Abstract
Pseudomonas putida DOT-T1E was used as a model to develop a “phenomics” platform to investigate the ability of P. putida to grow using different carbon, nitrogen, and sulfur sources and in the presence of stress molecules. Results for growth of wild-type DOT-T1E on 90 different carbon sources revealed the existence of a number of previously uncharted catabolic pathways for compounds such as salicylate, quinate, phenylethanol, gallate, and hexanoate, among others. Subsequent screening on the subset of compounds on which wild-type DOT-TIE could grow with four knockout strains in the global regulatory genes Δcrc, Δcrp, ΔcyoB, and ΔptsN allowed analysis of the global response to nutrient supply and stress. The data revealed that most global regulator mutants could grow in a wide variety of substrates, indicating that metabolic fluxes are physiologically balanced. It was found that the Crc mutant did not differ much from the wild-type regarding the use of carbon sources. However, certain pathways are under the preferential control of one global regulator, i.e., metabolism of succinate and d-fructose is influenced by CyoB, and l-arginine is influenced by PtsN. Other pathways can be influenced by more than one global regulator; i.e., l-valine catabolism can be influenced by CyoB and Crp (cyclic AMP receptor protein) while phenylethylamine is affected by Crp, CyoB, and PtsN. These results emphasize the cross talk required in order to ensure proper growth and survival. With respect to N sources, DOT-T1E can use a wide variety of inorganic and organic nitrogen sources. As with the carbon sources, more than one global regulator affected growth with some nitrogen sources; for instance, growth with nucleotides, dipeptides, d-amino acids, and ethanolamine is influenced by Crp, CyoB, and PtsN. A surprising finding was that the Crp mutant was unable to flourish on ammonium. Results for assayed sulfur sources revealed that CyoB controls multiple points in methionine/cysteine catabolism while PtsN and Crc are needed for N-acetyl-l-cysteamine utilization. Growth of global regulator mutants was also influenced by stressors of different types (antibiotics, oxidative agents, and metals). Overall and in combination with results for growth in the presence of various stressors, these phenomics assays provide multifaceted insights into the complex decision-making process involved in nutrient supply, optimization, and survival.
Pseudomonads are Gram-negative bacteria characterized by high metabolic versatility, aerobic respiration, and motility. Pseudomonas spp. are also ubiquitous environmental colonizers, and Pseudomonas putida inoculates have been used to promote plant growth and in bioremediation (3, 7, 29, 52, 53, 58). The key to the ubiquitous distribution of these bacteria is their use of highly sophisticated mechanisms to adapt to changes in the local environment. Understanding the physiological and genetic basis of bacterial responses to diverse energy sources or stressors is crucial to the development of beneficial applications such as the bioremediation of the environment, the delivery of ecological chemicals for plant disease control, and the mobilization of nutrients, such as insoluble phosphate, to promote plant growth (50, 61).
Catabolite repression control is generally defined as the ability of an organism to preferentially metabolize one carbon source over another when both carbon sources are present in the organism's environment (9, 11, 35, 55). This ordered assimilation of different compounds is regulated such that the expression of catabolic pathways for nonpreferred substrates remains inhibited until the preferred and often rapidly metabolizable substrate is consumed. This regimented selection of carbon source is chiefly made at the level of specific transcriptional induction as nearly all catabolic pathways for carbohydrates, organic acids, amino acids, and many aromatic compounds are controlled by specific regulatory proteins that require inducers for sufficient expression. The strict catabolite repression observed in Escherichia coli is best exemplified by the preferential consumption of glucose before lactose (20, 35). In P. putida preferential utilization of one carbon source over others has also been observed, for instance, in the inhibition exerted by lactate or pyruvate on the utilization of alkanes (11) or the preferential use of amino acids over aromatic carboxylic acids (27). In contrast to this, in P. putida another phenomenon called “crossed catabolite repression” has also been described in which the bacteria are able to use two carbon sources (e.g., glucose and toluene) simultaneously (10). This type of control is similar to that reported for Klebsiella oxytoca, which can use both sucrose and glycerol simultaneously (42). One of the most important signal molecules involved in catabolite repression in the Enterobacteriaceae (such as Escherichia and Salmonella spp.) is cyclic AMP (cAMP) (43). In E. coli when glucose, the preferential energy source, is consumed, the amount of cAMP increases, leading to its binding to the cAMP receptor protein (Crp); cAMP-Crp can then bind to numerous promoters, where it acts as a transcriptional activator or repressor (20). However, it is important to note here that in E. coli, cAMP-Crp-mediated regulation of catabolic gene expression mainly affects the duration of lag phase during diauxie while the true repression mechanism is mediated by inducer exclusion (20). This multifaceted regulation network (using global regulators and inducer exclusion) serves as an auto-regulatory mechanism to maintain sugar utilization at a level harmonized to the bacteria's metabolic capacities rather than to simply establish preferential utilization of certain carbon sources. Consequently, metabolic balance, rather than the term catabolite repression, perhaps better describes the physiological role of this regulatory network (6, 18).
In pseudomonads, however, catabolite repression does not involve cAMP; indeed, in P. putida and Pseudomonas aeruginosa, cAMP levels remain relatively constant, regardless of the growth conditions (41, 48). Catabolite repression in pseudomonads appears to integrate different signals, a characteristic which increases the complexity of the regulation system. Five potential regulators have been associated with catabolite repression in P. putida: Crc (19, 32), Crp (called Vfr in P. aeruginosa) (56, 60), CyoB (12, 33, 40), PtsN (2, 8), and RelA (57).
Despite the recent insights into the biochemistry and genetics of metabolism in pseudomonads, relatively little is known about the global strategies used to cope with the changing environment of growth substrates. There is also a modest understanding of the catabolic pathways that can be influenced by these global regulators when a single carbon, nitrogen, or sulfur source is present in the culture medium. The aim of the present study was to set up a “phenomics” screening platform to distinguish compounds that are metabolically useful to the bacteria from those that are nonmetabolizable. Clarifying this information will emphasize the importance and versatility of regulatory systems in the ecophysiological response of bacteria and provide possibilities for the fine-tuning of these systems in order to optimize beneficial applications such as plant disease control and bioremediation.
MATERIALS AND METHODS
Strains.
The P. putida strains and plasmids used in this study are listed in Table 1. Antibiotics used were kanamycin (Km; 50 μg/ml), rifampin (Rif; 10 μg/ml), gentamicin (Gm; 30 μg/ml), tetracycline (Tc; 20 μg/ml), and streptomycin (Sm; 50 μg/ml).
TABLE 1.
