subdivided into two biotypes, classical and El Tor, which differ biochemically and clinically (3). The first six cholera pandemics were caused by the classical biotype, but the current (seventh) pandemic has been caused by the El Tor biotype (33). Classical strains typically cause a more severe disease, while El Tor strains cause less severe and sometimes even asymptomatic cases. However, El Tor strains appear to have increased fitness in the environment, which may be why they have largely replaced classical strains as the cause of disease in recent years (49).
The subdivision into the classical or El Tor biotype is based on several laboratory tests (3). One of the commonly used tests is assessing sensitivity of the strain to the antimicrobial peptide polymyxin B. Classical strains are very sensitive to polymyxin B, while El Tor strains are relatively resistant. We hypothesize that differences in surface structures of the two biotypes are responsible for differential sensitivity. To test this and to determine the genetic basis of antimicrobial peptide resistance in V. cholerae, we carried out genetic screens to identify genes associated with resistance and sensitivity to polymyxin B in El Tor and classical V. cholerae, respectively. As a result of these screens, we chose to further characterize the role of msbB, a lipid IVA acyltransferase gene, with regard to antimicrobial peptide resistance and virulence in V. cholerae. We report that msbB contributes to resistance of El Tor strains to all antimicrobial peptides tested. Mutation of msbB in a classical strain led to significantly reduced innate resistance to several antimicrobial peptides, not including polymyxin B. While msbB mutants of both biotypes exhibit decreased colonization of infant mice, a more significant decrease was observed for the El Tor mutant. Mass spectrometry analysis confirmed that deletion of msbB from either biotype resulted in loss of an acyl chain, as expected. These results suggest that msbB from V. cholerae is required for wild-type antimicrobial peptide resistance and colonization. However, some biotype-specific phenotypes imply that the role of msbB may be different in each biotype.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The V. cholerae classical strain O395 and El Tor strains N16961 and C6706 were used in this study. The Escherichia coli strains JM101 and DH5αλpir were used for cloning, and SM10λpir was used for conjugation with V. cholerae. Plasmids used in this study included the suicide vector pKAS32 (39), the mariner transposon suicide vector pFD1 (32), and the arabinose-inducible expression vector pBAD18-Kan (15). Expression of transposase from pFD1 was induced by the addition of isopropyl-β-d-thiogalactopyranoside (Invitrogen, Carlsbad, CA) to a final concentration of 1 mM, and expression of pBAD was induced by the addition of l-arabinose to 0.1%. E. coli strains were transformed by standard methods (34), and plasmid DNA was introduced into V. cholerae by electroporation or by filter conjugation with SM10λpir. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and streptomycin, 100 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Invitrogen) was used in LB agar at 40 μg/ml.
Strain and plasmid construction.
El Tor (C6706) V. cholerae containing a transposon insertion in msbB was obtained from a nonredundant transposon insertion library (2). The majority of the transposon (including a kanamycin resistance cassette) was removed by FLP-mediated recombination, using the plasmid pFlpE, leaving behind a 192-bp scar. msbB in classical (O395) V. cholerae was disrupted by transduction of the transposon insertion from the El Tor background, using CP-T1ts phage (19). Again, the transposon was removed by FLP-mediated recombination for the purposes of complementation studies.
Full-length msbB was amplified from V. cholerae O395 chromosomal DNA by use of Expand Hi-Fidelity polymerase (Roche Molecular Biochemicals, Indianapolis, IN). After amplification, the PCR products were digested with EcoRI and XbaI and ligated into the arabinose-inducible expression vector pBAD18-Kan (15).
Peptide killing assays.
Antimicrobial peptide susceptibility assays were conducted as described previously (38). Briefly, overnight cultures of V. cholerae were subcultured 1:100 into LB and grown for 3 h at 37°C (to an optical density at 600 nm [OD600] of ∼0.5). Samples of 5 μl of peptide solution (at a concentration 10 times higher than the final concentration) were placed into wells of a 96-well polystyrene plate, and 45 μl of the bacterial culture was added. After 1 h of incubation at 37°C with shaking, serial dilutions of each culture were plated on LB agar plates. The number of CFU was counted after overnight incubation at 37°C. The percent survival was calculated as follows: survival (%) = (CFU[peptide treatment]/CFU[no treatment]) × 100.
Infant mouse colonization assay.
Three- to five-day-old CD1 mice were inoculated intragastrically with approximately 106 bacteria. The inocula were 50-50 mixtures of the strain of interest (O395 ΔmsbB or C6706 ΔmsbB ΔlacZ) and the appropriate control strain (O395 ΔlacZ or C6706, respectively). Inoculated mice were incubated at 30°C for 16 h, at which time they were sacrificed and their intestines were removed and homogenized. Serial dilutions of the intestinal homogenates were plated on LB agar plates containing streptomycin and X-Gal. Blue and white colonies were counted after overnight growth at 37°C. An aliquot of the inoculum was also serially diluted and plated for enumeration. As an in vitro control, the strain mixtures used to inoculate the mice were diluted 1:100 in fresh LB, grown overnight at 37°C, and plated onto LB plates containing streptomycin and X-Gal, and again the blue and white colonies were counted. The competitive index was calculated as the ratio of the wild type to the mutant in the input divided by the ratio of the wild type to the mutant in the output.
