Abstract
Campylobacter jejuni is a highly prevalent human pathogen for which pathogenic and stress survival strategies remain relatively poorly understood. We previously found that a C. jejuni strain 81-176 mutant defective for key virulence and stress survival attributes was also hyper-biofilm and hyperreactive to the UV fluorescent dye calcofluor white (CFW). We hypothesized that screening for CFW hyperreactive mutants would identify additional genes required for C. jejuni pathogenesis properties. Surprisingly, two such mutants harbored lesions in lipooligosaccharide (LOS) genes (waaF and lgtF), indicating a complete loss of the LOS outer core region. We utilized this as an opportunity to explore the role of each LOS core-specific moiety in the pathogenesis and stress survival of this strain and thus also constructed ΔgalT and ΔcstII mutants with more minor LOS truncations. Interestingly, we found that mutants lacking the LOS outer core (ΔwaaF and ΔlgtF but not ΔgalT or ΔcstII mutants) exhibited enhanced biofilm formation. The presence of the complete outer core was also necessary for resistance to complement-mediated killing. In contrast, any LOS truncation, even that of the terminal sialic acid (ΔcstII), resulted in diminished resistance to polymyxin B. The cathelicidin LL-37 was found to be active against C. jejuni, with the LOS mutants exhibiting modest but tiled alterations in LL-37 sensitivity. The ΔwaaF mutant but not the other LOS mutant strains also exhibited a defect in intraepithelial cell survival, an aspect of C. jejuni pathogenesis that has only recently begun to be clarified. Finally, using a mouse competition model, we now provide the first direct evidence for the importance of the C. jejuni LOS in host colonization. Collectively, this study has uncovered novel roles for the C. jejuni LOS, highlights the dynamic nature of the C. jejuni cell envelope, and provides insight into the contribution of specific LOS core moieties to stress survival and pathogenesis.
The Gram-negative pathogen Campylobacter jejuni is the leading cause of bacterial food-borne diarrheal disease in the developed world, affecting up to (and sometimes more than) 1% of the population of North America, Europe, Australia, and New Zealand each year (4, 9, 79). Acute symptoms of C. jejuni infection include severe watery to bloody diarrhea, fever, nausea, and vomiting (12). Postinfectious sequelae can also occur, including the highly debilitating and sometimes fatal acute ascending bilateral paralysis Guillain-Barré syndrome (GBS), thought to occur in ∼1 in 1,000 individuals infected with C. jejuni (35). Despite causing severe human disease, C. jejuni is a commensal in most other animal species (36). Up to 90% of commercial poultry products harbor live C. jejuni, and most cases of sporadic C. jejuni infection occur via consumption of undercooked poultry or cross-contamination of other food with raw poultry juice (36). C. jejuni is microaerophilic and capnophilic and requires rich media for growth and survival in the laboratory (81). Despite these fastidious attributes, C. jejuni can persist in unfavorable environments in nature, including water and milk, both of which are common sources of C. jejuni outbreaks (32, 48).
C. jejuni is polysaccharide rich, harboring four well-defined carbohydrate biosynthetic loci encoding proteins responsible for genesis of the lipooligosaccharide (LOS), capsular polysaccharide (CPS), O-linked flagellar sugars, and N-linked protein glycans (pgl) (21, 24, 46, 54, 65). The pgl system is well conserved among C. jejuni strains. In contrast, the LOS, CPS, and O-linked flagellar glycoproteins exhibit a high degree of interstrain variability, as evidenced by the extensive use of LOS and CPS antigens as strain serotyping systems. Many genes in these hypervariable regions are also subject to phase variation, further confounding immune system responses to C. jejuni infection.
The C. jejuni LOS comprises two main components: the hydrophobic lipid A anchor and an oligosaccharide consisting of a conserved inner core and a variable outer core (22). C. jejuni LOS lacks the O-antigen characteristic of lipopolysaccharides (LPS) found in other bacterial species. LPS and LOS participate in the pathogenesis of numerous Gram-negative bacteria, acting as endotoxins, adherence factors, factors that maintain the stability of the outer membrane and protect cells from environmental stresses, and host defense factors (19, 73, 86). Currently, the best-characterized contribution of the C. jejuni LOS to human disease is its relationship to the debilitating neuropathy GBS, in which antibodies mounted against certain ganglioside-mimicking LOS structures (21, 60, 82) cross-react with and attack gangliosides on peripheral nerves (3, 6, 85). LOS structures for many C. jejuni strains have now been elucidated, as have genes required for LOS biosynthesis (21). The LOS structure of our laboratory strain, 81-176, is shown in Fig. 1A; enzymes involved in its biosynthesis will be described in more detail in Results. Depending on phase variation of the cgtA gene, the 81-176 LOS can mimic GM2, GM3, GD1b, and GD2 gangliosides (21). Various C. jejuni LOS mutants have also been found to exhibit enhanced sensitivities to certain antimicrobial substances (37, 43, 53, 56), and two severely truncated mutants (a strain 81-176 ΔwaaC mutant and a strain 11168 mutant with a large LOS gene cluster deletion) are defective for host cell invasion (43, 56). However, the latter two mutants also produce altered CPS and lipid A, respectively (42, 56), and an invasion defect previously ascribed to a ΔwaaF mutant (42) was subsequently shown to be due to sensitivity of that mutant to the detergent used for host cell lysis (43). Thus, although previous work has shown that the C. jejuni LOS is important as a pathogenesis factor, the relative contributions of specific oligosaccharide core structures to particular pathogenesis-related properties have not been described, nor has a direct role for the LOS in host colonization been demonstrated.
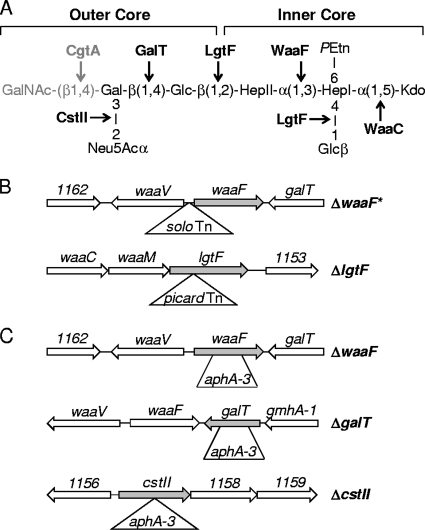
FIG. 1.
C. jejuni 81-176 lipooligosaccharide (LOS) structure and mutant strains used in this study. (A) Structure of the C. jejuni strain 81-176 LOS. Inner and outer core regions are delineated. Transferase genes responsible for the addition of each sugar residue are shown in bold. Abbreviations: GalNAc, N-acetylgalactosamine; NeuAc, N-acetylneuraminic acid (sialic acid); Gal, galactose; Glc, glucose; Hep, heptose; PEtn, phosphoethanolamine; Kdo, 2-keto-3-deoxymannooctulosonic acid. cgtA is phased off in our 81-176 strain; thus, CgtA and the terminal GalNAc are shown in gray. (B) LOS mutants isolated in the CFW screen. The ΔwaaF* mutant harbors the solo transposon, encoding kanamycin resistance, in the intergenic region between waaF and waaV. The ΔlgtF mutant harbors the picard transposon, encoding chloramphenicol resistance, upstream of the first glucosyltransferase domain, at the approximate site shown. Gene numbers are those annotated for strain 81-176 and are abbreviated versions of the “cjj81176_XXXX” nomenclature in the NCBI database. (C) Targeted LOS mutants. An aphA-3 cassette encoding kanamycin resistance but no downstream transcriptional terminator was used to disrupt waaF, galT, and cstII. Specifically, aphA-3 was used to remove 556 bp of waaF, including ∼75% of the single heptosyltransferase gene (ΔwaaF), and 446 bp of galT, including ∼75% of the single glycosyltransferase domain (ΔgalT). The ΔcstII mutant was constructed by inserting aphA-3 into cstII. Gene numbers are those annotated for strain 81-176 and are abbreviated versions of the “cjj81176_XXXX” nomenclature in the NCBI database.
Biofilms are surface-associated, dynamic consortia of microorganisms encased in a protective polymeric matrix (26). It is now thought that in nature, >99% of bacteria exist in biofilms rather than as free-swimming (planktonic) cells. Biofilm residents exhibit important survival differences from planktonic cells, including altered metabolism, physiology changes, and increased stress tolerance (67). The biofilm lifestyle is a key contributing factor to C. jejuni's prevalence and ability to withstand stressful environments in nature despite fastidious growth requirements (81). Our understanding of C. jejuni biofilms lags behind that of other bacteria; however, recent work has identified several genes involved in regulation and other aspects of biofilm dynamics (14, 18, 39, 40, 57, 75, 80). One of these genes controls a global stress response known as the stringent response (SR) (20): unexpectedly, a ΔspoT mutant incapable of mounting an SR also exhibited a dramatic hyper-biofilm phenotype (57). Biofilm upregulation in the ΔspoT mutant occurred concomitantly with increased reactivity to calcofluor white (CFW), a UV fluorescent dye that reacts with β-1,3- and β-1,4-linked polysaccharides on cell surfaces, suggesting that such a polysaccharide may be a component of the biofilm matrix.