P. putida strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| P. putida DOT-T1E | Tolr Rifr | 44 |
| P. putida DOT-T1EΔcrc | Gmr; DOT-T1E crc::Gm | This study |
| P. putida DOT-T1EΔcrp | Kmr; DOT-T1E crp::Km | This study |
| P. putida DOT-T1EΔcyoB | Tcr; DOT-T1E cyoB::Tc | 13 |
| P. putida DOT-T1EΔptsN | Kmr; DOT-T1E ptsN::Km | This study |
| E. coli HB101 | Smrhsd R− M+pro leu thi recA ara lacY galK xyl | 49 |
| E. coli CC118 | Δ(ara-leu)7697 ΔlacX74 ΔphoA20 galE galK thi rpsE rpoB argE recA1 | 26 |
| Plasmids | ||
| pRK600 | Cmrori ColE1 mobRK2 traRK2 | 24 |
| pKNG154 | Smr Kmr (with Km cassette inserted in ptsN) | 8 |
| pKNGcrc | Smr Gmr (with Gm cassette inserted in crc) | 2 |
| pKNGcrp | Smr Kmr (with Km cassette inserted in crp) | 2 |
Mutant strains generated in this study were produced by insertional inactivation of the desired gene. Plasmids pKNGcrp, pKNGcrc, and pKNG154 (Table 1) were derived from the suicide plasmid pKNG101 and were transferred from E. coli CC118 λpir into P. putida DOT-T1E by triparental mating using E. coli HB101 containing pRK600 to supply the transfer functions, as described previously (47). Transconjugants which incorporated the Km or Gm resistance cassettes were selected on M9 minimal medium with citrate as the sole carbon source (1) and Km or Gm, respectively. Bacteria able to grow on this medium were then counter-selected on LB medium (without NaCl and containing 7% [wt/vol] sucrose and either Km or Gm). Cells able to survive counter-selection with sucrose were expected to contain the disrupted allele in the chromosome of DOT-T1E. The absence of the wild-type allele and the presence of crc::Gmr, crp::Kmr, or ptsN::Kmr were confirmed by PCR. Two clones of each were retained, and the bona fide of the constructions was confirmed by Southern blotting. One of each correct mutant was used in further studies; the mutants were named DOT-T1EΔcrc, DOT-T1EΔcrp, and DOT-T1EΔptsN (Table 1). A mutant with a mini-Tn5-Km1 insertion in the cyoB gene was available in our group and had been isolated previously as a toluene-sensitive clone (13).
Pregrowth media and phenotypic assays in the phenotype microarrays.
Individual colonies of P. putida DOT-T1E and mutant strains were picked from the surface of cultures freshly grown LB medium plates supplemented with Rif, streaked onto M8 pregrowth (M8PG) medium plates (0.1% [wt/vol] glucose, 0.1 g/liter NH4Cl, 1 mM MgSO4, 0.6 mg/liter Fe-citrate, and micronutrients), and grown overnight at 30°C. Pregrowth of cells on M8PG medium was sufficient to deplete nutrient reserves such that the subsequent growth assays with different carbon, nitrogen, and sulfur sources were dependent on the nutritional sources provided. The biomass of the overnight plates described above was recovered from the plate surface and resuspended in 15 ml of M9 or M8 liquid medium to an optical density at 660 nm (OD660) of 0.2. The wells of the microplates were filled with 180 μl of the cellular suspension, and 20 μl of each carbon, nitrogen, or sulfur source was added to reach a final concentration of 5 mM. For sulfur source assays, the MgSO4 in the M9 medium was replaced with MgCl2. Positive-control wells consisted of full minimal medium containing glucose, NH4Cl, and MgSO4 as carbon, nitrogen, and sulfur sources, respectively; negative-control wells contained medium without cell inoculate.
All data recordings were performed using a type FP-1100-C Bioscreen C MBR analyzer system (OY Growth Curves Ab Ltd., Raisio, Finland) at 30°C, with continuous agitation. The turbidity was measured using a wideband filter at 420 to 580 nm every 60 min over a 24-h period. Each strain was assayed at least three times for each of the compounds tested, and plates were visually examined following each assay in order to verify the results. To validate the screening results, cultivations were also performed in 100-ml conical flacks with 20 ml of culture medium. Turbidities of cultures under these conditions were usually twice that seen in the microplates, which validates the high-throughput approach.
For stress experiments the strains were grown overnight on LB (1/5 dilution) plates, resuspended in M8 minimal medium as described above, and inoculated into microplate wells containing LB liquid medium, diluted 1/2, with the corresponding stressor concentration. Assays for the growth of DOT-T1E and the mutants in the presence of various aromatic compounds as sole carbon or nitrogen sources were performed in 100-ml flasks; compounds included propylbenzene, hexane, gallate, hexanoate, salicylate, 4-aminophenol, and phenol. Propylbenzene was supplied in the vapor phase.
Carbon sources for phenotype arrays.
The following carbon sources were used: aminobutyric acid, l-alanine, l-arginine, l-asparagine, l-aspartic acid, l-cysteine, l-glutamic acid, l-glutamine, l-glycine, l-histidine, l-isoleucine, l-leucine, l-lysine, l-methionine, l-phenylalanine, l-proline, l-serine, l-threonine, l-tryptophan, l-tyrosine, l-valine, 2-hydroxybutiric acid, 3,4-dihydroxyphenylacetic acid, 3-hydroxybenzoic acid, 4-hydroxyphenylpyruvate, 4-hydroxybenzoate, acetic acid, dl-mandelic acid, ferulic acid, formic acid, fumaric acid, glutaric acid, glycolic acid, homogentisate, ketobutyric acid, l-lactic acid, malonic acid, potassium phthalate, propionic acid, quinic acid, sodium acetate, sodium benzoate, sodium lactate, sodium malonate dibasic, sodium pyruvate, succinic acid, trisodium citrate, tropic acid, d-arabinose, d-fructose, d-fucose, d-galactose, d-glucose, d-glucuronic acid, d-lactose, d-mannitol, d-ribose, d-sorbitol, maltose, N-acetyl-d-glucosamine, sucrose, trehalose, xylose, 2-phenylethylamine, 3-hydroxybenzonitrile, 4-methylbenzoate, 4-nitrotoluene, 5 amino-n-valeric acid, adenosine, decanoic acid, glycerol, methylpyruvate, methylsalycilate, m-inositol, potassium thiocyanate, resorcinol, Tween 20, uridine, vaniline, propylbenzene, hexane, gallate, hexanoate, salicylate, 4-aminophenol, phenol, benzene, toluene, and ethylbenzene.
Nitrogen sources for phenotype arrays.
Nitrogen sources were the following: 2-amino benzoic acid, dl-homocysteine, dl-ornithine, d-alanine, d-arginine, d-asparagine, d-aspartic acid, d-glutamic acid, d-histidine, d-leucine, d-lysine, d-methionine, d-phenylalanine, d-proline, d-serine, d-threonine, d-tryptophan, d-tyrosine, d-valine, l-alanine, l-arginine, l-asparagine, l-aspartic acid, l-cysteine, l-glutamic acid, l-glutamine, l-glycine, l-histidine, l-homoserine, l-isoleucine, L-leucine, l-lysine, l-methionine, L-phenylalanine, l-proline, l-serine, l-threonine, l-tryptophan, l-tyrosine, l-valine, l-proline (trans-4-OH), Ala-Glu, Ala-Gly, Ala-His, Ala-Leu, Ala-Phe, Ala-Pro, Ala-Thr, Gly-Gln, Gly-Glu, Gly-Gly, Gly-Leu, Gly-Phe, Gly-Pro, Gly-Ser, Gly-Tyr, Gly-Val, Leu-Leu, Leu-Pro, Tyr-Ala, Ala-Ala-Ala, Gly-Gly-Ala, Gly-Gly-Gly, Gly-Gly-Leu, Ile-Pro-Ile, Leu-Gly-Gly, Lys-Lys-Lys, Val-Tyr-Val, 2,4-dinitrobenzoic acid, 3,4-dinitrobenzoic acid, 3,5-dinitrobenzoic acid, 3-amino benzoic acid, 4-amino benzoic acid, 2-nitrotoluene, 3-nitrotoluene, 4-nitrotoluene, adenine, adenosine 5′ diphosphate, agmatine sulfate, ammonium chloride, ammonium nitrate, d-(+)-glucosamine HCl, ethanolamine, ethylamine, formamide, hypoxanthine, inosine, m-toluidine, N,N-dimethylacetamide, N,N-dimethylformamide, N-acetyl-d-glucosamine, o-toluidine, phenylethylamine HCl, p-toluidine, putrescine, thiamine HCl, thymidine, triethylamine, uracyl, and urea.
Sulfur sources for phenotype arrays.