LPS purification.
Samples were prepared by growing 2 liters of each strain in LB medium overnight at 37°C. Stationary-phase cultures were dried and extracted according to the hot phenol-water procedure (46), using a specific protocol kindly supplied by Uwe Mamat. Briefly, the bacterial biomass from 2 liters of each stationary-phase culture was dried by sequential washing with phosphate-buffered saline (PBS), absolute ethanol, acetone, and diethyl ether. The dry biomass was homogenized in water and extracted three times with hot 90% phenol. The water phase containing the LPS was dialyzed for 1 week and lyophilized. Samples were further purified by sequential treatment with DNase I/RNase I and proteinase K and then pelleted by ultracentrifugation before 2 days of dialysis and lyophilization.
Mass spectrometry of V. cholerae LPS.
Electrospray ionization-Fourier transform ion cyclotron resonance mass spectrometry (ESI-FTICR MS) was performed using an Apollo II ion source and an APEX-Q instrument (Bruker Daltonics, Billerica, MA) in the negative-ion mode. Lipid samples were dissolved in a 49.95/49.95/0.1 (vol/vol/vol) mixture of isopropanol, water, and triethylamine (pH ∼9) and electrosprayed at a flow rate of 70 μl/h, with a capillary voltage of 3.7 kV and a drying gas temperature of 130°C.
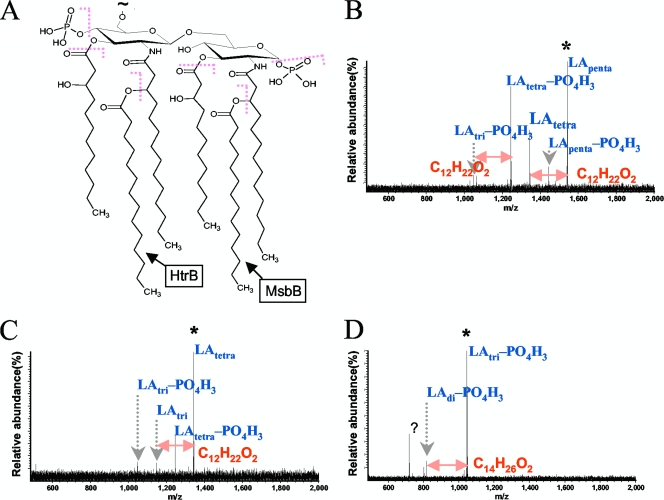
For tandem mass spectrometry (MS/MS) experiments, collision-activated dissociation (CAD) was performed with a collision cell direct current offset of 30 to 40 V, with Ar as the collision gas. All mass spectra were acquired with XMASS software (version 6.1; Bruker Daltonics) in broadband mode from m/z 200 to 5,000, using 256,000 data points, and were summed over 20 to 60 scans. Data processing was performed with MIDAS analysis software (35). LPS from E. coli K-235 (Sigma-Aldrich, St. Louis, MO) was used to perform external calibration based on two lipid A anionic species (m/z 1,796.212 for hexa-acylated lipid A and m/z 1,017.639 for triacylated lipid A lacking one phosphate group). Mass spectral peak assignments were based on accurate masses (better than 10 ppm) calculated from the V. cholerae lipid A structure (45) (see Fig. 5A) and verified by tandem mass spectrometry.
FIG. 5.
Structure of lipid A from O1 V. cholerae (A) and CAD tandem mass spectra of LApenta-PO4H3 (B), LAtetra (C), and LAtri (D) from C6706 ΔmsbB. A collision cell voltage of 30 to 40 V was used to induce fragmentation of mass-selectively accumulated precursor ions. Observed bond cleavages are summarized in panel A. Similar results were obtained from tandem mass spectrometry of C6706, O395, and O395 ΔmsbB LPS anions (data not shown). Precursor ions are marked with asterisks.
NF-κB induction.
HEK-Blue-4 cells (HEK293 cells stably transfected with Toll-like receptor 4 [TLR4], MD2, and CD14 as well as a secreted alkaline phosphatase [SEAP] reporter gene under the control of an NF-κB-inducible promoter) were obtained from Invivogen (San Diego, CA) and were maintained according to the vendor-supplied protocols. Stable expression of SEAP under the control of the NF-κB promoter is induced by LPS in this cell line, resulting in accumulation of extracellular SEAP proportional to the level of NF-κB induction. Tenfold dilutions of purified LPS samples were added to wells of a 96-well plate, and approximately 25,000 HEK-Blue-4 cells were added to each well. The plates were incubated at 37°C in a CO2 incubator overnight. SEAP was assayed spectrophotometrically by using an alkaline phosphatase (AP)-specific chromogen present in the vendor-supplied HEK detection medium and reading the absorbance of the culture at 620 nm.
RESULTS
Genetic screens for mutants demonstrating altered resistance to polymyxin B.
The level of inhibition by polymyxin B is one of the standard tests used for biotyping V. cholerae strains, with El Tor strains classified as resistant (3). Since the genetic basis for this differential sensitivity is currently unknown, we developed two genetic screens aimed at identifying mutants of the two biotypes demonstrating altered resistance to this antimicrobial peptide.