Despite its prevalence, much less is understood about the pathogenesis of C. jejuni than that of prototypical enteric bacteria such as Escherichia coli and Salmonella spp. This is in part due to the lack of a tractable small animal model of disease (55). As such, in vitro assessments such as invasion and intracellular survival have become key virulence markers for strains and genetic mutants, as has determination of stress survival attributes contributing to overall fitness. C. jejuni is also more difficult to manipulate genetically than E. coli and Salmonella spp. Furthermore, although several well-annotated C. jejuni genome sequences have been published, in silico efforts to identify pathogenesis determinants have been frustrating, with none of the sequenced C. jejuni genomes revealing obvious pathogenicity islands, type III “injection-like” secretion systems, or other hallmark virulence determinants (31, 72). As such, screens and selections are critical for identifying C. jejuni genes important for modulating pathogenesis-associated properties.
As noted, we previously found that the ΔspoT mutant exhibited a dramatic hyper-biofilm phenotype (57). Although the precise nature of the concomitantly upregulated CFW-reactive polymer remains unknown, initial studies suggested that it was not composed of previously described polysaccharides (57). We also previously demonstrated that the SR is important for specific C. jejuni virulence- and stress-related attributes (20). Collectively, this led us to rationalize that screening a random transposon (Tn) library for C. jejuni mutants exhibiting a CFW-hyperreactive phenotype would uncover genes important for biofilm dynamics and other key aspects of C. jejuni pathogenesis and survival. Somewhat unexpectedly, two such mutants harbored insertions suggesting complete loss of the LOS outer core. We took advantage of this finding to generate mutants defective for each of the other LOS core-specific enzymes in 81-176, with the goal of assigning specific roles in pathogenesis properties to distinct moieties of the LOS. Consequently, this study has identified novel roles for LOS sugars in modulating biofilm dynamics, complement sensitivity, resistance to two classes of antimicrobial peptides, and intracellular survival. We also present the first direct evidence of a role for the C. jejuni LOS in host colonization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni strain 81-176, originally isolated from an outbreak of campylobacteriosis after consumption of raw milk (48), was the wild-type strain for these studies. All C. jejuni strains were grown at 38°C on Mueller-Hinton (MH) agar or broth (Oxoid) supplemented with vancomycin (10 μg/ml) and trimethoprim (5 μg/liter) (MH-TV) under microaerobic and capnophilic conditions (6% O2 and 12% CO2). E. coli DH5α was used for plasmid construction and was grown on Luria-Bertani broth (LB; Sigma) at 37°C. When necessary, media were supplemented with kanamycin (50 μg/ml), chloramphenicol (30 μg/ml), or ampicillin (100 μg/ml). C. jejuni mutant strains used in this study are listed in Table 1.
TABLE 1.
C. jejuni strains used in this study
| Strain | Descriptiona | Reference |
|---|---|---|
| 81-176 | Wild type | 48 |
| ΔwaaF* mutant | 81-176 waaV-waaF::Tn solo | This study |
| ΔwaaF mutant | 81-176 waaF::aphA-3 | This study |
| ΔlgtF mutant | 81-176 lgtF::Tn picard | This study |
| ΔgalT mutant | 81-176 galT::aphA-3 | This study |
| ΔcstII mutant | 81-176 cstII::aphA-3 | This study |
| ΔwaaF-c mutant | 81-176 waaF::aphA-3 rrn-cat::waaF | This study |
| ΔlgtF-c mutant | 81-176 lgtF::aphA-3 rrn-aphA-3::lgtF | This study |
| ΔgalT-c mutant | 81-176 galT::aphA-3 rrn-cat::galT | This study |
Tn solo, aphA-3, and rrn-aphA-3 confer kanamycin resistance. Tn picard and rrn-cat confer chloramphenicol resistance.
Random in vitro Tn mutagenesis of C. jejuni using the mariner transposon. (i) Purification of MBP-Himar1.
The MBP-Himar1 transposase was purified according to a modified protocol from Akerley and Lampe (1) and instructions from New England Biolabs (NEB) (63a).
An overnight culture of E. coli TB1 containing the plasmid pMALC9 (1) grown in LB and ampicillin (100 μg/ml) at 37°C was subcultured 1/50 into 100 ml fresh LB containing ampicillin and 0.2% (wt/vol) glucose, and growth at 37°C was continued. At an approximate optical density at 600 nm (OD600) of 0.5, protein expression was induced with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubation was continued for 2 h. Bacterial cells were harvested by centrifugation and frozen. The cell pellet was resuspended in column buffer (CB; 20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA) and sonicated for 2 min (10-s pulse on/10-s pulse off). Cellular debris was removed by centrifugation for 10 min at 13,000 rpm, and protease inhibitors (Roche complete, mini EDTA-free protease inhibitor cocktail tablets) were added to the supernatant.
The amylose resin (NEB) was prepared by washing with transposase wash buffer (TWB; 20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol [DTT], 10% glycerol), as described by the manufacturer. The lysate was diluted to a final volume of 5 ml using CB. The amylose resin was added to the lysate and incubated with shaking overnight at 4°C. The resin and lysate were then added to an empty 5-ml column (Qiagen), and the flowthrough was collected by gravity flow. The column was washed four times with 2 ml TWB. A total of 0.4 ml of transposase elution buffer (TEB; 20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 2 mM DTT, 10% glycerol, 10 mM maltose) was added to the column and incubated for 5 min, and the elution fraction containing the purified transposase was collected. The transposase was aliquoted in 10-μl volumes and frozen at −80°C. Protein concentration was determined by the Bradford assay (Bio-Rad) (approximately 0.16 mg/ml), and purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
(ii) In vitro transposon mutagenesis.
In vitro transposition reactions were performed as described by Hendrixson et al. (28) and Akerley and Lampe (1), except that DNA was purified using Qiagen DNeasy columns following transposition and prior to ligation. The DNA from the ligation reaction was dialyzed on a 0.025-μm hydrophobic filter floating on distilled water (dH2O) for 20 min and then transformed by natural transformation into C. jejuni 81-176. Kanamycin-resistant (solo Tn from pFalcon) and chloramphenicol-resistant (picard Tn from pEnterprise) clones were selected on MH plates supplemented with the appropriate antibiotics. Approximately 3,000 to 4,000 single colonies from each round of mutagenesis were directly harvested from the plates. A total of 4 rounds of mutagenesis were carried out to construct the library. To confirm random Tn insertion, 10 of the colonies from each of the solo and picard Tn libraries were screened by Southern analyses (data not shown).
CFW screening.
Wild-type C. jejuni 81-176, 81-176::solo Tn library, and 81-176::picard Tn library strains were taken directly from the freezer and grown for 2 days on MH-TV plates. Strains were diluted to an OD600 of 0.3. One hundred microliters of 10−5 dilutions were plated on brain heart infusion (BHI; BD) agar plates supplemented with antibiotics and 0.002% CFW (fluorescent brightener 28; Sigma) and incubated for 2 days in the dark. Plates were inspected under long-wave UV light, and hyper-fluorescent colonies were patched onto MH plates and grown for 1 day. Each clone was then again patched onto BHI-CFW plates and incubated for 2 days in the dark, and hyper-fluorescence was confirmed under long-wave UV light. To confirm linkage of the CFW hyperreactive phenotype with the transposon insertion, genomic DNA was prepared from the mutant strains using the Wizard genomic DNA purification kit (Promega) and reintroduced into wild-type 81-176 by natural transformation and selection on the appropriate antibiotic. The retransformed strains were grown in shaking MH broth culture to early log phase and diluted to an OD600 of 0.2, and 10 μl was spotted onto BHI-CFW plates. To avoid any impact of the screening process on phenotypic analyses, the ΔlgtF retransformed strain generated during linkage analysis was used for all subsequent assays and complementation. The spotting technique described above was also used to generate the CFW plate data described in Results.
Transposon mapping via random PCR.
Using the genomic DNA described above, the region flanking the transposon insertion sites were amplified using the CEKG technique discussed by Salama et al. (76). First, the primers CEKG2A, CEKG2B, and CEKG2C were used with the mariner-2 or mariner-3 primer to amplify transposon-flanking sites (see Table S1 for all primer sequences except the CEKG primers, which are found in reference 76). A second PCR was performed on the product of the first PCR using the primers CEKG4 and mariner-IR-1. The amplicons were purified and sequenced using the primer MarOut3. Fine mapping to determine the precise location of the transposon for the ΔwaaF* and ΔlgtF mutants was done by sequencing the insert region amplified using the primers brt1-Tn-F and brt1-Tn-R for the ΔwaaF* mutant and brt28-Tn-F and brt28-Tn-R for the ΔlgtF mutant.
Construction of ΔwaaF, ΔgalT, and ΔcstII mutants and ΔwaaF-c, ΔlgtF-c, and ΔgalT-c complemented strains.