Sulfur sources were the following: dl-ethionine, dl-homocysteine, d-methionine, l-cysteine, l-methionine, sodium sulfate, taurine, 2-thiohydantoin, agmatine sulfate, cysteamine, dl-alpha-lipoamide, N-acetyl-l-cysteamine, sodium sulfite, sodium taurocholate, thioflavin T, thiouracil, and thiourea.
Stress compounds for phenotype arrays.
The following stress compounds were used AgNO3 (3 μM), CdCl2 (0.156 mM), CoCl2 (0.156 mM), CuSO4 (1 mM), HgCl2 (0.4 μg/ml), LiCl (0.25 M), MnSO4 (1 mM), NiCl2 (1 mM), RbCl (6.2 μM), K2TeO3 (0.975 μg/ml), ZnCl2 (0.5 mM), nondetergent sulfobetaine (NDSB-201; 1%), cetyltrimethylammonium bromide (CTAB; 0.001%), N-lauryl sarcosine (NLS; 0.078%), sodium dodecyl sulfate (SDS; 0.06%), deoxycholate (DOC; 1%), Triton X-100 (1%), EDTA (0.125 mM), 2,2′-bipyridine (0.5 mM), NaCl (0.5 M), K2Cr2O7 (12.5 μg/ml), H2O2 (0.004%), NH2OH (30 μg/ml), methyl viologen (100 μM), tert-butyl hydroperoxide (TBH; 0.0015%), ethidium bromide (EtBr; 0.1 mg/ml), KSCN (80 mM), KCN (0.325 mg/ml), K2HAsO4 (0.9 mg/ml), NaBr (0.25 M), ampicillin (Ap; 10 μg/ml), carbenicillin (Cb; 80 μg/ml), chloramphenicol (Cm; 15 μg/ml), cefotaxime (Ctx; 0.375 μg/ml), erythromycin (Ery; 15 μg/ml), gentamicin (2 μg/ml), kanamycin (0.195 μg/ml), nalidixic acid (Nal; 0.0125 mg/ml), neomycin (Neo; 1 μg/ml), norfloxacin (Nor; 0.05 μg/ml), novomycin (Nov; 100 μg/ml), piperacillin (Pip; 10 μg/ml), streptomycin (Sm; 2 μg/ml), spectinomycin (Sp; 0.1 mg/ml), tetracycline (Tc; 0.05 μg/ml).
RESULTS AND DISCUSSION
Reference growth conditions for DOT-T1E in carbon, nitrogen, and sulfur sources.
As an initial approach we tested the wild-type strain DOT-T1E for the ability to grow in the presence of 80 different carbon sources, 101 nitrogen sources, and 17 sulfur sources using the phenomics platform. The reference conditions were set for the wild-type strain using ammonium chloride as the nitrogen source (carbon and sulfur assays), glucose as the carbon source (nitrogen and sulfur assays), and MgSO4 as the sulfur source (carbon and nitrogen assays) (see Materials and Methods for complete medium descriptions). The results for the growth of DOT-T1E in the various nutrient sources are presented in the supplemental material. Under the conditions used, DOT-T1E could not grow in the presence of some of the nutrient sources tested; and in a series of comparative assays with mutant strains, the compounds on which the wild-type could not grow were omitted. Thus, from these results a phenomics platform was developed (5).
These preliminary results confirmed the limited ability of P. putida to use sugars as a C source, which is restricted to glucose, fructose, and glucuronic acid (a uronic acid structurally related to glucose), as well as its inability to assimilate C1 compounds (i.e., formaldehyde and formic acid). The results also confirm the ability of P. putida to use organic acids (such as acetic, citric, glutaric, quinic, lactic, and succinic), certain l-amino acids (Ala, Arg, Asn, Glu, His, Ile, Lys, Pro, Tyr, and Val), various amino organic compounds, and aromatic compounds (3,4-dihydroxyphenylacetate [14], 4-hydroxybenzoate, homogentisate, sodium benzoate, 2-phenylethylamine, toluene, ethylbenzene, propylbenzene, gallate [37], 4-aminophenol, and salicylate [51]). Importantly, a number of the carbon source compounds on which wild-type P. putida DOT-T1E can grow have not been previously described, including decanoate, 3,4-dihydroxyphenylacetate, 4-hydroxyphenylpyruvate, phenylethylamine, phenylpyruvate, quinate, salicylate, and gallate (see Fig. S1 and Table S1 in the supplemental material). Potential pathways for these compounds are the focus of other studies, leading to a more comprehensive understanding of the metabolic pathways present within P. putida DOT-T1E (our unpublished results).
The wild-type strain was assayed for its ability to utilize 101 different nitrogen sources for growth (Fig. S2 and Table S1). Different results were obtained for the nitrogen source assays versus the carbon source results. We found that the wild-type DOT-T1E was able to use a number of amino acids (ornithine, Ala, and Phe), dipeptides, ethanolamine, and adenine as an N source but not as a C source. Of note is the observation that DOT-T1E used as an N source a good number of either d- or l-amino acids when they were supplied as the sole nitrogen sources (i.e., d-ornithine, d-alanine, d-arginine, d-asparagine, d-lysine, and d-valine). d-Met was an amino acid that allowed better growth than the l-isomer. A recent article by Lam et al. (25) showed that certain d-amino acids modulate the composition of the peptidoglycan cell wall and are used by bacteria to better adapt to environmental conditions. Our results support the idea that DOT-T1E may rely directly on natural sources of these d-amino acids in response to environmental cues. Alternatively, l-amino acids, supplied as nitrogen sources, that allowed DOT-T1E to flourish (while d-forms did not) include l-Glu, l-His, l-Phe, l-Ser, and l-Tyr. These results suggest that DOT-T1E does not express racemases that can convert the d-forms of these amino acids into the corresponding l-isomers. Other compounds that did not support growth of wild-type DOT-T1E when used as sole nitrogen sources include all eight tripeptides tested (see Fig. S2 in the supplemental material).
Of the 17 sulfur sources tested, the wild-type strain was able to grow on 15. These include amino acids and organic acids, as well as other compounds such as cysteamine and thiouracil (see Fig. S3 in the supplemental material).
Comparative growth of DOT-T1E, Δcrc, Δcrp, ΔcyoB, and ΔptsN on distinct carbon sources.
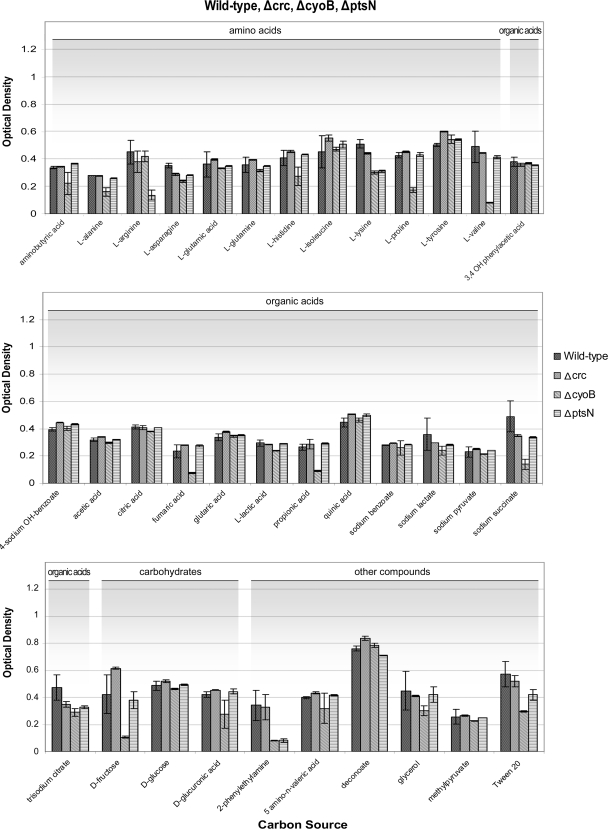
In order to assess the ability of the strains to adapt and use available carbon sources, we tested the four global regulatory mutants using 45 distinct compounds as carbon sources (Fig. 1 and 2; see also tables in the supplemental material). The utilization of most C sources by the global regulatory mutants and the parental strain were similar although some differences were noted that are described below. The results suggest that P. putida DOT-T1E is able to balance metabolic fluxes to overcome the lack of a global regulator. The most extreme case was that of the Δcrc mutant, which behaved most like the wild-type strain in the presence of different sources of carbon; this strain grew to turbidities similar to those of DOT-T1E in the presence of all the assayed carbon sources (Fig. 1 and Table 2), suggesting that, under these conditions, the Crc global regulator is not essential for the ability of wild-type P. putida DOT-T1E to metabolize carbon.