The first screen was a straightforward transposon mutagenesis of O395 (classical) V. cholerae with subsequent selection of the mutants on plates containing an inhibitory concentration of polymyxin B. O395 cannot grow on solid media containing 12 μg/ml polymyxin B, and therefore any mutants that survive are predicted to have a transposon insertion in a gene that results in increased resistance to this antimicrobial peptide. O395 V. cholerae was mutagenized using a mariner-based transposon (32), and the resulting pool of mutants was plated on LB agar containing 12 μg/ml polymyxin B. From this screen, 16 colonies were obtained, and the locations of the transposon insertions were identified by random-primed sequencing. The insertions mapped to four genes, one of which was VC0212, annotated msbB. In E. coli and Shigella flexneri, MsbB transfers a myristate group to penta-acylated lipid A, which is the final step in the synthesis of lipid A (6, 40).
The second screen was designed to identify transposon insertions in El Tor V. cholerae strains showing decreased resistance to polymyxin B. El Tor V. cholerae strain N16961 forms colonies on solid media containing at least 50 μg/ml polymyxin B, and therefore mutants that do not grow well at this concentration likely have lesions in genes required for the increased resistance phenotype of the El Tor biotype. V. cholerae N16961 was mutagenized with a mariner transposon, and the pool of mutants was plated on LB agar containing kanamycin to select for insertion of the transposon. The resulting colonies were picked into 96-well plates containing liquid medium, generating a library of approximately 5,000 mutants. This mutant library was replica plated onto LB agar plates, with and without 50 μg/ml polymyxin B. Mutants unable to form colonies in the presence of the peptide were sequenced to identify the locations of the transposon insertions. A much larger pool of mutants answered this screen than answered the screen for classical mutants that had become resistant to polymyxin B, which is not a surprising result, as the classical screen was for a gain of function. Among 35 mutants that were sequenced, we identified insertions mapping to 26 different genes. Of particular interest, one of the transposon insertions from this screen also mapped to msbB; therefore, we chose to further investigate the role of this gene in the differential resistance of the two biotypes to polymyxin B.
Strains of V. cholerae lacking msbB are sensitive to several antimicrobial peptides.
To characterize the role of MsbB in the two biotypes of V. cholerae, we constructed deletions of msbB in classical and El Tor V. cholerae and also cloned msbB into an inducible plasmid for use in complementation studies. For the El Tor studies, we used strain C6706, as this strain was used to generate the nonredundant library in V. cholerae (2). We identified the library clone carrying the transposon insertion in msbB and removed the transposon sequence by recombination (as it contained a kanamycin cassette). We then transduced the transposon insertion into the classical (O395) background, using the CP-T1ts transducing phage, and also removed the sequence of the transposon. This enabled the use of pBAD18-Kan-msbB for subsequent complementation studies.
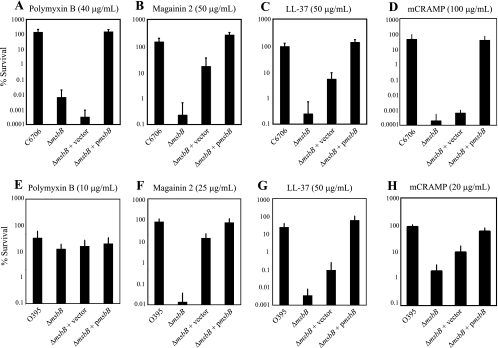
To verify that MsbB contributes to polymyxin B sensitivity/resistance in the two biotypes of V. cholerae, we examined the susceptibility of the msbB mutants to the antimicrobial peptide. When exposed to 40 μg/ml polymyxin B in liquid medium for 1 h, El Tor V. cholerae strain C6706 demonstrated no loss of plating efficiency compared to cells exposed to medium lacking the peptide. This is consistent with the classification of El Tor V. cholerae as resistant to polymyxin B. The C6706 msbB mutant demonstrated a plating efficiency >4 orders of magnitude less than that of the wild type, a phenotype that could be complemented fully by expression of msbB from a plasmid in this background (Fig. 1A). We tested classical strain O395 and its corresponding msbB deletion derivative, using a much lower concentration of polymyxin B given the more sensitive phenotype of classical strains. After exposure to 10 μg/ml polymyxin B in liquid medium for 1 h, the wild-type strain plated at nearly 100% efficiency, and this was not significantly altered by mutation of msbB (Fig. 1E). Thus, although the classical strain was generally more sensitive to polymyxin B than the El Tor strain was, the level of resistance it did exhibit was independent of MsbB, unlike what was observed with the El Tor strain.
FIG. 1.