The ΔwaaF mutant was constructed by PCR amplification of waaF from 81-176 genomic DNA using the primers waaF-F and waaF-R and by cloning the PCR product into the commercial vector pGEM-T (Promega). Inverse PCR was performed on the resulting vector using the primers pGEM-waaF-F and pGEM-waaF-R. The resulting amplicon and the plasmid pUC18K-2 (58), carrying a nonpolar kanamycin resistance (aphA-3) cassette, were each digested with XbaI and KpnI enzymes and ligated to form the plasmid pGEM-waaF-K. This plasmid was delivered into 81-176 via natural transformation, and kanamycin-resistant colonies were isolated. ΔwaaF mutants were confirmed via PCR analysis and sequencing.
The ΔgalT mutant was constructed in the same manner as the ΔwaaF mutant except that the initial primers were galT-F and galT-R, and inverse PCR was performed using the primers pGEM-galT-F and pGEM-galT-R.
The ΔcstII mutant was constructed as a control for a previous study. Briefly, the cstII gene from C. jejuni strain ATCC 43446 was amplified in two stages using the primers CJ-131, CJ-269, CJ-132, and CJ-268. CJ-131 and -269 were the primers used to amplify the 5′ region of cstII, and CJ-132 and -268 were the primers used to amplify the 3′ region of cstII. This was done to insert KpnI and SacI sites in the middle of the gene. A final PCR using CJ-131 and CJ-132 was performed to amplify the full-length cstII gene, containing two restriction sites in the middle, an NdeI site at the 5′ end, and a SalI site at the 3′ end. The amplicon was then inserted into the plasmid pCWori+(-lacZ), giving the plasmid pCST-60. The aphA-3 gene, encoding a kanamycin resistance cassette, was introduced into pCST-60 using the KpnI and SacI sites. The resulting plasmid, designated pCST-72, was electroporated into C. jejuni strain 81-176, and ΔcstII colonies were isolated. Positive clones were verified by PCR and sequencing.
For complemented strains, the waaF gene was PCR amplified from 81-176 genomic DNA using the primers pR-waaF-F and pR-waaF-R. The waaF amplicon was digested with MfeI and XbaI and ligated into pRRC (45), which had been digested with the same enzymes, to produce pRRC-waaF. The galT gene was PCR amplified from 81-176 genomic DNA using the primers pR-galT-F and pR-galT-R, and the lgtF gene was amplified using the primers pR-lgtF-F and pR-lgtF-R. The resulting galT and lgtF amplicons were digested with XbaI and ligated into XbaI-digested pRRC and pRRK, respectively, to produce the pRRC-galT and pRRK-lgtF plasmids. The correct orientation of the genes in the plasmids was confirmed, after which the plasmids were naturally transformed into the respective mutants, the ΔwaaF, ΔgalT, and ΔlgtF mutants. Recombination was confirmed via PCR. The pRRC and pRRK plasmids were kindly provided by Brendan Wren (45) and Julian Ketley.
C. jejuni LOS analysis by SDS-PAGE.
LOS samples were prepared from whole-cell lysates using a modified method described by Hitchcock and Brown (29). Briefly, cells, standardized to an OD600 of 5.0, were resuspended in lysis buffer (2% SDS, 4% β-mercaptoethanol, 10% glycerol, 1.0 M Tris [pH 6.8], bromophenol blue) and heated for 5 min at 95°C. Samples were then treated with proteinase K and incubated overnight at 55°C. For silver stain analyses, LOS preparations were heated for 5 min at 95°C and 10 μl of each sample was separated via 15% SDS-PAGE. The resulting gels were stained with silver as described previously (83) and developed with Bio-Rad silver stain developer (Bio-Rad).
Isolation of LOS and MS analysis.
A half plate of confluent bacteria was resuspended in 0.3 ml of phosphate-buffered saline (PBS). One milliliter of 100% ethanol was then added, mixed, and allowed to stand for 1 h at room temperature. Cells were harvested, and the pellet was washed twice in 100% ethanol, twice in 100% acetone, and allowed to air dry. The intact LOS of the C. jejuni strains were prepared and analyzed by electrophoresis-assisted open-tubular liquid chromatography-mass spectrometry (EA-OTLC-MS) as described previously (17).
Biofilm formation assay and biofilm quantification.
The ability of C. jejuni strains to form biofilms was assayed using a modified version of a previously described method (57, 68). Briefly, 100-μl samples of mid-log-phase overnight cultures were diluted to an OD600 of 0.002, inoculated into 96-well microtiter polypropylene plates, and incubated for 24, 48, or 72 h at 38°C under standard C. jejuni growth conditions. At the specified time points, 25 μl of a 1% crystal violet (CV) solution in 100% ethanol was added to the wells, and the solution was incubated at room temperature for 15 min. The wells were then rinsed thoroughly with distilled water five times. Biofilms were quantified by dissolving the remaining CV with a solution composed of 30% methanol and 10% acetic acid. Absorbance was measured at 550 nm using a spectrophotometer (Thermo Electron Co.).
Serum sensitivity assay.
Sensitivity to complement-mediated killing was assayed by a slightly modified method of Guerry et al. (23). Mid-log-phase overnight bacterial cultures were diluted in PBS to a concentration of 106 CFU/ml and incubated in pooled human serum (10% serum as the final concentration) for 40 or 80 min at 38°C under standard C. jejuni growth conditions. At the specified time points, bacterial survival was assessed via CFU enumeration on MH plates. Strains were also incubated with pooled human serum that was heat inactivated at 60°C for 1 h as a control.
Sensitivity to antimicrobial peptides, SDS, and EDTA.
The MICs of LL-37, polymyxin B (Sigma), SDS (Fisher Scientific), and EDTA (Sigma) for the strains were determined using a microtiter broth dilution method (52) in MH broth and an initial inoculum of 106 cells/ml (diluted from overnight mid-log-phase cultures). Polypropylene microtiter plates containing bacterial strains with the various substances were incubated for 48 h at 38°C under standard C. jejuni growth conditions, and dilutions were spotted on MH plates for survivability. LL-37 was kindly provided by R. E. W. Hancock. Shown are the results of a representative experiment of at least three independent repeats on different days.
Adherence, invasion, and intracellular survival assay in Caco-2 cells.
Bacterial infections in Caco-2 intestinal epithelial cells were performed as previously described (20), except that shaking MH broth mid-log-phase bacterial cultures were used instead of biphasic cultures.
Mouse colonization.
BALB/cByJ mice from Jackson Laboratories (Bar Harbor, ME) were maintained in ABSL-2 housing in the Division of Lab Animal Services at the Medical College of Georgia, with five mice per experimental group. Each mouse was infected with a mixture of 5 × 109 CFU of the wild type and either the C. jejuni ΔlgtF mutant or ΔwaaF mutant via oral gavage as previously described (69). C. jejuni organisms shed in fecal pellets from each mouse at 7, 14, and 21 days postinfection were homogenized and enumerated on MH agar containing 5% (vol/vol) sheep's blood and 20 μg/ml cefoperazone, 10 μg/ml vancomycin, and 2 μg/ml amphotericin B, plus chloramphenicol or kanamycin at 15 μg/ml or 30 μg/ml, respectively, as warranted. The level of detection was 1 × 102 CFU/g fecal pellet. All animal treatments were carried out in accordance with NIH guidelines for the care and use of laboratory animals, using procedures approved by the Medical College of Georgia Institutional Care and Use Committee (protocol AUP 07-03-923, approved 12 April 2007).
Statistical analysis.
Results obtained were assessed for statistical significance using a two-tailed unpaired Student t test. P values of less than 0.05 were considered statistically significant (see the figure legends for specific values).
RESULTS
Isolation and construction of LOS mutants of Campylobacter jejuni strain 81-176.
To identify C. jejuni genes involved in the planktonic-biofilm switch and potentially other pathogenesis attributes, transposon libraries were constructed using previously described solo and picard Mariner transposons (28, 50) and screened for mutants exhibiting CFW hyperreactivity. Somewhat surprisingly, two mutants mapped to loci predicted to be involved in LOS biosynthesis. The structure of the LOS for strain 81-176 and transferases responsible for LOS biosynthesis are shown in Fig. 1A (21, 23, 25, 41-43, 47). The sites of transposon (Tn) insertions are shown in Fig. 1B. One mutant mapped to an intergenic site approximately equidistant from the start codons of waaF and a gene annotated as waaV. WaaF is a heptosyltransferase responsible for the addition of HepII onto HepI (Fig. 1A) (41, 43, 66). waaV is uncharacterized in C. jejuni but exhibits homology to various putative bacterial glycosyltransferases. A targeted C. jejuni ΔwaaV deletion strain was unaffected for CFW reactivity, LOS structure (by silver stain and mass spectrometry analyses [see Table S2 in the supplemental material]), and other tested phenotypes (data not shown); because of this, and based on data described below, we have designated the Tn mutant the ΔwaaF* strain. Nonetheless, because of the intergenic nature of the ΔwaaF* strain, a targeted ΔwaaF strain was constructed and used for subsequent analyses (Fig. 1C). The second mutant harbored an insertion in lgtF, encoding a two-domain glucosyltransferase responsible for the addition of β-1,4 glucose to HepI and β-1,2 glucose to HepII (Fig. 1A and B) (43).