FIG. 1.
Growth of P. putida DOT-T1E and its isogenic Δcrc, ΔcyoB, and ΔptsN strains with different C sources. The wild-type and mutant strains were assayed for their ability to grow on 35 defined carbon sources at 5 mM over a 24-h period. Results represent the average of at least three different experiments. Final turbidities of less than 0.2 are considered to indicate a lack of growth ability on that compound. The Δcrp mutant is absent from this analysis because it does not grow in NH4Cl, which is the nitrogen source used in these assays.
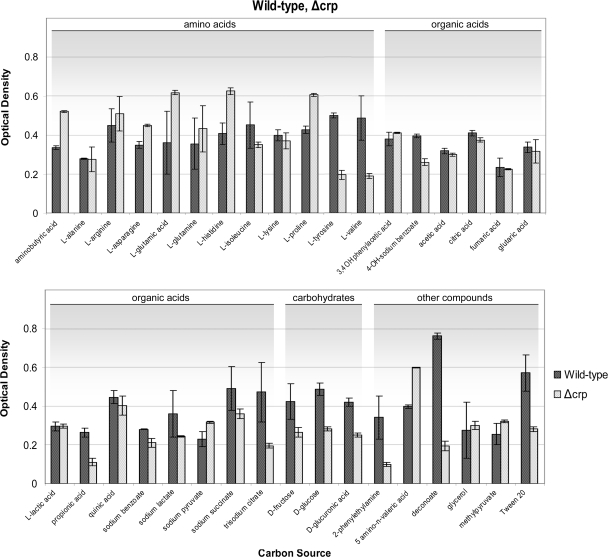
FIG. 2.
Growth of P. putida DOT-T1E and its isogenic Δcrp mutant with different C sources. The wild-type and Δcrp mutant strains were assayed for their ability to grow in defined carbon sources at 5 mM over a period of 24 h, using Ala-Thr as a nitrogen source. Results presented are the average of three assays standard deviations.
TABLE 2.
Summary of phenomics assay compounds on which the global regulator mutant strains could not flourisha
| Nutrient group and type or growth conditionb | Compound(s) on which the indicated strain showed deficient growthc |
|||
|---|---|---|---|---|
| Δcrc | Δcrp | ΔcyoB | ΔptsN | |
| Carbon sources (n = 35) | ||||
| Amino acid(s) | l-Tyrosine, l-valine | l-Proline, l-valine | l-Arginine | |
| Organic acid(s) | Propionic acid, trisodium citrate | Propionic acid, sodium succinate | ||
| Carbohydrate | d-Fructose | |||
| Other compound(s) | 2-Phenylethylamine, decanoate | 2-Phenylethylamine | 2-Phenylethylamine | |
| Nitrogen sources (n = 40) | ||||
| Amino acid(s) | d-Valine | d-Amino acids, l-asparagine, l-aspartic, l-glutamine, l-serine, l-tyrosine, l-valine, l-glycine, l-lysine | d-Amino acids, l-tyrosine, l-valine, l-serine, l-histidine, l-glycine | d-Amino acids, l-lysine, l-glycine, l-histidine |
| Dipeptides | Ala-Glu, Ala-His, Ala-Leu | Ala-Glu, Ala-His, Ala-Leu, Gly-Gly, Gly-Ser | Ala-Glu, Ala-His, Ala-Leu | |
| Other compound(s) | Adenine, agmatine sulfate, ammonium chloride, ammonium nitrate, ethanolamine, hypoxanthine, urea | Adenine, ethanolamine, hypoxanthine, urea | Adenine, ethanolamine, hypoxanthine, urea | |
| Sulfur sources (n = 15) | ||||
| Amino acid(s) | dl-Ethionine, d- and l-methionine | |||
| Other compound(s) | N-Acetylcysteamine | Cysteamine, N-acetylcysteamine | N-Acetylcysteamine | |
| Stressor(s) (n = 45) | 2,2′-Bipyridine cobalt, CTAB, EDTA, H2O2, methyl viologen | Cadmium, cobalt, methyl viologen, nickel, streptomycin, spectinomycin | 2,2′-Bipyridine, cadmium, manganese | 2,2′-Bipyridine, mercury |
In regards to phenomics assays for the Δcrp mutant, our initial results were surprising and in stark contrast to results for the Δcrc mutant in that the strain appeared unable to grow on all carbon sources tested. Upon further analysis, these results were found to be an effect of the inability of Δcrp to grow on the chosen reference condition nitrogen source, which we initially set to ammonium chloride. Therefore, the assays were repeated using Ala-Thr (the preferred nitrogen source of the Δcrp strain) (Fig. 2). The Crp mutant grew deficiently with l-valine, l-tyrosine, decanoate, propionate, and 2-phenylethylamine. The PstN mutant exhibited reduced growth with l-arginine and 2-phenylethylamine (Fig. 1) while the CyoB mutant exhibited diminished growth with l-proline, l-valine, fumaric acid, propionate, succinate, d-fructose, and 2-phenylethylamine (Fig. 1 and Table 2).
In looking over the results from all the tested knockout strains (summarized in Table 2), we see that for a number of compounds two or more regulators are required for proper growth. These metabolites may represent important pathway controls that require diverse inputs in order to be activated. These compounds include l-valine and propionate (requires Crp and CyoB) and 2-phenylethylamine (requires Crp, CyoB, and PtsN).
In order to flourish on l-valine as a carbon source, both the Crp and CyoB global regulators were needed (Table 2). The pathways for the degradation of the branched chain amino acids (BCAA) valine, leucine, and isoleucine have many enzymes in common (21, 22, 23, 28). Because l-valine metabolism is compromised while l-leucine metabolism was left intact, our results suggest that CyoB and Crp may exert control over one of the few enzymes present within the valine pathway that is missing from the leucine pathway, such as CDS (3-hydroxyacyl-coenzyme A [CoA] dehydrogenase family protein; KEGG code 1.1.1.35). The mechanism through which Crp and CyoB enable growth on BCAA, surprisingly, may also involve the Crc regulator. This is because the Crc global regulator of P. putida KT2440 has been previously shown to repress a number of enzymes involved in BCAA metabolism (34). Hence, we postulate that while Crc may exert inhibitory control over BCAA metabolism, the positive influence of Crp and CyoB trumps this inhibition. Therefore, it may be only in the Crp and CyoB knockout strains that the Crc regulator is allowed free reign such that, in their absence, Crc is able to shut down the valine arm of the BCAA catabolism pathway.