MsbB contributes to antimicrobial peptide resistance in V. cholerae. El Tor (A to D) and classical (E to H) strains were grown to mid-logarithmic phase and then treated with antimicrobial peptides for 1 h. C6706, C6706 ΔmsbB, C6706 ΔmsbB + pBAD18-Kan (vector), and C6706 ΔmsbB + pBAD18-Kan-msbB (pmsbB) were incubated with 40 μg/ml polymyxin B (A), 50 μg/ml magainin 2 (B), 50 μg/ml LL-37 (C), and 100 μg/ml mCRAMP (D). O395, O395 ΔmsbB, O395 ΔmsbB + pBAD18-Kan (vector), and O395 ΔmsbB + pBAD18-Kan-msbB (pmsbB) were incubated with 10 μg/ml polymyxin B (E), 25 μg/ml magainin 2 (F), 50 μg/ml LL-37 (G), and 20 μg/ml mCRAMP (H). After treatment with the peptides, serial dilutions of each culture were plated for enumeration. Data represent the means and standard deviations for at least three independent experiments performed in duplicate.
To determine if the phenotypes of the msbB mutants were specific to polymyxin B or were representative of a general response to antimicrobial peptides, we examined survival of the mutants in the presence of three other antimicrobial peptides, magainin 2, LL-37, and mCRAMP. Magainin 2 is a 23-amino-acid alpha-helical peptide isolated from frog skin that has been used widely to investigate the mechanism of action of cationic antimicrobial peptides. LL-37, or human cathelicidin, is a 37-amino-acid antimicrobial peptide expressed by neutrophils and epithelial cells. mCRAMP (cathelin-related antimicrobial peptide) is the mouse version of LL-37 (28). This antimicrobial peptide is the primary innate immune defense mechanism of the neonatal mouse intestine. Wild-type C6706 was resistant to exposure to 50 μg/ml magainin 2, 50 μg/ml LL-37, and 100 μg/ml mCRAMP in liquid medium for 1 h. When C6706 lacking msbB was exposed to the antimicrobial peptides at the same concentrations, survival was decreased by >2 orders of magnitude for magainin 2 and LL-37 (Fig. 1B and C) and by 5 orders of magnitude for mCRAMP (Fig. 1D). Again, wild-type resistance to these peptides could be restored by expressing msbB from a plasmid. It is notable that the presence of the vector inexplicably increased resistance to magainin 2 and LL-37. This effect was not due to the presence of the arabinose used to induce msbB expression, as the addition of arabinose did not increase resistance of C6706 ΔmsbB (no plasmid) to the peptides (data not shown). The effect was also not due to the presence of the empty vector per se, as it was the same vector used in all of the assays and we did not observe increased resistance to polymyxin B when cells carried the vector alone (Fig. 1A). Therefore, this increased resistance was likely due to inhibition of the peptide activities in the presence of kanamycin selection.
Wild-type classical strain O395 was tested for plating efficiency in 25 μg/ml magainin 2, 50 μg/ml LL-37, and 20 μg/ml mCRAMP. These concentrations range from 20 to 100% of those used for the El Tor strain. Plating efficiencies of O395 in the presence of these concentrations of peptide were very high, although for magainin 2 and LL-37 they did not quite reach 100% (Fig. 1F to H). However, when O395 lacking msbB was exposed to these antimicrobial peptides at the same concentrations used with the wild type, the plating efficiency was decreased by >2 to 3 orders of magnitude (Fig. 1F to H). Again, the presence of the empty vector increased resistance to these peptides. Overall, it appears that MsbB contributes to antimicrobial peptide resistance in El Tor and classical strains of V. cholerae but does not contribute significantly to polymyxin B resistance in classical strains.
MsbB contributes to mouse colonization in both biotypes of V. cholerae.
Resistance to antimicrobial peptides has been associated with virulence in several bacterial pathogens (11, 29). Specifically, msbB mutants of S. flexneri, pathogenic E. coli, Salmonella enterica serovar Typhimurium, and Klebsiella pneumoniae are all attenuated for virulence (4, 6, 24, 40). To determine if deletion of msbB affects V. cholerae colonization in either of the two biotypes, we inoculated infant mice with a mixture of wild-type V. cholerae of each biotype and the respective msbB deletion derivative and assessed their competitive indices. In vitro experiments comparing the growth rates of the wild-type and deletion strains at 37°C indicated that the msbB deletion did not cause a growth defect in either background (data not shown). O395 ΔmsbB showed a small but significant colonization defect compared to wild-type O395 (P = 0.015). C6706 ΔmsbB showed a more dramatic colonization defect than did C6706 (P < 0.0001) (Fig. 2). In most of the El Tor V. cholerae-inoculated mice, the mutant strain was significantly lower in the output mixture (∼10×). The average colonization defects were not significantly different between the two biotypes due to the presence of an outlier in the El Tor competition. We concluded that lipid A acylation by MsbB contributes to host colonization in both biotypes of V. cholerae, with a more significant impact observed in the El Tor background.
FIG. 2.
MsbB is required for wild-type colonization levels in V. cholerae. The indicated strains were mixed 1:1 and inoculated into infant mice. Each data point represents one mouse. The ratios of the wild-type to the mutant strains in the intestinal homogenates were divided by the ratios of the strains in the inocula to calculate the competitive indices.
LPS stimulation of TLR4.