Many studies of the role of C. jejuni LOS on pathogenic properties have been conducted using a single glycosyltransferase mutant. While such studies have been useful for defining general functions of the LOS, we decided to take advantage of the mutants in hand to embark on a comparative analysis of sequentially truncated LOS strains on specific aspects of C. jejuni pathogenesis. Previous work demonstrated that the ΔwaaC mutant is pleiotropic and also affects CPS production (42). Mass spectrometry also verified that cgtA is phased off in our 81-176 background, a phenomenon not uncommon in this strain of C. jejuni (25). Thus, to complete our repertoire of LOS-specific mutants, we generated targeted deletions in galT and cstII, in addition to the aforementioned ΔwaaF strain, for further analyses (Fig. 1C). GalT transfers the β-1,4 galactose to the β-1,2 glucose added by LgtF (43), while CstII adds the sialic acid (Neu5Acα) to the β-1,4 galactose (Fig. 1A) (21, 23).
Confirmation of sequential LOS truncations.
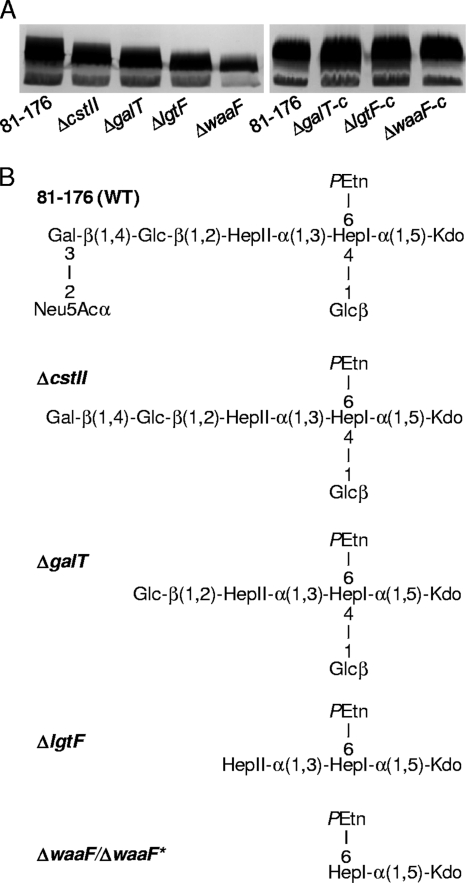
To verify the specific LOS disruptions, LOS profiles of each mutant were analyzed by SDS-PAGE/silver staining and mass spectrometry. Each mutant displayed faster-migrating LOS species than the 81-176 wild type by SDS-PAGE and silver stain analyses (Fig. 2A), with the ΔcstII, ΔgalT, ΔlgtF, and ΔwaaF mutants exhibiting decreasing sizes of their LOSs in a stepwise manner, as predicted. Complemented strains were generated for three of the four mutants (ΔgalT-c, ΔlgtF-c, and ΔwaaF-c strains), each of which restored normal LOS migration (Fig. 2A). Mass spectrometry likewise confirmed predicted LOS structures for the ΔcstII, ΔgalT, and ΔlgtF mutants, with complementation restoring wild-type profiles (Fig. 2B, Tables S2 and S3 in the supplemental material, and data not shown). Interestingly, neither the ΔwaaF nor ΔwaaF* mutant expressed any sugar residues beyond HepI, including the β-1,4 glucose previously shown to be linked to HepI in a ΔwaaF mutant of 81-176 (41). This suggests either that the addition of HepII is required for the addition of both Glc residues to the inner core or that the β-1,4-linked glucose species is a minor component of the LOS in the absence of WaaF function.
FIG. 2.
LOS profile and structures of the 81-176 mutant and complemented strains. (A) The LOS of C. jejuni strains was resolved by SDS-PAGE and visualized by silver staining. Complemented strains are denoted by “-c”. (B) Using mass spectrometry analyses, the core LOS structure of each mutant strain was deduced by comparing the observed mass species of the mutant strains to those of the wild type, each other, and the wild-type 81-176 LOS core structure reported by Guerry et al. and Kanipes et al. (25, 43). Abbreviations: GalNAc, N-acetylgalactosamine; NeuAc, N-acetylneuraminic acid (sialic acid); Gal, galactose; Glc, glucose; Hep, heptose; PEtn, phosphoethanolamine; Kdo, 2-keto-3-deoxymannooctulosonic acid.
Biofilm formation is enhanced in the absence of the LOS outer core.
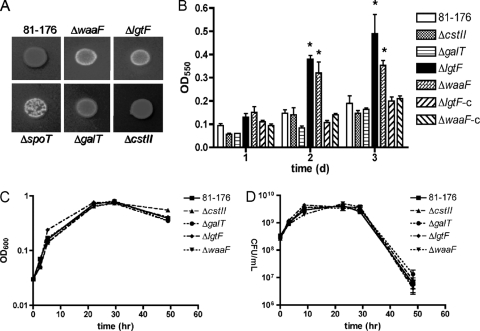
As noted, we were initially interested in identifying CFW-hyperreactive mutants because of their potential link with C. jejuni biofilm formation. For reference, the CFW profile of each mutant is shown in Fig. 3A, with the 81-176 wild type and the ΔspoT CFW hyperreactive strains shown as controls. Although the ΔgalT mutant was very modestly CFW hyperreactive and the ΔcstII mutant was modestly hyporeactive, only the ΔwaaF and ΔlgtF mutants consistently exhibited significant differences from the wild type, with each mutant displaying clear CFW hyperreactivity (Fig. 3A).
FIG. 3.
CFW reactivity, biofilm formation, and broth growth properties of LOS mutant strains. (A) CFW reactivity after 48 h of growth on BHI plates containing 0.002% CFW was visualized under long-wave UV light. All strains were assessed on the same plate, with spot rearrangement necessary for presentation purposes. (B) Biofilm formation was assessed after 1, 2, and 3 days of incubation in MH broth in polypropylene plates. Biofilms were stained with crystal violet, dissolved, and quantified by measuring the absorbance at 550 nm. Complemented strains are denoted by “-c”. Error bars were calculated from triplicate readings and are representative of three independent assays. The asterisk (*) indicates statistically significant differences from wild-type 81-176 (P < 0.01). (C, D) Growth in shaking MH broth cultures was assessed by absorbance at 600 nm (C) and plating of serial dilutions to determine CFU/ml of culture (D). (D) Points represent means derived from triplicate readings and are representative of three independent assays.
Biofilm formation was assessed using a previously established standing culture assay followed by crystal violet staining and spectrophotometric quantification of triplicate biofilms (54, 68). Shown are results from a representative experiment from multiple experimental trials (Fig. 3B). Consistent with CFW reactivity profiles, the ΔlgtF and ΔwaaF mutants also exhibited a statistically significant increase in biofilm formation compared to that of the wild type at 2 and 3 days postinoculation. In contrast, the ΔgalT and ΔcstII mutants did not exhibit a difference in biofilm formation from the wild type, nor did the ΔlgtF-c and ΔwaaF-c complemented strains. To investigate whether growth differences might account for the biofilm observations and other attributes described below, we also assessed the growth of each mutant strain in standard shaking broth cultures. None of the mutants exhibited growth or survival defects compared to the wild type by either OD600 or CFU/ml analyses during normal growth curves in vitro (Fig. 3C and D).
The LOS outer core is important for protecting C. jejuni from complement-mediated killing.
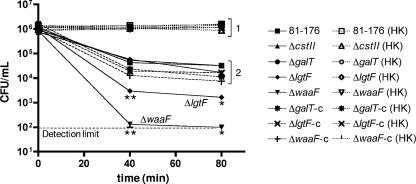
To begin to assess the role of specific LOS moieties on host-related properties, we investigated the ability of our mutant and complemented strains to survive 40- and 80-min exposures to 10% pooled human serum. Only the ΔlgtF and ΔwaaF mutants exhibited statistically significant defects in serum resistance compared to the wild type (Fig. 4), with ΔwaaF mutant recovery near the detection limit after 40 min of incubation. Serum preincubated at 60°C (heat killed) abrogated all killing (Fig. 4, bracketed group 1). The ΔgalT and ΔcstII mutants and all complemented strains exhibited sensitivity levels similar to that of the wild type (Fig. 4, bracketed group 2).
FIG. 4.
Complement-mediated killing of LOS mutant strains. Strains were incubated in PBS containing 10% pooled human serum or 10% heat-killed human serum (HK) for 0, 40, and 80 min, and survival was assessed by plating serial dilutions to determine numbers of CFU/ml. Complemented strains are denoted by the “-c”. Bracketed group “1” contains data points for all strains incubated with heat-killed serum. Bracketed group “2” contains data points for, from the top down, wild-type 81-176 and the ΔcstII, ΔgalT, ΔlgtF-c, ΔgalT-c, and ΔwaaF-c strains. The ΔlgtF and ΔwaaF mutants in 10% normal pooled human serum are noted on the graph as well as in the symbol key. Error bars are present but in most cases are too small to see. The asterisks represent statistically significant differences from wild-type 81-176, with the double asterisk (**) representing a P of <0.005 and the single asterisk (*) representing a P of <0.02.
LOS truncations result in modest, tiled sensitivities to LL-37 and hypersensitivity to polymyxin B.