2-Phenylethylamine is unique in that all three Crp, CyoB, and PtsN regulators were separately required in order for DOT-T1E to flourish with this compound as the sole carbon source (Fig. 1 and 2 and Table 2). Since phenylethylamine catabolism is related to the phenylacetate regulon (3), we tested whether other compounds within the phenylacetate metabolism pathway are equally dependent upon multiple global regulators, we assayed the growth phenotype with three other compounds from the phenylacetate metabolome, namely, phenylpyruvate, phenylethanol, and phenylacetate. Wild-type DOT-T1E and all mutants were able to grow on phenylpyruvate, while the Δcrp strain grew deficiently on phenylacetate; all three mutants had important limitations for growth on phenylethanol as a carbon source (see Table S2 in the supplemental material). Interestingly, Crc was also required for growth on phenylethanol, showing the complexity of the interplay between all four regulators within a single pathway while identifying both phenylethanol and 2-phenylethylamine as key points of control.
An interesting finding regarding the Δcrp mutant, which becomes clear upon close inspection of Fig. 2, is that this strain shows significantly higher yields than the wild type when grown on a number of amino acids (l-Glu, l-His, and l-Pro), aminobutyrate, and 5-amino-n-valeric acid as carbon sources. Therefore, Crp, along with being required for growth on l-Val, also appears to inhibit metabolic pathways for other amino acids—a trademark of the process of carbon control, in which global regulators function dually to repress certain pathways and activate others in order to optimize growth.
Comparative growth of DOT-T1E, Δcrc, Δcrp, ΔcyoB, and ΔptsN on distinct nitrogen sources.
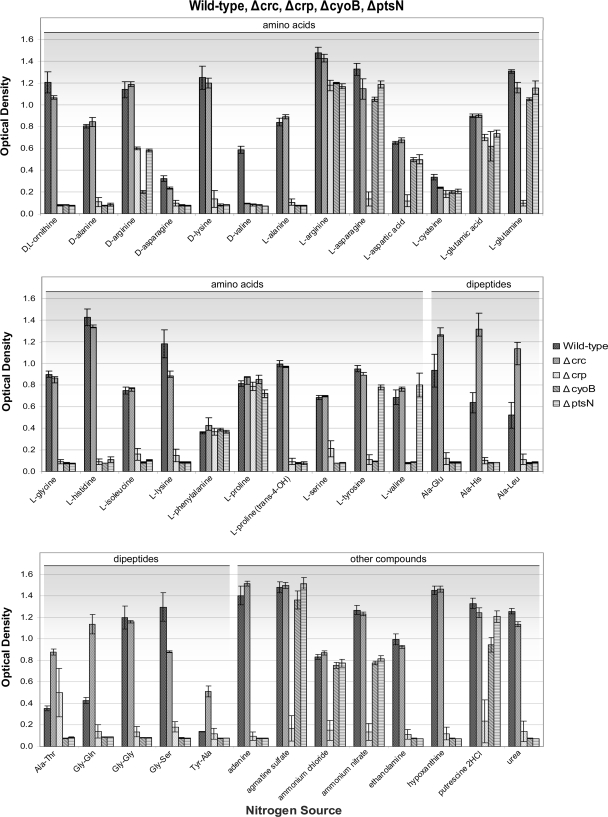
In order to assess the ability of the strains to adapt and use available nitrogen sources, we tested the four global regulatory mutants using 41 distinct compounds as nitrogen sources (Fig. 3 and Table S1). Growth assays for Δcrc on the N sources indicate that the Δcrc mutant was the least compromised of all mutants and grew similarly to the wild-type strain on many of the compounds tested. The only compound on which the Δcrc mutant was unable to grow was d-valine (Table 2). Another observation that separates the Δcrc mutant from the other mutants is that it was able to grow better than the wild type on some of the sources tested, particularly dipeptides (Ala-Glu, Ala-His, Ala-Leu, Ala-Thr, and Gly-Gln) (Fig. 3). A recent publication from Fernando Rojo's group included an extensive proteomic and transcriptomic analysis of a P. putida KT2440 Δcrc mutant strain that had been grown in rich medium (34). It was found that up to 134 genes were differentially expressed in the absence of Crc, with 80% increased in expression and 20% showing a decrease. Interestingly, further analysis revealed that most of these genes were implicated in the transport or metabolism of amino acids and sugars. Based on our results, we hypothesize that the Crc global regulator exerts an inhibitory role in the regulation of either dipeptide transport systems or dipeptide assimilation pathways and their overall use as a source of energy.
FIG. 3.
Growth with different N sources of P. putida DOT-T1E and isogenic mutants deficient in a global regulator. The wild-type and mutant strains were assayed for their ability to grow on 40 defined nitrogen sources at 5 mM over a period of 24 h. Results presented are the averages and standard deviations of at least three assays. Interestingly the Δcrp mutant could not utilize NH4Cl as a nitrogen source. This observation led us to use Ala-Thr the nitrogen source which was preferred by the Δcrp mutant strain in future assays for with this mutant.
One of the most striking results obtained from the nitrogen source phenomics assays was that the Δcrp mutant was unable to utilize ammonium chloride as a nitrogen source (Fig. 3). Bacteria assimilate nitrogen through ammonia via two distinct pathways, depending on the concentration of the source material. In E. coli, when there are high concentrations of ammonia, the enzyme glutamate dehydrogenase (GDH) catalyzes a reductive amination of 2-oxoglutarate to glutamate; when ammonium concentrations are low (<1 mM), a two-step reaction occurs involving the enzymes glutamine synthetase (GS) and glutamine:2-oxoglutarate aminotransferase (GOGAT) (16, 17). In P. putida the latter pathway is used for the assimilation of nitrogen from reduced nitrate or nitrite and from trinitrotoluene (TNT) (7, 45). It is therefore possible that the GS/GOGAT pathway might be affected in the Δcrp mutant, and/or that in the mutant the transport of inorganic nitrogen sources is compromised. Regardless of the specific action sites, these results suggest that Crp is relevant in the regulation of nitrogen metabolism in DOT-T1E.
The Δcrp strain was unable to use certain amino acids and nitrogenated organic compounds as nitrogen sources (Fig. 3 and Table 2). The compounds with which this mutant could grow include the Ala-Thr, Gly-Gln, Gly-Gly, and Gly-Ser dipeptides; putrescine; l-arginine; and l-serine. However, in addition to the apparent importance of Crp, both ΔcyoB and ΔptsN mutants showed compromised nitrogen metabolism and were unable to flourish on a number of nitrogen sources. Amino acids that all three mutants (Δcrp, ΔcyoB, and ΔptsN) were unable to metabolize were d-Ala, d-Asn, d-Lys, and dl-ornithine and a number of l-amino acids, (l-Ala, l-Gly, l-His, l-Ile, l-Lys, and l-Ser) as well as trans-4-hydroxyproline. Additionally, ΔcyoB and ΔptsN strains were unable to use certain dipeptides, along with other select compounds, as nitrogen sources (adenine and ethanolamine), suggesting that these three global regulators (Crp, CyoB, and PtsN) work together to coordinate the activation of the various pathways required for optimal nitrogen metabolism.
Comparative growth of DOT-T1E, Δcrc, Δcrp, ΔcyoB, and ΔptsN on distinct sulfur sources.