Previous work on MsbB from V. cholerae demonstrated that it is not a functional acyltransferase when expressed in an E. coli background lacking secondary acyltransferases (17). However, based on the phenotypes observed in our previous experiments, we would predict that MsbB is a functional acyltransferase when expressed in V. cholerae. To investigate this further, we assayed stimulation of TLR4 by V. cholerae LPS preparations in HEK-Blue-4 cells. This cell line is transfected with the human TLR4, MD2, and CD14 genes as well as a secreted alkaline phosphatase reporter gene under the control of an NF-κB-inducible promoter. LPS stimulation of TLR4 in this cell line leads to activation of NF-κB and secretion of alkaline phosphatase into the growth medium. HEK-Blue-4 cells are highly sensitive to hexa-acylated LPS, and therefore we should be able to crudely determine the acylation patterns of the lipid A species in our strains by stimulating these cells with LPS preparations (43). To this end, we purified LPSs from O395, C6706, and their respective msbB deletion derivatives. Different concentrations of purified LPS were mixed with HEK-Blue-4 cells in the vendor's detection growth medium (Invivogen) and then incubated overnight. After incubation, stimulation of TLR4 was indicated by a change in color of the medium from pink to blue. The differences between the strains were determined spectrophotometrically by reading the absorbance of the cultures at 620 nm (Fig. 3). LPSs from both of the msbB deletion strains did not induce NF-κB expression in the reporter cell line at any concentration tested, whereas LPSs from the wild-type strains were active at concentrations as low as 0.01 μg/ml (Fig. 3). Notably, it appeared that LPS from the El Tor strain stimulated a greater TLR4 response than did LPS from the classical strain. This was an unexpected result, as the lipid A structures from all O1 strains are predicted to be identical. These results suggest that the lipid A species of both biotypes of V. cholerae require modification by MsbB to be recognized by TLR4.
FIG. 3.
LPS from V. cholerae lacking msbB does not stimulate TLR4. HEK-Blue-4 cells were treated overnight with 10-fold dilutions of LPS prepared from classical (O395 and O395 ΔmsbB) and El Tor (C6706 and C6706 ΔmsbB) V. cholerae. SEAP was measured colorimetrically by reading the absorbance of the wells at 620 nm. Data represent the means and standard deviations for three independent experiments performed in duplicate.
Lipid A acyltransferase activity of MsbB in classical and El Tor V. cholerae.
Based on all of our previous experiments, it appears that MsbB is a functional acyltransferase in V. cholerae, in spite of the previous reports on its activity in E. coli (17). To determine the acylation patterns of the msbB mutants in both V. cholerae biotypes, we analyzed LPS preparations from the strains using mass spectrometry. The spectra from the LPSs of both wild-type strains contained a series of expected peaks, including those representing tetra-, penta-, and hexa-acylated lipid A (Fig. 4A and C). The spectra from both msbB deletion strains (Fig. 4B and D) had similar patterns of peaks, but they were missing the peak corresponding to the mass of hexa-acylated lipid A (m/z 1,766.165 for the singly deprotonated anion), as shown in the insets of Fig. 4B and D. This result suggests that the lipid A moiety of the msbB mutants is underacylated compared to wild-type lipid A, due to loss of the final myristate group. Underacylation of LPS from msbB deletion strains is consistent with the proposed acyltransferase activity of MsbB and explains the inability of LPS from these mutants to efficiently stimulate TLR4 in the HEK-Blue-4 cell assay. The V. cholerae lipid A structure (45) is shown in Fig. 5A, with observed bond cleavages indicated in red. The CAD tandem mass spectra for lipid A species with five, four, and three (minus one phosphate) acyl chains are shown in Fig. 5B to D.
FIG. 4.
FTICR mass spectra of LPSs from strains C6706 (A), C6706 ΔmsbB (B), O395 (C), and O395 ΔmsbB (D). Peak assignments were confirmed by accurate masses and by tandem mass spectrometry as shown in Fig. 5. Unassigned peaks differ from the assigned lipid A peaks mainly by the mass of water, phosphate, or ethylene. Lipid A containing six acyl chains (m/z 1,766.165) is labeled LAhexa, lipid A containing five acyl chains (m/z 1,539.971) is labeled LApenta, lipid A containing four acyl chains (m/z 1,341.809) is labeled lipid Atetra, and lipid A containing three acyl chains (m/z 1,045.671) is labeled LAtri.
DISCUSSION
In spite of longtime use of polymyxin B resistance as a test to differentiate between the classical and El Tor biotypes of V. cholerae, the genetic basis of this difference has been unexplored. Polymyxin B and other antimicrobial peptides bind to lipid A, and many bacteria have evolved systems to modify their lipid A structure in order to increase resistance to polymyxin B and other antimicrobial peptides. In this study, we used transposon mutagenesis of both biotypes of V. cholerae to identify mutants with altered sensitivity to polymyxin B. Surprisingly, we identified msbB insertions in both screens, in spite of the fact that we were screening for sensitivity to polymyxin B in one and for resistance in the other. Due to this unexpected result, we decided to further investigate the role of MsbB in antimicrobial peptide resistance in V. cholerae. We found that MsbB is required for wild-type resistance to polymyxin B and several other antimicrobial peptides in the El Tor background but does not significantly contribute to resistance to polymyxin B in the classical background.