Because the LOS is a major component of the C. jejuni cell envelope and a target of antimicrobial peptides (AMPs), we next explored the sensitivity of the LOS mutants to AMPs. Determination of the MICs of LL-37 and polymyxin B (PxB) for wild-type C. jejuni 81-176 and the LOS mutants revealed several unexpected findings. First, while the MIC of LL-37 for our wild-type strain was 5.68 μg/ml, the serially truncated LOS mutants yielded tiled LL-37 MICs, with the ΔwaaF (1.42 μg/ml) and ΔlgtF (2.40 μg/ml) mutants exhibiting modestly lower MICs than the wild type and the ΔgalT (8.08 μg/ml) and ΔcstII (12.13 μg/ml) mutants unexpectedly exhibiting modestly higher MICs than the wild type (Table 2). In contrast, all of the mutants displayed a significant (>15-fold) decrease in MIC for PxB compared to that of the wild type (3.13 μg/ml), with the various LOS mutants displaying modest differences from each other in a trend opposite to that observed for LL-37 (ranging from 0.21 μg/ml for the ΔwaaF mutant to 0.06 μg/ml for the ΔcstII mutant) (Table 2).
TABLE 2.
MICs of LL-37, polymyxin B, SDS, and EDTA
| Strain | MIC of: |
|||
|---|---|---|---|---|
| LL-37 (μg/ml) | Polymyxin B (μg/ml) | SDS (μg/ml) | EDTA (mM) | |
| 81-176a | 5.68 | 3.13 | 248 | 0.39 |
| ΔcstII mutant | 12.13 | 0.06 | 157 | 0.39 |
| ΔgalT mutant | 8.08 | 0.09 | 157 | 0.39 |
| ΔlgtF mutant | 2.40 | 0.16 | 157 | 0.39 |
| ΔwaaF mutant | 1.42 | 0.21 | 157 | 0.39 |
Complemented strains had MICs equivalent to that of wild-type 81-176.
As the AMP data suggested general alterations in the cell wall, we also explored the sensitivity of the mutants to detergents, salts, and chelators and investigated cell surface hydrophobicity and general profiles of outer membrane proteins. No differences in sensitivity to Tween 20, deoxycholate, or sodium chloride were observed (data not shown); however, each LOS mutant exhibited an ∼2-fold decrease in its MIC of SDS (Table 2). No overt differences were observed for EDTA sensitivity (Table 2), outer membrane protein profiles (data not shown), or surface hydrophobicity utilizing hexadecane- and ammonium sulfate-based assays (11, 37; data not shown).
The ΔwaaF mutant exhibits a defect in intracellular survival.
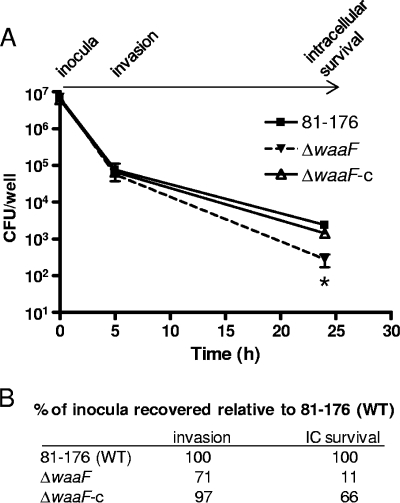
Previous work indicated that mutants defective specifically for the LOS (i.e., the ΔwaaF, ΔlgtF, ΔgalT, and ΔcstII mutants) were not defective for invasion of host cells in vitro (25, 43). As intracellular survival provides another recently recognized measure of C. jejuni pathogenic potential, we also assessed the ability of each of our mutant strains to survive inside Caco-2 intestinal epithelial cells for 24 h postinoculation. Consistent with previous observations, none of our LOS mutants exhibited an invasion defect (Fig. 5 and data not shown). Furthermore, the ΔcstII, ΔgalT, and ΔlgtF mutants did not display differences from the wild type in intracellular survival (data not shown). However, the ΔwaaF mutant exhibited a reproducible and statistically significant defect (P < 0.05) for intracellular survival (Fig. 5A), with only 11% of the bacteria recovered relative to wild-type levels 24 h after inoculation (Fig. 5B). Complementation restored the intracellular survival defect to near-wild-type levels (Fig. 5A and B).
FIG. 5.
Invasion and intracellular survival. A gentamicin protection assay was used to assess invasion and intracellular (IC) survival in Caco-2 (intestinal epithelial) cells for all LOS mutant strains constructed. Only the ΔwaaF mutant differed from wild-type 81-176 and is the only strain shown. The “invasion” time point represents 3 h of infection followed by 2 h of gentamicin treatment to kill extracellular bacteria. For “intracellular survival,” the cells were incubated in fresh medium for an additional 19 h following the gentamicin treatment prior to bacterial enumeration. (A) Numbers of CFU/well recovered at each time point. The asterisk (*) denotes a statistically significant difference for the ΔwaaF mutant compared to both wild-type 81-176 and the ΔwaaF-c strain at the “intracellular survival” time point (P < 0.05), as assessed by both total numbers of bacteria recovered and the amounts of inocula recovered as percentages of the wild-type inoculum recovered. (B) The percentage of the inoculum recovered was calculated for each strain at each time point. Numbers shown represent the percent recovered relative to the wild-type (WT).
A mouse competition model identifies a role for the C. jejuni LOS in colonization in vivo.
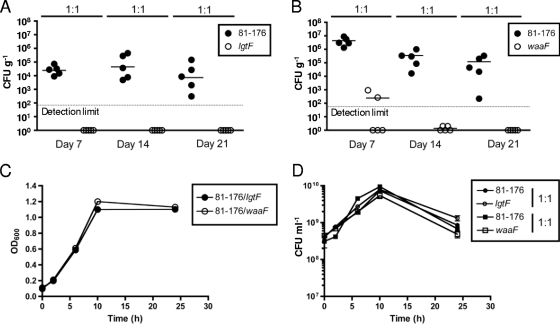
To date, little is known regarding participation of the C. jejuni LOS in host colonization. To explore this, we tested our two most truncated mutants, the ΔlgtF and ΔwaaF mutants, in a recently described mouse competition model for C. jejuni colonization (69). BALB/c ByJ mice were infected orogastrically with a mixture of bacteria containing equal doses of the wild-type and mutant C. jejuni strains. Fecal pellets were harvested at 7, 14, and 21 days postinfection and plated for bacterial counts on selective and nonselective plates. Both the ΔlgtF mutant (Fig. 6A) and the ΔwaaF mutant (Fig. 6B) exhibited a striking and statistically significant colonization defect evident from 7 days postinfection. Both the ΔwaaF and ΔlgtF mutants grew comparably on selective and nonselective plates used to assess levels of mutant versus that of the wild type following colonization, and each mutant exhibited wild-type motility (data not shown). Furthermore, when mutant and wild-type strains were grown together in shaking broth culture, neither the ΔlgtF nor the ΔwaaF mutant exhibited a growth defect in competition with the wild type (Fig. 6C and D), indicating that the colonization defect likely reflects in vivo-specific phenomena.
FIG. 6.
In vivo colonization of the ΔlgtF and ΔwaaF mutants. (A, B) BALB/c ByJ mice were fed a 1:1 mixture of wild-type 81-176 and either the ΔlgtF (A) or ΔwaaF (B) mutant, and colonization levels were determined at 7, 14, and 21 days postinfection. Mean colonization levels are denoted by horizontal bars. Both the ΔwaaF and ΔlgtF mutants colonized at levels that were statistically significantly different from the wild-type 81-176 level (P < 0.01 for both strains at all time points). Symbols located on the x axis represent colonization levels below the limit of detection. (C, D) Shaking MH broth cultures were inoculated with a 1:1 mixture of wild-type 81-176 and either the ΔlgtF or ΔwaaF mutant. Growth and recovery were assessed by measuring the absorbance at 600 nm (C) and plating for CFU/ml (D) on plates identical to those used to assess colonization.
DISCUSSION
This study, initiated following an unbiased screen to identify C. jejuni genes likely to be important for biofilm formation, stress survival, and virulence-associated attributes, has resulted in the delineation of novel roles for the C. jejuni LOS in a number of key pathogenesis properties. Our in-depth comparison of serially truncated LOS core mutations in a single highly invasive and virulent strain, 81-176, has also established for the first time clear cutoff points for the LOS in modulating certain pathogenic traits (i.e., biofilms, complement resistance, and intracellular survival) and both all-or-nothing and graded effects of specific LOS moieties on other attributes (i.e., resistance to different AMPs). We also present the first direct evidence of a role for the C. jejuni LOS in colonization.