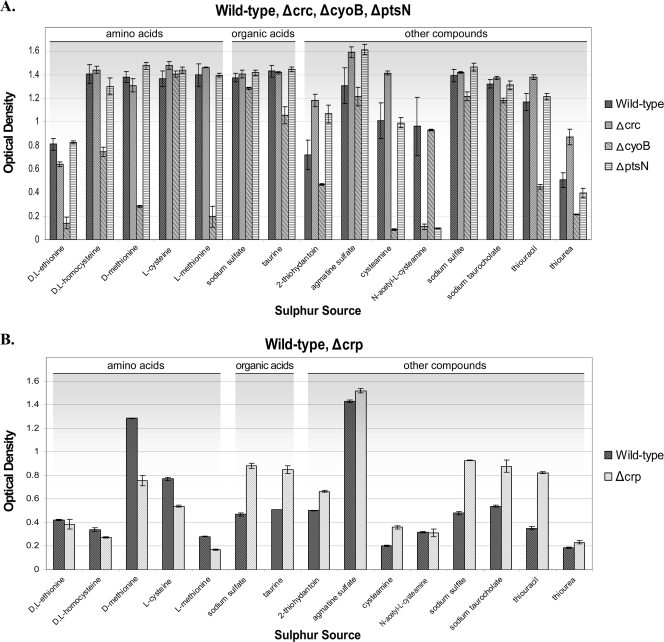
Of the 17 sulfur sources initially tested, the wild-type strain could use 15 of them for growth. When tested in the presence of these sulfur sources, the wild-type consistently grew to a turbidity of ≥0.5 during a 24-h period (Fig. 4A).
FIG. 4.
(A) The wild-type and mutant strains were assayed for their ability to grow on 15 defined sulfur sources at 5 mM over a 24-h period. An average from three independent experiments is represented. The Δcrp mutant is absent from this analysis because it does not grow in NH4Cl, which is the nitrogen source used in these assays. (B) The wild-type and Δcrp mutant strains were assayed for their ability to grow in defined sulfur sources at 5 mM over a period of 24 h, using Ala-Thr as a nitrogen source. Results presented are the average of three assays and standard deviations.
In general, the results obtained from assays using sulfur sources with different mutants were consistent (summarized in Table 2) in that most of the mutants grew similarly to the wild-type strain on the sulfur sources tested (Fig. 4A and B). In particular, the Δcrp strain was able to flourish on all of the tested compounds when assayed alongside the DOT-T1E wild-type strain using Ala-Thr as a nitrogen source (Fig. 4B) while, of all the compounds tested, there was only one, N-acetyl-l-cysteamine (Fig. 4A), on which the ΔptsN and Δcrc mutants could not grow. The ΔcyoB mutant could not use cysteamine and did not grow well on thiourea or d- or l-methionine. These three compounds are metabolized, as described by Vermeij and Kertesz (59), as part of the pathways that convert l-cysteine and l-methionine through methanethiol into sulfite. Insight into these pathways has been provided by transcriptomic arrays that were carried out with a P. putida KT2440 ΔcyoB strain (34). The results showed that an enzyme within this pathway known as methionine γ-lyase (mdeA; PP1308) has a 2.16-fold decreased expression in the ΔcyoB mutant versus the wild type. This suggests that CyoB may activate this pathway. Our results concur, suggesting that in DOT-T1E, CyoB is implicated in the regulation of the pathways involved in methionine and cysteine metabolism.
Comparative growth of DOT-T1E, Δcrc, Δcrp, ΔcyoB, and ΔptsN in the presence of stress molecules.
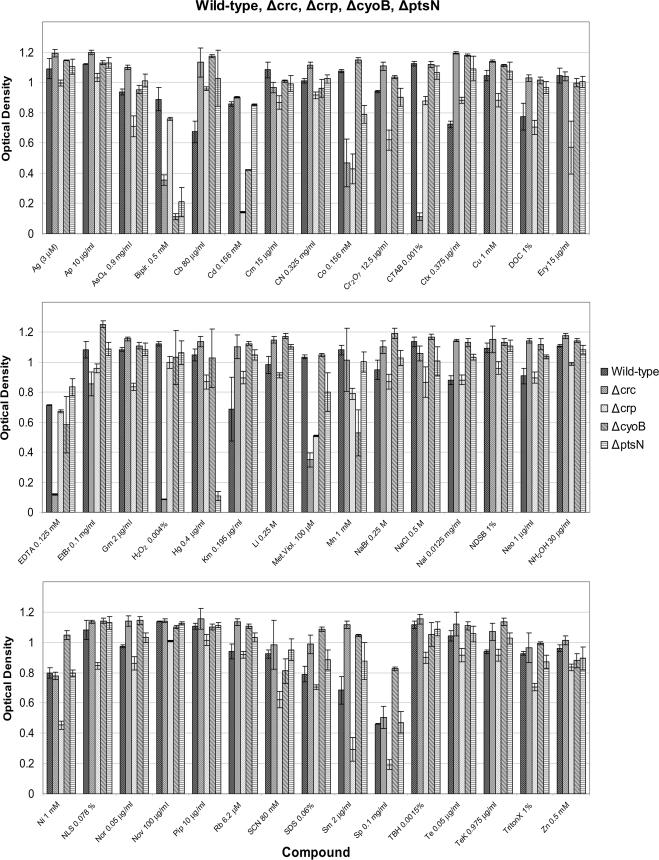
Global regulators are often involved in the control of multiple unrelated cellular functions. The above set of results indicated that the four global regulator systems tested influence bacterial growth on a number of nutrient sources; it is also well known that in pseudomonads Crc is involved in biofilm formation and may influence cell-to-cell communication (38). For this reason we decided to test the response of the wild-type strain and the mutants to a series of deleterious chemicals, including metal ion chelators, oxidative stress agents, detergents, metals, and antibiotics, among others, that may be important to these cellular functions. Comparative growth analysis was performed for the DOT-T1E wild-type and mutant strains in the presence of 45 distinct stress molecules (Fig. 5). The wild-type strain was able to grow in the presence of all compounds at the concentrations tested; however, some of the mutants were severely affected in their ability to grow (Fig. 5 and Table 2).
FIG. 5.
Effect on growth of P. putida DOT-T1E and its isogenic mutants in the presence of organic and inorganic stressors. The wild-type and all mutant strains were assayed for their ability to grow in the presence of 45 different stress compounds over a period of 24 h. Results presented are the average of three assays and standard deviations.
Three of the mutants, namely, ΔcyoB and ΔptsN and, to a lesser extent, Δcrc, were compromised in their ability to survive in the presence of 2,2′-bipyridine. This compound is a bidentate chelating agent able to chelate transition metals, including Cr, Mn, Fe, Co, Ni, Cu, and Zn. Previous transcriptome studies using the P. putida KT2440 ΔcyoB mutant indicated reduced expression of iron assimilation genes (34). The genes PP2582 (putative heme oxygenase), PP2193 (outer membrane ferric siderophore receptor), PP3330 (putative outer membrane ferric siderophore receptor), and PP5307 (exbD Fe3+-siderophore transport system inner membrane protein) all showed a 2-fold reduction in expression in the ΔcyoB mutant. Therefore, our phenomics results support the notion that CyoB is important to in vivo iron assimilation while the results appear also to implicate PtsN and Crc global regulators in the ability of DOT-T1E to survive under the low availability of transition metals.
One of the most important survival mechanisms for bacteria exposed to harmful compounds, such as toxic metal cations, involves expulsion of the compounds via efflux pumps. In pseudomonads one such efflux pump, known as CzcCBA, has been shown to impart resistance to not only cadmium but also cobalt (36). Our results show that while Δcrc is sensitive to cobalt and ΔcyoB is sensitive to cadmium, the Δcrp mutant is sensitive to both cobalt and cadmium. These results suggest that the Crp regulator may be involved in the upregulation of efflux pump expression in the presence of these toxic metal cations. This effect could be mediated directly by changes in expression of the czcCBA operon or indirectly by changes in the expression of the two-component regulatory genes czcR-czcS (39). Both the czcCBA operon and the czcR-czcS regulatory system genes are present on the chromosome of DOT-T1E and can be localized to the same ∼14-kb fragment as the ydiB quinate/shikimate dehydrogenase (A. Segura, L. Molina, and J. L. Ramos, unpublished results). It is also of interest that the crc mutant grew deficiently in the presence of the CTAB detergent and the chelating agent EDTA. The TtgABC efflux pump seems to be relevant in the extrusion of these compounds (P. Godoy and C. Daniels, unpublished results), but whether or not expression of this pump is compromised in this background has not yet been analyzed.