An unanswered question from this study is why the transposon insertion in msbB answered the classical screen for increased resistance to polymyxin B. From the antimicrobial peptide killing assays, it is clear that the constructed insertion in msbB in O395 does not dramatically increase or decrease resistance to polymyxin B. One explanation for the observed difference is that the growth conditions used for the initial screen were different from those used for the subsequent peptide killing assays. The screen selected for growth on solid medium containing an inhibitory concentration of polymyxin B, and the other assays were conducted using liquid medium with only 1 h of peptide treatment. Bacteria in the screen were therefore exposed to polymyxin B for a longer time, meaning that the screen may have enabled unidentified mutations to arise, leading to polymyxin B resistance. Complementation of the original transposon insertion with plasmid-expressed MsbB did not significantly alter plating efficiency in the presence of polymyxin B (data not shown). Thus, the transposon insertion in msbB may not in fact be the source of the polymyxin B resistance. Another possibility is that a suppressor mutation may be acquired more easily in the absence of msbB. Additionally, it is possible that the transposon insertion influences expression of adjacent genes in the V. cholerae genome, resulting in increased resistance to polymyxin B. Nevertheless, the identification of this mutant in both screens directed us to further investigate the role of msbB in polymyxin B resistance in both V. cholerae biotypes.
Antimicrobial peptide resistance is often associated with virulence in bacterial pathogens, and msbB mutants in other species have demonstrated decreased virulence (4, 6, 11, 24, 29, 40). These observations led us to test whether the msbB mutants showed decreased colonization levels in the infant mouse model of V. cholerae infection. We found that msbB is necessary for wild-type colonization in both backgrounds, with a more dramatic impact on colonization by the El Tor biotype. The fact that the msbB mutants exhibited decreased levels of infant mouse colonization compared to those of the wild type is intriguing. Adin et al. showed that deletion of msbB and other secondary acyltransferases in Vibrio fischeri results in formation of unusually shaped cells, chains of cells, and reduced motility (1). We did not find any unusual cell morphology in the V. cholerae mutants compared to the wild-type strains by electron microscopy, and motility was not affected (data not shown). Furthermore, the msbB mutants did not have a growth defect and were not outcompeted when grown with their respective wild-type strains as an in vitro control for our mouse experiments (data not shown). The most likely cause of the reduced colonization by the El Tor msbB mutant is its demonstrated sensitivity to antimicrobial peptides. Epithelial cells in the intestinal lumen of the adult mouse produce large quantities of antimicrobial peptides termed cryptdins, the mouse version of human defensins, in response to bacterial infection (7). In neonatal mice (less than 2 weeks old), the primary innate immune defense mechanism is the production of CRAMP (26). CRAMP is the murine equivalent of human LL-37 (28). The El Tor msbB mutant was far more sensitive to CRAMP than the wild type was (>5 orders of magnitude) (Fig. 1G). The classical msbB mutant was also more sensitive to CRAMP than the wild type was, but the reduction was modest, at <2 orders of magnitude. The differences in colonization observed for the two biotypes may be a reflection of their differences in sensitivity to CRAMP present in the infant mouse intestinal tract. We also cannot exclude the possibility that disruption of the outer membrane structure by loss of a lipid A acyl group results in negative secondary effects on other surface organelles. Changes in the outer membrane may lead to problems such as anchoring of the flagella or the toxin-coregulated pilus (TCP) in the membrane, both of which could have detrimental effects on bacterial colonization.
Extensive research has been carried out on V. cholerae, aimed at developing a live oral vaccine strain to stimulate long-lived immunity. The prototypical platform for such strains is one that maintains expression of toxin-coregulated pilus and the cholera toxin B subunit—a potent mucosal adjuvant in its own right—but lacks expression of the cholera toxin A subunit, which is responsible for the toxin activity that leads to the massive diarrhea associated with natural infection by V. cholerae (42). Such strains would still colonize well and induce a mucosal response against protective epitopes—including LPS—but would not produce cholera disease due to the lack of subunit A production. Several candidates with this basic design have been tested in various models, including human volunteers, and the results have been frustrated by an unacceptable level of reactogenicity, in which the vaccine strain causes residual symptoms such as nausea or diarrhea (non-cholera-like). Reactogenicity may be related to an inflammatory response induced by the vaccine strains, perhaps induced in part by stimulation of Toll-like receptors by components of the vaccine strain, such as flagellins (18, 47). Given our data showing that the msbB mutant V. cholerae strains described here were not completely deficient for colonization and also failed to stimulate TLR4 to any significant extent, we suggest that it is worth considering inclusion of msbB mutations in future V. cholerae vaccine strains, as has been done for vaccine formulations against other Gram-negative pathogens (8, 22, 23, 30, 44). Obviously, in order to try such an approach, it will be necessary to determine whether the nonstimulatory LPS produced by msbB mutant strains retains sufficient antigenicity to serve as a protective target of vaccination.
Acknowledgments
This work was supported in part by NIH grant R01AI045125 to V.J.D. and by an NSF Career Award (CHE-05-47699) to K.H. J.S.M. was a trainee of the MMMP Training Program at University of Michigan.