As noted, bacterial biofilms play significant roles in infectious disease and confer advantages over bacteria in the planktonic state, including enhanced stress survival and antibiotic resistance. For a fastidious yet prevalent organism like C. jejuni, a biofilm lifestyle is especially important for surviving unfavorable conditions (81). Among our LOS mutants, only the ΔlgtF and ΔwaaF mutants exhibited a hyper-biofilm phenotype, while the ΔgalT and ΔcstII mutants were similar to the wild type, suggesting a role for the LOS outer core in maintenance of planktonic growth and/or biofilm dispersal. Although this is a new finding for C. jejuni, the LPSs and LOSs of other bacteria have also been shown to interface with biofilm production. Some groups reported findings similar to ours, whereby LPS/LOS mutations result in enhanced biofilms and/or exopolysaccharides involved in biofilm formation (59, 71, 78), while others reported that LPS defects result in diminished biofilms (10, 63). Still others observed mixed biofilm results for LPS mutants depending on the incubation conditions (7). Together with our prior observation that a C. jejuni ΔkpsM (CPS export) mutant is also CFW hyperreactive and hyper-biofilm forming (39, 57), this supports a model in which the cell envelope is highly dynamic, with compensatory changes such as upregulation or increased presentation of different surface polysaccharides in response to truncation or elimination of others likely occurring under certain conditions and throughout the bacterial life cycle (57). Although our understanding of C. jejuni biofilm dynamics still lags behind that of other bacteria, our findings also clearly place the LOS outer core in the growing list of factors influencing the planktonic-biofilm switch in C. jejuni (14, 18, 20, 39, 40, 75, 80) and suggest that future studies of other genes arising from our screen are likely to contribute even more to this relatively nascent area of C. jejuni research.
The LOS outer core was also found to confer to C. jejuni protection against complement-mediated killing. Although our ΔcstII sialyltransferase mutant was not defective for serum resistance, deletion of neuC1, involved in sialic acid biosynthesis, from a noninvasive C. jejuni strain was previously shown to cause increased serum sensitivity (23). However, non-LOS sialylated structures such as flagellin also occur in that strain (23), suggesting pleiotropic effects of the neuC1 mutation. The C. jejuni CPS has also been shown to be important for complement resistance (13): a ΔkpsM mutant was hypersensitive to serum (8), and diminished CPS production in response to exposure to host cells yielded reduced serum resistance (15). Complement activation can occur via three distinct pathways, including the lectin pathway, whereby carbohydrate ligands on bacterial cell surfaces are recognized by pattern recognition receptors such as mannose-binding lectin (MBL) (49). Although the precise means by which C. jejuni surface polysaccharides contribute to serum resistance are not yet known, our LOS results are consistent with studies of Neisseria spp., with which it has been shown that MBL binds bacteria more strongly when the LOS is truncated (34) and that loss of LOS phosphoethanolamine (PEtn) results in enhanced complement killing (51). Similar mechanisms may occur in C. jejuni and await further study.
Although the entire outer core was required for optimal protection from complement, the ΔwaaF mutant was more serum sensitive than the ΔlgtF mutant, suggesting an important role for the inner core HepII moiety in this attribute. Interestingly, we have also now found that the ΔwaaF deep rough mutant is unique among our LOS core-specific mutants in exhibiting a defect in long-term survival of C. jejuni inside epithelial cells, an increasingly recognized in vitro marker for C. jejuni pathogenesis (14, 20, 86). Several previous studies had suggested a role for the LOS in the C. jejuni-host cell interaction, but concrete evidence for a role of the core oligosaccharide had not been elucidated. For instance, one study assessing the invasion and colonization potential of different C. jejuni strains suggested the enhanced presence of LOS-relevant genes cgtB and wlaN in invasive strains (61); however, mutants harboring gene deletions were not examined. Other studies indicated that three LOS mutants exhibited diminished invasion capacities, but these likely reflect pleiotropic and non-LOS core-specific effects. For example, one study involved a mutant deleted for a large LOS locus containing htrB (56). HtrB is involved in lipid A biosynthesis, and the large deletion mutant exhibited a growth defect (56) suggesting significant phenotypic and genotypic differences from our targeted core mutants. Another study reported that a ΔwaaC mutant is defective for invasion (41); however, waaC also participates in biosynthesis of the CPS, which itself influences invasion of C. jejuni into epithelial cells in vitro (8, 42). A third published invasion defect was subsequently shown to be due to the detergent sensitivity of the ΔwaaF mutant studied (41, 43). We routinely utilize a water-based lysis procedure for harvesting bacteria from cell infections (20) which does not affect survival of the ΔwaaF mutant (43; our unpublished observations). Our ΔwaaF mutant intracellular survival findings are also not likely due to defects in CPS production, as (i) Western blots with Penner and CPS antisera yielded wild-type profiles (unpublished observations), (ii) WaaF was previously shown to be independent of the CPS biosynthesis pathway (66), and (iii) CPS alterations are expected to yield invasion defects (8) absent from our LOS mutants. Together, these findings indicate that WaaF joins other C. jejuni factors recently identified as important for impacting intraepithelial cell survival, which now also include the SR, the FeoB iron uptake protein, the enzyme PPK1, the CprS sensor kinase, and anaerobic adaptation (14, 20, 62, 80, 84).
AMPs are a critical component of the host innate immune defense against invading pathogens. PxB binds negatively charged structures such as the LPS/LOS, displaces calcium and magnesium ions, disrupts the outer membrane, and promotes self-uptake (87). LL-37 is a human cathelicidin active against Gram-negative and Gram-positive bacteria, binding the cell surface via electrostatic interactions to promote membrane leakage (23, 64, 77). Each of our LOS mutants was significantly defective for PxB resistance. This included the very modestly truncated ΔcstII mutant, implicating a role for the sialic acid component not only in GBS but also in protection from PxB. In contrast, the LOS mutants exhibited only moderate (4-fold or lower) differences in LL-37 MICs compared to the wild type, with an interesting tiled pattern observed for the truncation series. These observations suggest cell envelope perturbations despite the fact that we were unable to detect overt changes in outer membrane protein profiles or cell surface hydrophobicity. One potential explanation for the latter is that C. jejuni 81-176 harbors an extensive CPS, including an additional α-glucan capsule (70), that may interfere with detection of hydrophobicity alterations associated with LOS truncations. Nonetheless, consistent with envelope perturbations, our LOS mutants exhibited SDS sensitivities similar to those reported for the ΔwaaF mutant and the large LOS deletion mutants of strain 11168 (37, 56), and our AMP observations are consistent with a recent E. coli study hypothesizing that negative cell surface charges normally buried by the LPS/LOS are exposed in LPS/LOS mutants and thereby more available to interact with cationic molecules (5). Previous studies of very severe LOS truncations in C. jejuni also implicated roles for the LOS in PxB resistance (37, 53, 56); however, our work provides the first evidence of the importance of LgtF, GalT, and CstII in this aspect of innate immunity and the first demonstration of an antimicrobial effect of LL-37 toward C. jejuni. These and other findings described above also again reflect our increasing appreciation for cell envelope dynamics and the likely involvement of feedback loops and as-yet-unidentified regulatory mechanisms in these processes. For instance, C. jejuni AMP resistance also involves efflux pumps (2, 27), surface expression or function of which may be altered due to secondary or tertiary effects of the LOS mutations. The CFW observations presented here and previously (57) also illustrate that alterations to envelope polysaccharides can cause compensatory changes in other envelope components, which in turn may affect attributes like AMP resistance, electrostatic interactions, and surface hydrophobicity.
Finally, this study provides the first evidence of a role for the C. jejuni LOS in host colonization. Previous work suggested a correlation between the presence of an LOS gene (cgtB, encoding a β-1,3-galactosyltransferase) and the colonization potential of clinical C. jejuni isolates (61); however, as noted above, targeted gene deletions were not tested, and the structure of the 81-176 LOS does not suggest the activity of CgtB in LOS biosynthesis (21). Our observations now clearly establish that, as with other C. jejuni polysaccharides like the CPS (38), pgl (44), and flagellar glycosylation (33) systems, the LOS is an important component in host colonization. The mouse data also highlight the utility of mouse competition models to assess the ability of C. jejuni both to colonize the intestinal tract (31, 69) and to disseminate systemically into deeper tissues (30). The utility of these models is additionally important given a recent study showing unpredictable variability in competition studies in the more traditional chicken model of colonization (16). Future work to explore additional LOS mutants of 81-176 and other C. jejuni strains should lend even more insight into the role of this structure in colonization. The animal and biofilm data also touch on the question of whether C. jejuni biofilms are important in vivo. While evidence presented here might suggest not, it should also be noted that every C. jejuni hypo- or hyper-biofilm mutant identified to date exhibits planktonic growth sensitivities and/or motility defects, as well as colonization and/or host cell interaction defects (14, 18, 20, 39, 40, 57, 74, 75, 80). Thus, the relevance of C. jejuni biofilms to colonization might be addressable only via studies of mutant strains in which altered biofilm formation is the only observable difference from the wild type.
Collectively, this work has yielded novel insight into the importance of the C. jejuni LOS in a number of pathogenesis-associated properties. Analysis of our truncation series also identified the relevance of specific LOS moieties and their respective transferases to specific aspects of stress survival and/or pathogenicity. This study further highlights the dynamic nature of the C. jejuni cell envelope and, through use of a mouse competition model, provides the first direct evidence for a role of the LOS in colonization. Future work stemming from this platform should lend even more insight into the pathogenesis of this important food-borne organism.