The only global regulator mutant that exhibited compromised growth in the presence of mercury was the ΔptsN strain. Mercury resistance in bacteria is normally mediated through reduction of Hg++ to Hg0 by a mercuric reductase known as MerA. Mercury resistance operons have previously been reported in Pseudomonas species, such as the genes (merRTPADE) described on the Tn501 of P. aeruginosa (54). In this system the products of merR and merD normally regulate the expression of the operon (4); no reports of the involvement of global regulators have been described for this system. However, our ΔptsN mutant clearly behaves differently from the wild type and the other three mutants in the presence of this toxic cation. Interestingly, the genes encoding merRTPADE could not be found on the chromosome of DOT-T1E, so it is likely that this strain uses an alternative pathway to mediate resistance to mercury.
Conclusion.
P. putida is a ubiquitous bacterium capable of thriving in many different environments, an ability which is in part due to its metabolic versatility. In natural environments, catabolite repression is likely of great importance in the success of bacteria when they compete for available nutrients with other microorganisms (15). Bacteria are able to adapt to changes in their environment via numerous signaling pathways, and this adaptation commonly involves the alteration of metabolic gene expression in order to meet the current growth requirements (46). Global regulatory genes are frequently involved in this decision-making process as they allow for substrate-specific responses and direct regulation of the metabolic pathway in question.
P. putida has at least five potential global regulators related to metabolism (Crc, Crp, CyoB, PtsN, and RelA). In the present study we investigated the role of four of these regulators in the response of the bacteria when it is subjected to different growth conditions and stresses. A relevant feature revealed in this study is that global catabolite repression systems impose order in the way that nutrients are assimilated, whereas, in general, they have little effect on the pattern of nutrients utilized by DOT-T1E. The extreme case regarding carbon and nitrogen utilization is the mutant deficient in Crc, which exhibited a pattern of carbon and nitrogen source utilization almost identical to that of the parental strain. Transcriptomic and proteomic analysis with a Δcrc mutant of the closely-related KT2440 strain revealed that Crc is the key global regulator in control of assimilation of amino acids and organic acids. This effect is notorious in cells grown in LB medium, as revealed in transcriptomic/proteomic analysis by Moreno et al. (34). In contrast to this ordered consumption, when the chemicals are present as single nutrients, thereby eliminating the possibility of preferences, no effects are observed.
Another relevant finding is that the utilization of some compounds as C sources and N sources is influenced by more than one global regulatory system, which emphasizes the interplay required by bacteria to thrive with certain nutrients. This is the case for the utilization of propionate (that is under CyoB and Crp control), 2-phenylethalonamine, adenine, and certain dipeptides that are under the control of Crp, CyoB, and PtsN. This indicates that control can be exerted at more than one catabolic point and that a number of critical steps exist as regulatory check points. The results obtained are multifaceted, and each experimental condition provides a different insight into the function of the global regulators.
In summary, we have used a comprehensive phenomics analysis to examine the importance of global regulatory factors under diverse conditions, including the availability of distinct sources of carbon, nitrogen, and sulfur and in the presence of stress molecules. Our data corroborate the complexity of the regulatory systems in P. putida DOT-T1E and provide important information in regard to the catabolic nature of pseudomonads. These results provide insight into the functioning of global regulators in P. putida and will aid in the expansion of system biology models of in silico pathways that are being developed as part of the PSYSMO systems analysis of P. putida (30, 31; http://www.psysmo.org/home/).
Supplementary Material
Acknowledgments
The work in the manuscript was supported by grants from projects BIO2006-05668 and CSD2007-00005 from the Spanish Ministry of Science and Education, and project CVI344 from Junta de Andalucía and ERANET (GEN2006-27750-C5-5-E/SYS). C.D. is a recipient of an I3P fellowship from Consejo Superior de Investigaciones Científicas.
We thank Benjamin J. Pakuts for critical reading of the manuscript and M. Mar Fandila and Carmen D. Lorente for secretarial assistance.
Footnotes
Published ahead of print on 5 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abril, M. A., C. Michán, K. N. Timmis, and J. L. Ramos. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda-Olmedo, I., J. L. Ramos, and S. Marqués. 2005. Integration of signals through Crc and PtsN in catabolite repression of Pseudomonas putida TOL plasmid pWW0. Appl. Environ. Microbiol. 71:4191-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias, S., E. R. Olivera, M. Arcos, G. Naharro, and J. M. Luengo. 2008. Genetic analyses and molecular characterization of the pathways involved in the conversion of 2-phenylethylamine and 2-phenylethanol into phenylacetic acid in Pseudomonas putida U. Environ. Microbiol. 10:413-432. [DOI] [PubMed] [Google Scholar]
- 4.Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355-384. [DOI] [PubMed] [Google Scholar]
- 5.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 7.Caballero, A., A. Esteve-Nuñez, G. J. Zylstra, and J. L. Ramos. 2005. Assimilation of nitrogen from nitrite and trinitrotoluene in Pseudomonas putida JLR11. J. Bacteriol. 187:396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cases, I., J. Pérez-Martín, and V. de Lorenzo. 1999. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the σ54-dependent Pu promoter of the TOL plasmid. J. Biol. Chem. 274:15562-15568. [DOI] [PubMed] [Google Scholar]
- 9.Collier, D. N., P. W. Hager, and P. V. Phibbs, Jr. 1996. Catabolite repression control in the Pseudomonads. Res. Microbiol. 147:551-561. [DOI] [PubMed] [Google Scholar]
- 10.del Castillo, T., and J. L. Ramos. 2007. Simultaneous catabolite repression between glucose and toluene metabolism in Pseudomonas putida is channeled through different signaling pathways. J. Bacteriol. 189:6602-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinamarca, M. A., I. Aranda-Olmedo, A. Puyet, and F. Rojo. 2003. Expression of the Pseudomonas putida OCT plasmid alkane degradation pathway is modulated by two different global signals: evidence from continuous cultures. J. Bacteriol. 185:4772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinamarca, M. A., A. Ruíz-Manzano, and F. Rojo. 2002. Inactivation of cytochrome o ubiquinol oxidase relieves catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 184:3785-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duque, E., V. García, J. de la Torre, P. Godoy, P. Bernal, and J. L. Ramos. 2004. Plasmolysis induced by toluene in a cyoB mutant of Pseudomonas putida. Environ. Microbiol. 6:1021-1031. [DOI] [PubMed] [Google Scholar]
- 14.García, B., E. R. Olivera, B. Miñambres, M. Fernández-Valverde, L. M. Cañedo, M. A. Prieto, J. L. García, M. Martínez, and J. M. Luengo. 1999. Novel biodegradable aromatic plastics from a bacterial source. Genetic and biochemical studies on a route of the phenylacetyl-CoA catabolon. J. Biol. Chem. 274:29228-29241. [DOI] [PubMed] [Google Scholar]
- 15.Görke, B., and J. Stülke. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613-624. [DOI] [PubMed] [Google Scholar]