We thank Uwe Mamat for the detailed LPS purification protocol and Andrew Camilli for the CP-T1ts transducing phage. We also thank Ewen Cameron and John Mekalanos for the nonredundant Vibrio cholerae transposon library.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Adin, D. M., N. J. Phillips, B. W. Gibson, M. A. Apicella, E. G. Ruby, M. J. McFall-Ngai, D. B. Hall, and E. V. Stabb. 2008. Characterization of htrB and msbB mutants of the light organ symbiont Vibrio fischeri. Appl. Environ. Microbiol. 74:633-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cameron, D. E., J. M. Urbach, and J. J. Mekalanos. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 105:8736-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatterjee, S. N., and K. Chaudhuri. 2003. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim. Biophys. Acta 1639:65-79. [DOI] [PubMed] [Google Scholar]
- 4.Clements, A., D. Tull, A. W. Jenney, J. L. Farn, S. H. Kim, R. E. Bishop, J. B. McPhee, R. E. Hancock, E. L. Hartland, M. J. Pearse, O. L. Wijburg, D. C. Jackson, M. J. McConville, and R. A. Strugnell. 2007. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J. Biol. Chem. 282:15569-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darveau, R. P. 1998. Lipid A diversity and the innate host response to bacterial infection. Curr. Opin. Microbiol. 1:36-42. [DOI] [PubMed] [Google Scholar]
- 6.D'Hauteville, H., S. Khan, D. J. Maskell, A. Kussak, A. Weintraub, J. Mathison, R. J. Ulevitch, N. Wuscher, C. Parsot, and P. J. Sansonetti. 2002. Two msbB genes encoding maximal acylation of lipid A are required for invasive Shigella flexneri to mediate inflammatory rupture and destruction of the intestinal epithelium. J. Immunol. 168:5240-5251. [DOI] [PubMed] [Google Scholar]
- 7.Eisenhauer, P. B., S. S. Harwig, and R. I. Lehrer. 1992. Cryptdins: antimicrobial defensins of the murine small intestine. Infect. Immun. 60:3556-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feodorova, V. A., L. N. Pan'kina, E. P. Savostina, L. V. Sayapina, V. L. Motin, S. V. Dentovskaya, R. Z. Shaikhutdinova, S. A. Ivanov, B. Lindner, A. N. Kondakova, O. V. Bystrova, N. A. Kocharova, S. N. Senchenkova, O. Holst, G. B. Pier, Y. A. Knirel, and A. P. Anisimov. 2007. A Yersinia pestis lpxM-mutant live vaccine induces enhanced immunity against bubonic plague in mice and guinea pigs. Vaccine 25:7620-7628. [DOI] [PubMed] [Google Scholar]
- 9.Gmeiner, J., O. Luderitz, and O. Westphal. 1969. Biochemical studies on lipopolysaccharides of Salmonella R mutants. 6. Investigations on the structure of the lipid A component. Eur. J. Biochem. 7:370-379. [DOI] [PubMed] [Google Scholar]
- 10.Groisman, E. A., J. Kayser, and F. C. Soncini. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179:7040-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groisman, E. A., C. Parra-Lopez, M. Salcedo, C. J. Lipps, and F. Heffron. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11939-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunn, J. S. 2001. Bacterial modification of LPS and resistance to antimicrobial peptides. J. Endotoxin Res. 7:57-62. [PubMed] [Google Scholar]
- 13.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 14.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock, R. E., and G. Diamond. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8:402-410. [DOI] [PubMed] [Google Scholar]
- 17.Hankins, J. V., and M. S. Trent. 2009. Secondary acylation of Vibrio cholerae lipopolysaccharide requires phosphorylation of Kdo. J. Biol. Chem. 284:25804-25812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison, L. M., P. Rallabhandi, J. Michalski, X. Zhou, S. R. Steyert, S. N. Vogel, and J. B. Kaper. 2008. Vibrio cholerae flagellins induce Toll-like receptor 5-mediated interleukin-8 production through mitogen-activated protein kinase and NF-kappaB activation. Infect. Immun. 76:5524-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hava, D. L., and A. Camilli. 2001. Isolation and characterization of a temperature-sensitive generalized transducing bacteriophage for Vibrio cholerae. J. Microbiol. Methods 46:217-225. [DOI] [PubMed] [Google Scholar]
- 20.Heine, H., E. T. Rietschel, and A. J. Ulmer. 2001. The biology of endotoxin. Mol. Biotechnol. 19:279-296. [DOI] [PubMed] [Google Scholar]
- 21.Jin, T., M. Bokarewa, T. Foster, J. Mitchell, J. Higgins, and A. Tarkowski. 2004. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 172:1169-1176. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. H., K. S. Kim, S. R. Lee, E. Kim, M. S. Kim, E. Y. Lee, Y. S. Gho, J. W. Kim, R. E. Bishop, and K. T. Chang. 2009. Structural modifications of outer membrane vesicles to refine them as vaccine delivery vehicles. Biochim. Biophys. Acta 1788:2150-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, T., R. Konig, J. Sha, S. L. Agar, C. T. Tseng, G. R. Klimpel, and A. K. Chopra. 2008. Immunological responses against Salmonella enterica serovar Typhimurium Braun lipoprotein and lipid A mutant strains in Swiss-Webster mice: potential use as live-attenuated vaccines. Microb. Pathog. 44:224-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Low, K. B., M. Ittensohn, T. Le, J. Platt, S. Sodi, M. Amoss, O. Ash, E. Carmichael, A. Chakraborty, J. Fischer, S. L. Lin, X. Luo, S. I. Miller, L. Zheng, I. King, J. M. Pawelek, and D. Bermudes. 1999. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat. Biotechnol. 17:37-41. [DOI] [PubMed] [Google Scholar]