Supplementary Material
Acknowledgments
We thank all members of the Gaynor lab for helpful discussions throughout the course of this work and Jenny Vermeulen for excellent technical support. We thank Bob Hancock for providing us with LL-37 and expert input on antimicrobial peptides, Dave Hendrixson and Dave Lampe for providing reagents and advice for the transposon mutagenesis, Dave Hendrixson for 81-176 capsule antiserum, Marie-France Karwaski and Denis Brochu for technical help, Monica Dzieciatkowska for help with the mass spectrometry analysis, and David Speert, Kelly MacDonald, and the Hancock laboratory for preparing pooled human serum.
J.A.F. and S.A.T. were supported by U.S. National Institutes of Health grants AI055715, AI058284, and AI061026 (to S.A.T.). E.F. is supported by postdoctoral fellowships from the Canadian Institutes for Health Research (CIHR) and the Michael Smith Foundation for Health Research (MSFHR). E.C.G. is supported by a Canada Research Chair award, the MSFHR, and a Burroughs Wellcome Fund Career Development Award in the Biomedical Sciences. This work was funded by CIHR grant MOP-68981 to E.C.G. and the Burroughs Wellcome Fund.
Footnotes
Published ahead of print on 5 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akerley, B. J., and D. J. Lampe. 2002. Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol. 358:100-108. [DOI] [PubMed] [Google Scholar]
- 2.Akiba, M., J. Lin, Y. W. Barton, and Q. Zhang. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemother. 57:52-60. [DOI] [PubMed] [Google Scholar]
- 3.Allos, B. M., F. T. Lippy, A. Carlsen, R. G. Washburn, and M. J. Blaser. 1998. Campylobacter jejuni strains from patients with Guillain-Barre syndrome. Emerg. Infect. Dis. 4:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amini, S., H. Goodarzi, and S. Tavazoie. 2009. Genetic dissection of an exogenously induced biofilm in laboratory and clinical isolates of E. coli. PLoS Pathog. 5:e1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ang, C. W., J. D. Laman, H. J. Willison, E. R. Wagner, H. P. Endtz, M. A. De Klerk, A. P. Tio-Gillen, N. Van den Braak, B. C. Jacobs, and P. A. Van Doorn. 2002. Structure of Campylobacter jejuni lipopolysaccharides determines antiganglioside specificity and clinical features of Guillain-Barre and Miller Fisher patients. Infect. Immun. 70:1202-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anriany, Y., S. N. Sahu, K. R. Wessels, L. M. McCann, and S. W. Joseph. 2006. Alteration of the rugose phenotype in waaG and ddhC mutants of Salmonella enterica serovar Typhimurium DT104 is associated with inverse production of curli and cellulose. Appl. Environ. Microbiol. 72:5002-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-777. [DOI] [PubMed] [Google Scholar]
- 9.Baker, M., N. Wilson, and R. Edwards. 2007. Campylobacter infection and chicken: an update on New Zealand's largest ‘common source outbreak.’ N. Z. Med. J. 120:1261. [http://www.nzma.org.nz/journal/120-1261/2717/content.pdf]. [PubMed] [Google Scholar]
- 10.Balestrino, D., J. M. Ghigo, N. Charbonnel, J. A. Haagensen, and C. Forestier. 2008. The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ. Microbiol. 10:685-701. [DOI] [PubMed] [Google Scholar]
- 11.BellonFontaine, M. N., J. Rault, and C. J. van Oss. 1996. Microbial adhesion to solvents: A novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf. B Biointerfaces 7:47-53. [Google Scholar]
- 12.Blaser, M. J., and J. Engberg. 2008. Clinical aspects of Campylobacter jejuni and Campylobacter coli infections, p. 99-121. In I. Nachamkin, C. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 13.Blaser, M. J., P. F. Smith, and P. F. Kohler. 1985. Susceptibility of Campylobacter isolates to the bactericidal activity of human serum. J. Infect. Dis. 151:227-235. [DOI] [PubMed] [Google Scholar]
- 14.Candon, H. L., B. J. Allan, C. D. Fraley, and E. C. Gaynor. 2007. Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J. Bacteriol. 189:8099-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corcionivoschi, N., M. Clyne, A. Lyons, A. Elmi, O. Gundogdu, B. W. Wren, N. Dorrell, A. V. Karlyshev, and B. Bourke. 2009. Campylobacter jejuni cocultured with epithelial cells reduces surface capsular polysaccharide expression. Infect. Immun. 77:1959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coward, C., P. M. van Diemen, A. J. Conlan, J. R. Gog, M. P. Stevens, M. A. Jones, and D. J. Maskell. 2008. Competing isogenic Campylobacter strains exhibit variable population structures in vivo. Appl. Environ. Microbiol. 74:3857-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dzieciatkowska, M., D. Brochu, A. van Belkum, A. P. Heikema, N. Yuki, R. S. Houliston, J. C. Richards, M. Gilbert, and J. Li. 2007. Mass spectrometric analysis of intact lipooligosaccharide: direct evidence for O-acetylated sialic acids and discovery of O-linked glycine expressed by Campylobacter jejuni. Biochemistry 46:14704-14714. [DOI] [PubMed] [Google Scholar]
- 18.Fields, J. A., and S. A. Thompson. 2008. Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190:3411-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frirdich, E., and C. Whitfield. 2005. Lipopolysaccharide inner core oligosaccharide structure and outer membrane stability in human pathogens belonging to the Enterobacteriaceae. J. Endotoxin Res. 11:133-144. [DOI] [PubMed] [Google Scholar]
- 20.Gaynor, E. C., D. H. Wells, J. K. MacKichan, and S. Falkow. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56:8-27. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, M., C. T. Parker, and A. P. Moran. 2008. Campylobacter jejuni lipooligosaccharides: structures and biosynthesis, p. 483-504. In I. Nachamkin, C. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 22.Golec, M. 2007. Cathelicidin LL-37: LPS-neutralizing, pleiotropic peptide. Ann. Agric. Environ. Med. 14:1-4. [PubMed] [Google Scholar]
- 23.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68:6656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerry, P., and C. M. Szymanski. 2008. Campylobacter sugars sticking out. Trends Microbiol. 16:428-435. [DOI] [PubMed] [Google Scholar]
- 25.Guerry, P., C. M. Szymanski, M. M. Prendergast, T. E. Hickey, C. P. Ewing, D. L. Pattarini, and A. P. Moran. 2002. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 27.Hannula, M., and M. L. Hanninen. 2008. Effect of putative efflux pump inhibitors and inducers on the antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 57:851-855. [DOI] [PubMed] [Google Scholar]
- 28.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 29.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofreuter, D., V. Novik, and J. E. Galan. 2008. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4:425-433. [DOI] [PubMed] [Google Scholar]
- 31.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galan. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74:4694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holme, R. 2003. Drinking water contamination in Walkerton, Ontario: positive resolutions from a tragic event. Water Sci. Technol. 47:1-6. [PubMed] [Google Scholar]
- 33.Howard, S. L., A. Jagannathan, E. C. Soo, J. P. Hui, A. J. Aubry, I. Ahmed, A. Karlyshev, J. F. Kelly, M. A. Jones, M. P. Stevens, S. M. Logan, and B. W. Wren. 2009. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect. Immun. 77:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jack, D. L., A. W. Dodds, N. Anwar, C. A. Ison, A. Law, M. Frosch, M. W. Turner, and N. J. Klein. 1998. Activation of complement by mannose-binding lectin on isogenic mutants of Neisseria meningitidis serogroup B. J. Immunol. 160:1346-1353. [PubMed] [Google Scholar]
- 35.Jacobs, B. C., A. van Belkum, and H. P. Endtz. 2008. Guillain-Barre syndrome and Campylobacter infection, p. 245-261. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 36.Jacobs-Reitsma, W., U. Lyhs, and J. A. Wagenaar. 2008. Campylobacter in the food supply, p. 627-644. In I. Nachamkin, C. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 37.Jeon, B., W. Muraoka, A. Scupham, and Q. Zhang. 2009. Roles of lipooligosaccharide and capsular polysaccharide in antimicrobial resistance and natural transformation of Campylobacter jejuni. J. Antimicrob. Chemother. 63:462-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, M. A., K. L. Marston, C. A. Woodall, D. J. Maskell, D. Linton, A. V. Karlyshev, N. Dorrell, B. W. Wren, and P. A. Barrow. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72:3769-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joshua, G. W., C. Guthrie-Irons, A. V. Karlyshev, and B. W. Wren. 2006. Biofilm formation in Campylobacter jejuni. Microbiology 152:387-396. [DOI] [PubMed] [Google Scholar]