- 16.Helling, R. B. 1994. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J. Bacteriol. 176:4664-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helling, R. B. 1998. Pathway choice in glutamate synthesis in Escherichia coli. J. Bacteriol. 180:4571-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrera, M. C., E. Duque, J. J. Rodríguez-Herva, A. Fernández-Escamilla, and J. L. Ramos. January 2010. Identification and characterization of the PhhR regulon in Pseudomonas putida. Environ. Microbiol. doi: 10.1111/j.1462-2920.02124.x. [DOI] [PubMed]
- 19.Hester, K. L., J. Lehman, F. Najar, L. Song, B. A. Roe, C. H. MacGregor, P. W. Hager, P. V. Phibbs, Jr., and J. R. Sokatch. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inada, T., K. Kimata, and H. Aiba. 1996. Mechanism responsible for glucose/lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells 1:293-301. [DOI] [PubMed] [Google Scholar]
- 21.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa, M., S. Goto, M. Hattori, K. F. Aoki-Kinoshita, M. Itoh, S. Kawashima, T. Katayama, M. Araki, and M. Hirakawa. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa, M., S. Goto, M. Furumichi, M. Tanabe, and M. Hirakawa. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 24.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 25.Lam, H., D. C. Oh, F. Cava, C. N. Takacs, J. Clardy, M. A. de Pedro, and M. K. Waldor. 2009. d-Amino acids govern stationary phase cell wall remodelling in bacteria. Science 325:1552-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. U. S. A. 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marqués, S., I. Aranda-Olmedo, and J. L. Ramos. 2006. Controlling bacterial physiology for optimal expression of gene construct. Curr. Opin. Biotechnol. 17:50-56. [DOI] [PubMed] [Google Scholar]
- 28.Massey, L. K., J. R. Sokatch, and R. S. Conrad. 1976. Branched-chain amino acid catabolism in bacteria. Bacteriol. Rev. 40:42-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matilla, M. A., M. Espinosa-Urgel, J. J. Rodríguez-Herva, J. L. Ramos, and M. I. Ramos-González. 2007. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 8:R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews, L., G. Gopinath, M. Gillespie, M. Caudy, D. Croft, B. de Bono, P. Garapati, J. Hemish, H. Hermjakob, B. Jassal, A. Kanapin, S. Lewis, S. Mahajan, B. May, E. Schmidt, I. Vastrik, G. Wu, E. Birney, L. Stein, and P. D'Eustachio. 2009. Reactome knowledgebase of biological pathways and processes. Nucleic Acids Res. 37:D619-D622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews, L., P. D'Eustachio, M. Gillespie, C. Croft, B. de Bono, G. Gopinath, B. Jassal, S. Lewis, E. Schmidt, I. Vastrik, G. Wu, E. Birney, and L. Stein. 2007. An introduction to the Reactome knowledgebase of human biological pathways and processes. NCI/Nature Pathway Interaction Database. doi: 10.1038/pid.2007.3. [DOI]
- 32.Morales, G., J. F. Linares, A. Beloso, J. P. Albar, J. L. Martínez, and F. Rojo. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186:1337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales, G., A, Ugidos, and F. Rojo. 2006. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 8:1764-1774. [DOI] [PubMed] [Google Scholar]
- 34.Moreno, R., M. Martínez-Gomariz, L. Yuste, C. Gil, and F. Rojo. 2009. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9:2910-2928. [DOI] [PubMed] [Google Scholar]
- 35.Nakada, D., and B. Magasanik. 1964. The roles of inducer and catabolite repressor in the synthesis of beta-galactosidase by Escherichia coli. J. Mol. Biol. 8:105-127. [DOI] [PubMed] [Google Scholar]
- 36.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 37.Nogales, J., A. Canales, J. Jiménez-Barbero, J. L. García, and E. Díaz. 2005. Molecular characterization of the gallate dioxygenase from Pseudomonas putida KT2440. J. Biol. Chem. 280:3582-35390. [DOI] [PubMed] [Google Scholar]
- 38.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perron, K., O. Caille, C. Rossier, C. Van Delden, J. L. Dumas, and T. Köhler. 2004. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 279:8761-8768. [DOI] [PubMed] [Google Scholar]
- 40.Petruschka, L., G. Burchhardt, C. Muller, C. Weihe, and H. Herrmann. 2001. The cyo operon of Pseudomonas putida is involved in carbon catabolite repression of phenol degradation. Mol. Genet. Genomics 266:199-206. [DOI] [PubMed] [Google Scholar]
- 41.Phillips, A. T., and L. M. Mulfinger. 1981. Cyclic adenosine 3′,5′-monophosphate levels in Pseudomonas putida and Pseudomonas aeruginosa during induction and carbon catabolite repression of histidase synthesis. J. Bacteriol. 145:1286-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piñar, G., K. Kovárová, T. Egli, and J. L. Ramos. 1998. Influence of carbon source of nitrate removal by nitrate-tolerant Klebsiella oxytoca CECT 4460 in batch and chemostat cultures. Appl. Environ. Microbiol. 64:2970-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180:3323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramos, J. L., M. M. González-Pérez, A., Caballero, and van P. van Dillewijn. 2005. Bioremediation of polynitrated aromatic compounds: plants and microbes put up a fight. Curr. Opin. Biotechnol. 16:275-281. [DOI] [PubMed] [Google Scholar]
- 46.Reva, O. N., C. Weinel, M. Weinel, K. Bohm, D. Stjepandic, J. D. Hoheisel, and B. Tümmler. 2006. Functional genomics of stress response in Pseudomonas putida KT2440. J. Bacteriol. 188:4079-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodríguez-Herva, J. J., M. I. Ramos-González, and J. L. Ramos. 1996. The Pseudomonas putida peptidoglycan-associated outer membrane lipoprotein is involved in maintenance of the integrity of the cell envelope. J. Bacteriol. 178:1699-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rojo, F., and A. Dinamarca. 2004. Catabolite repression and physiological control, p. 365-387. In J. L. Ramos (ed.), Pseudomonas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Sashidhar, B., and A. R. Podile. 2009. Transgenic expression of glucose dehydrogenase in Azotobacter vinelandii enhances mineral phosphate solubilization and growth of sorghum seedlings. Microb. Biotechnol. 2:521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sazonova, O. I, T. TI. Izmalkova, I. A. Kosheleva, and A. M. Boronin. 2008. Salicylate degradation by Pseudomonas putida strains not involving the “classical” nah2 operon. Mikrobiologia 77:798-804. [PubMed] [Google Scholar]
- 52.Segura, A., S. Rodríguez-Conde, C. Ramos, and J. L. Ramos. 2009. Bacterial responses and interactions with plants during rhizoremediation. Microb. Biotechnol. 2:452-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segura, A., P. de Wit, and G. M. Preston. 2009. Life of microbes that interact with plants. Microb. Biotechnol. 2:412-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanisich, V. A., P. M. Bennett, and N. H. Richmond. 1977. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J. Bacteriol. 129:1227-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stulke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 56.Suh, S. J., L. J. Runyen-Janecky, T. C. Maleniak, P. Hager, C. H. MacGregor, N. A. Zielinski-Mozny, P. V. Phibbs, Jr., and S. E. West. 2002. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology 148:1561-1569. [DOI] [PubMed] [Google Scholar]
- 57.Sze, C. C., and V. Shingler. 1999. The alarmone (p) ppGpp mediates physiological-responsive control at the sigma 54-dependent Po promoter. Mol. Microbiol. 31:1217-1228. [DOI] [PubMed] [Google Scholar]
- 58.Van Dillewijn, P., A. Caballero, J. A. Paz, M. M. González-Pérez, J. M. Oliva, and J. L. Ramos. 2007. Bioremediation of 2,4,6-trinitrotoluene under field conditions. Environ. Sci. Technol. 41:1378-1383. [DOI] [PubMed] [Google Scholar]
- 59.Vermeij, P., and M. A. Kertesz. 1999. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J. Bacteriol. 181:5833-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.West, S. E., A. K. Sample, and J. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, C. H., S. M. Bernard, G. L. Andersen, and W. Chen. 2009. Developing microbe-plant interactions for applications in plant-growth promotion and disease control, production of useful compounds, remediation and carbon sequestration, Microb. Biotechnol. 2:428-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.