- 25.Luderitz, O., A. M. Staub, and O. Westphal. 1966. Immunochemistry of O and R antigens of Salmonella and related Enterobacteriaceae. Bacteriol. Rev. 30:192-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menard, S., V. Forster, M. Lotz, D. Gutle, C. U. Duerr, R. L. Gallo, B. Henriques-Normark, K. Putsep, M. Andersson, E. O. Glocker, and M. W. Hornef. 2008. Developmental switch of intestinal antimicrobial peptide expression. J. Exp. Med. 205:183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Netea, M. G., M. van Deuren, B. J. Kullberg, J. M. Cavaillon, and J. W. Van der Meer. 2002. Does the shape of lipid A determine the interaction of LPS with Toll-like receptors? Trends Immunol. 23:135-139. [DOI] [PubMed] [Google Scholar]
- 28.Pestonjamasp, V. K., K. H. Huttner, and R. L. Gallo. 2001. Processing site and gene structure for the murine antimicrobial peptide CRAMP. Peptides 22:1643-1650. [DOI] [PubMed] [Google Scholar]
- 29.Poyart, C., E. Pellegrini, M. Marceau, M. Baptista, F. Jaubert, M. C. Lamy, and P. Trieu-Cuot. 2003. Attenuated virulence of Streptococcus agalactiae deficient in d-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 49:1615-1625. [DOI] [PubMed] [Google Scholar]
- 30.Ranallo, R. T., R. W. Kaminski, T. George, A. A. Kordis, Q. Chen, K. Szabo, and M. M. Venkatesan. 2010. Virulence, inflammatory potential, and adaptive immunity induced by Shigella flexneri msbB mutants. Infect. Immun. 78:400-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robey, M., W. O'Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sack, D. A., R. B. Sack, G. B. Nair, and A. K. Siddique. 2004. Cholera. Lancet 363:223-233. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Senko, M. W., J. D. Canterbury, S. Guan, and A. G. Marshall. 1996. A high-performance modular data system for Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 10:1839-1844. [DOI] [PubMed] [Google Scholar]
- 36.Shafer, W. M., X. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shai, Y. 2002. Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236-248. [DOI] [PubMed] [Google Scholar]
- 38.Shi, Y., M. J. Cromie, F. F. Hsu, J. Turk, and E. A. Groisman. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53:229-241. [DOI] [PubMed] [Google Scholar]
- 39.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 40.Somerville, J. E., Jr., L. Cassiano, and R. P. Darveau. 1999. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67:6583-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stumpe, S., R. Schmid, D. L. Stephens, G. Georgiou, and E. P. Bakker. 1998. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J. Bacteriol. 180:4002-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tacket, C. O., G. Losonsky, J. P. Nataro, S. J. Cryz, R. Edelman, A. Fasano, J. Michalski, J. B. Kaper, and M. M. Levine. 1993. Safety and immunogenicity of live oral cholera vaccine candidate CVD 110, a delta ctxA delta zot delta ace derivative of El Tor Ogawa Vibrio cholerae. J. Infect. Dis. 168:1536-1540. [DOI] [PubMed] [Google Scholar]
- 43.Teghanemt, A., D. Zhang, E. N. Levis, J. P. Weiss, and T. L. Gioannini. 2005. Molecular basis of reduced potency of underacylated endotoxins. J. Immunol. 175:4669-4676. [DOI] [PubMed] [Google Scholar]
- 44.van der Ley, P., L. Steeghs, H. J. Hamstra, J. ten Hove, B. Zomer, and L. van Alphen. 2001. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect. Immun. 69:5981-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villeneuve, S., H. Souchon, M. M. Riottot, J. C. Mazie, P. Lei, C. P. Glaudemans, P. Kovac, J. M. Fournier, and P. M. Alzari. 2000. Crystal structure of an anti-carbohydrate antibody directed against Vibrio cholerae O1 in complex with antigen: molecular basis for serotype specificity. Proc. Natl. Acad. Sci. USA 97:8433-8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides, extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5:83-91. [Google Scholar]
- 47.Xicohtencatl-Cortes, J., S. Lyons, A. P. Chaparro, D. R. Hernandez, Z. Saldana, M. A. Ledesma, M. A. Rendon, A. T. Gewirtz, K. E. Klose, and J. A. Giron. 2006. Identification of proinflammatory flagellin proteins in supernatants of Vibrio cholerae O1 by proteomics analysis. Mol. Cell. Proteomics 5:2374-2383. [DOI] [PubMed] [Google Scholar]
- 48.Yeaman, M. R., and N. Y. Yount. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27-55. [DOI] [PubMed] [Google Scholar]
- 49.Yoon, S. S., and J. J. Mekalanos. 2006. 2,3-Butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect. Immun. 74:6547-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]