- 40.Kalmokoff, M., P. Lanthier, T. L. Tremblay, M. Foss, P. C. Lau, G. Sanders, J. Austin, J. Kelly, and C. M. Szymanski. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanipes, M. I., L. C. Holder, A. T. Corcoran, A. P. Moran, and P. Guerry. 2004. A deep-rough mutant of Campylobacter jejuni 81-176 is noninvasive for intestinal epithelial cells. Infect. Immun. 72:2452-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanipes, M. I., E. Papp-Szabo, P. Guerry, and M. A. Monteiro. 2006. Mutation of waaC, encoding heptosyltransferase I in Campylobacter jejuni 81-176, affects the structure of both lipooligosaccharide and capsular carbohydrate. J. Bacteriol. 188:3273-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanipes, M. I., X. Tan, A. Akelaitis, J. Li, D. Rockabrand, P. Guerry, and M. A. Monteiro. 2008. Genetic analysis of lipo-oligosaccharide core biosynthesis in Campylobacter jejuni 81-176. J. Bacteriol. 190:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlyshev, A. V., O. L. Champion, C. Churcher, J. R. Brisson, H. C. Jarrell, M. Gilbert, D. Brochu, F. St. Michael, J. Li, W. W. Wakarchuk, I. Goodhead, M. Sanders, K. Stevens, B. White, J. Parkhill, B. W. Wren, and C. M. Szymanski. 2005. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55:90-103. [DOI] [PubMed] [Google Scholar]
- 45.Karlyshev, A. V., and B. W. Wren. 2005. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl. Environ. Microbiol. 71:4004-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlyshev, A. V., B. W. Wren, and A. P. Moran. 2008. Campylobacter jejuni capsular polysaccharide, p. 505-522. In I. Nachamkin, C. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 47.Klena, J. D., S. A. Gray, and M. E. Konkel. 1998. Cloning, sequencing, and characterization of the lipopolysaccharide biosynthetic enzyme heptosyltransferase I gene (waaC) from Campylobacter jejuni and Campylobacter coli. Gene 222:177-185. [DOI] [PubMed] [Google Scholar]
- 48.Korlath, J. A., M. T. Osterholm, A. Judy, J. C. Forgang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 49.Lambris, J. D., D. Ricklin, and B. V. Geisbrecht. 2008. Complement evasion by human pathogens. Nat. Rev. Microbiol. 6:132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lampe, D. J., B. J. Akerley, E. J. Rubin, J. J. Mekalanos, and H. M. Robertson. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. U. S. A. 96:11428-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis, L. A., B. Choudhury, J. T. Balthazar, L. E. Martin, S. Ram, P. A. Rice, D. S. Stephens, R. Carlson, and W. M. Shafer. 2009. Phosphoethanolamine substitution of lipid A and resistance of Neisseria gonorrhoeae to cationic antimicrobial peptides and complement-mediated killing by normal human serum. Infect. Immun. 77:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin, J., Y. Wang, and K. V. Hoang. 2009. Systematic identification of genetic loci required for polymyxin resistance in Campylobacter jejuni using an efficient in vivo transposon mutagenesis system. Foodborne Pathog. Dis. 6:173-185. [DOI] [PubMed] [Google Scholar]
- 54.Logan, S., I. C. Schoenhofen, and P. Guerry. 2008. O-linked flagellar glycosylation in Campylobacter, p. 471-482. In I. Nachamkin, C. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 55.Mansfield, L. M., D. B. Schauer, and J. G. Fox. 2008. Animal models of Campylobacter jejuni infections, p. 367-379. In I. Nachamkin, C. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 56.Marsden, G. L., J. Li, P. H. Everest, A. J. Lawson, and J. M. Ketley. 2009. Creation of a large deletion mutant of Campylobacter jejuni reveals that the lipooligosaccharide gene cluster is not required for viability. J. Bacteriol. 191:2392-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLennan, M. K., D. D. Ringoir, E. Frirdich, S. L. Svensson, D. H. Wells, H. Jarrell, C. M. Szymanski, and E. C. Gaynor. 2008. Campylobacter jejuni biofilms up-regulated in the absence of the stringent response utilize a calcofluor white-reactive polysaccharide. J. Bacteriol. 190:1097-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meredith, T. C., U. Mamat, Z. Kaczynski, B. Lindner, O. Holst, and R. W. Woodard. 2007. Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J. Biol. Chem. 282:7790-7798. [DOI] [PubMed] [Google Scholar]
- 60.Moran, A. P. 1997. Structure and conserved characteristics of Campylobacter jejuni lipopolysaccharides. J. Infect. Dis. 176(Suppl. 2):S115-S121. [DOI] [PubMed] [Google Scholar]
- 61.Muller, J., B. Meyer, I. Hanel, and H. Hotzel. 2007. Comparison of lipooligosaccharide biosynthesis genes of Campylobacter jejuni strains with varying abilities to colonize the chicken gut and to invade Caco-2 cells. J. Med. Microbiol. 56:1589-1594. [DOI] [PubMed] [Google Scholar]
- 62.Naikare, H., K. Palyada, R. Panciera, D. Marlow, and A. Stintzi. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74:5433-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63a.New England Biolabs Inc. 2008. Protein expression and analysis. pMAL protein purification and expression system instruction manual. New England Biolabs Inc., Ipswich, MA. http://www.neb.com/nebecomm/ManualFiles/manualE8200.pdf.
- 64.Nijnik, A., and R. E. Hancock. 2009. The roles of cathelicidin LL-37 in immune defences and novel clinical applications. Curr. Opin. Hematol. 16:41-47. [DOI] [PubMed] [Google Scholar]
- 65.Nothaft, H., S. Amber, M. Aebi, and C. M. Szymanski. 2008. N-linked protein glycosylation in Campylobacter, p. 447-470. In I. Nachamkin, C. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 66.Oldfield, N. J., A. P. Moran, L. A. Millar, M. M. Prendergast, and J. M. Ketley. 2002. Characterization of the Campylobacter jejuni heptosyltransferase II gene, waaF, provides genetic evidence that extracellular polysaccharide is lipid A core independent. J. Bacteriol. 184:2100-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 68.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 69.Pajaniappan, M., J. E. Hall, S. A. Cawthraw, D. G. Newell, E. C. Gaynor, J. A. Fields, K. M. Rathbun, W. A. Agee, C. M. Burns, S. J. Hall, D. J. Kelly, and S. A. Thompson. 2008. A temperature-regulated Campylobacter jejuni gluconate dehydrogenase is involved in respiration-dependent energy conservation and chicken colonization. Mol. Microbiol. 68:474-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papp-Szabo, E., M. I. Kanipes, P. Guerry, and M. A. Monteiro. 2005. Cell-surface alpha-glucan in Campylobacter jejuni 81-176. Carbohydr. Res. 340:2218-2221. [DOI] [PubMed] [Google Scholar]
- 71.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 73.Plant, L., J. Sundqvist, S. Zughaier, L. Lovkvist, D. S. Stephens, and A. B. Jonsson. 2006. Lipooligosaccharide structure contributes to multiple steps in the virulence of Neisseria meningitidis. Infect. Immun. 74:1360-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quinones, B., W. G. Miller, A. H. Bates, and R. E. Mandrell. 2009. Autoinducer-2 production in Campylobacter jejuni contributes to chicken colonization. Appl. Environ. Microbiol. 75:281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reeser, R. J., R. T. Medler, S. J. Billington, B. H. Jost, and L. A. Joens. 2007. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl. Environ. Microbiol. 73:1908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salama, N. R., B. Shepherd, and S. Falkow. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186:7926-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Senyurek, I., M. Paulmann, T. Sinnberg, H. Kalbacher, M. Deeg, T. Gutsmann, M. Hermes, T. Kohler, F. Gotz, C. Wolz, A. Peschel, and B. Schittek. 2009. Dermcidin-derived peptides show a different mode of action than the cathelicidin LL-37 against Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2499-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shelud'ko, A. V., O. V. Kulibiakina, A. A. Shirokov, L. P. Petrova, L. Matora, and E. I. Katsy. 2008. The effect of mutations in the synthesis of lipopolysaccharides and calcofluor-binding polysaccharides on biofilm formation by Azospirillum brasilense. Mikrobiologiia 77:358-363. (In Russian.) [PubMed] [Google Scholar]
- 79.Stafford, R. J., P. J. Schluter, A. J. Wilson, M. D. Kirk, G. Hall, and L. Unicomb. 2008. Population-attributable risk estimates for risk factors associated with Campylobacter infection, Australia. Emerg. Infect. Dis. 14:895-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Svensson, S. L., L. M. Davis, J. K. MacKichan, B. J. Allan, M. Pajaniappan, S. A. Thompson, and E. C. Gaynor. 2009. The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol. Microbiol. 71:253-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svensson, S. L., E. Frirdich, and E. C. Gaynor. 2008. Survival strategies of Campylobacter jejuni: stress responses, the viable but nonculturable state, and biofilms, p. 571-590. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 82.Szymanski, C. M., F. S. Michael, H. C. Jarrell, J. Li, M. Gilbert, S. Larocque, E. Vinogradov, and J. R. Brisson. 2003. Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278:24509-24520. [DOI] [PubMed] [Google Scholar]
- 83.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 84.Watson, R. O., and J. E. Galan. 2008. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 4:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuki, N. 2001. Infectious origins of, and molecular mimicry in, Guillain-Barre and Fisher syndromes. Lancet Infect. Dis. 1:29-37. [DOI] [PubMed] [Google Scholar]
- 86.Zarantonelli, M. L., M. Huerre, M. K. Taha, and J. M. Alonso. 2006. Differential role of lipooligosaccharide of Neisseria meningitidis in virulence and inflammatory response during respiratory infection in mice. Infect. Immun. 74:5506-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zavascki, A. P., L. Z. Goldani, J. Li, and R. L. Nation. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60:1206-1215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.