Abstract
Inactivation of sll0861 in Synechocystis sp. strain PCC 6803 or the homologous gene alr2432 in Anabaena sp. strain PCC 7120 had no effect on the growth of these organisms at a light intensity of 30 μmol photons m−2 s−1 but reduced their growth at a light intensity of 5 or 10 μmol photons m−2 s−1. In Anabaena, inactivation of the gene also significantly reduced the rate of heterocyst differentiation under low-light conditions. The predicted products of sll0861 and alr2432 and homologs of these genes showed similarity to N-acetylmuramic acid 6-phosphate etherase (MurQ), an enzyme involved in peptidoglycan recycling, in Escherichia coli. E. coli murQ and the cyanobacterial homologs could functionally substitute for each other. We hypothesize that murQ in cyanobacteria promotes low-light adaptation through reutilization of peptidoglycan degradation products.
Cyanobacteria are procaryotes that perform oxygenic photosynthesis and have a Gram-negative cell wall structure (7). They are found in oceans, bodies of freshwater, and the soil surface and contribute significantly to global primary productivity (33). In many environments, light often is a limiting factor for their growth.
The efficiency of light harvesting and the distribution of excitation energy in photosystems are important in low-light adaptation. In Prochlorococcus marinus, high- and low-light-adapted ecotypes differ in the number of pcb genes that encode light-harvesting antenna proteins (3, 11). In Synechocystis sp. PCC 6803, rpaC, a gene required for the transition state, can promote growth in white light at an intensity of 2 μmol photons m−2 s−1 (10, 22). On the other hand, reutilization of secreted substances or degradation products may promote growth under light-limiting conditions. For example, low-light conditions can stimulate the uptake of amino acids in the cyanobacterium Planktothrix rubescens (31).
Bacteria can break down peptidoglycan (PG) and reutilize the degradation products to synthesize new PG. This process is called PG recycling. In cyanobacteria and other Gram-negative bacteria, PG forms a continuous layer completely surrounding the cell between the cytoplasmic membrane and the outer membrane (12). The net-like layer consists of glycan strands cross-linked by short peptides with GlcNAc-anhydro-N-acetylmuramic acid (anhMurNAc)-l-Ala-d-Glu-meso-diaminopimelic acid-d-Ala as the repeating unit (23). In Escherichia coli, PG is degraded to GlcNAc-anhMurNAc-peptides or GlcNAc-anhMurNAc and peptides in the periplasmic space, and the GlcNAc-anhMurNAc-peptides and GlcNAc-anhMurNAc are then imported into the cytoplasm by the permease AmpG (13). GlcNAc-anhMurNAc-peptides are processed into GlcNAc-anhMurNAc and tripeptides by AmpD (anhydro-N-acetylmuramyl-l-Ala amidase) and LdcA (LD-carboxypeptidase) in the cytoplasm and reutilized (13, 26). PG accounts for about 2% of the cell mass in Gram-negative bacteria. The reutilization of PG degradation products may promote growth under nutrient-limiting conditions. However, so far, no experimental evidence directly supports this hypothesis. For example, inactivation of ampG or other genes involved in PG recycling apparently does not affect the normal growth rate of E. coli (8, 13, 14, 27, 30), except that it results in autolysis during the stationary growth phase in an ldcA mutant (26).
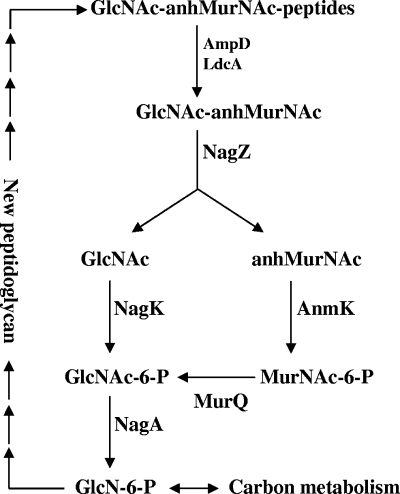
Cyanobacteria have a PG structure similar to that of Gram-negative bacteria, except for small differences, such as the thickness, degree of cross-linking, and covalent linkage of the polysaccharide (15, 16). In the present study, we found that a gene that is highly conserved in cyanobacteria has a function similar to that of murQ, a gene involved in reutilization of GlcNAc-anhMurNAc in E. coli. As shown in Fig. 1, GlcNAc-anhMurNAc is processed into GlcNAc and anhMurNAc by NagZ (β-N-acetylglucosaminidase) (8), and then GlcNAc is phosphorylated by NagK (GlcNAc kinase), producing GlcNAc-6-P (24), while anhMurNAc is phosphorylated by AnmK (anhMurNAc kinase), producing MurNAc-6-P (28), and is converted by MurQ (MurNAc-6-P etherase) into GlcNAc-6-P (14, 29). GlcNAc-6-P deacetylase (NagA) further converts GlcNAc-6-P to GlcN-6-P, which can be used in synthesis of new PG or enter carbohydrate metabolism (24). We show here that murQ and its homologs in cyanobacteria can promote growth under light-limiting conditions. Also, in a filamentous N2-fixing cyanobacterium, Anabaena sp. strain PCC 7120, the murQ homolog enhances heterocyst differentiation at a low light intensity.
FIG. 1.
Schematic diagram showing the PG recycling pathway described by Uehara et al. (29). anhMurNAC, anhydro-N-acetylmuramic acid; GlcN-6-P, glucosamine 6-phosphate; GlcNAc, N-acetylglucosamine; GlcNAc-6-P, N-acetylglucosamine 6-phosphate; MurNAC-6-P, N-acetylmuramic acid 6-phosphate.
MATERIALS AND METHODS
Strains and culture conditions.
Synechocystis sp. strain PCC 6803 was obtained from J. Zhao, Beijing University. Anabaena sp. strain PCC 7120 and Microcystis aeruginosa PCC 7806 were obtained from the Freshwater Algae Culture Collection of the Institute of Hydrobiology, Chinese Academy of Sciences.
The cyanobacterial strains were grown as previously described (37, 38) using light intensities of 5, 10, and 30 μmol photons m−2 s−1. For light-activated heterotrophic growth (LAHG), Synechocystis strains were cultured in BG11 medium with 5 mM glucose and exposed to light at an intensity of 5 μmol photons m−2 s−1 for 5 min per day but otherwise were kept in the dark. Specific growth rates (day−1) were calculated as follows: [(log ODt2−log ODt1)/log2]/ (t2 − t1), where ODt1is the turbidity (optical density at 730 nm) after t1 days and ODt2is the turbidity (optical density at 730 nm) after t2 days. For induction of heterocyst formation, Anabaena PCC 7120 grown in BG11 medium was collected, washed twice in BG110 medium (BG11 medium without NaNO3), and incubated in the same medium with different light intensities for 24, 48, and 72 h. Heterocyst frequencies were calculated by determining the number of heterocysts in 1,000 cells. The turbidity of cyanobacteria and heterocyst frequencies were calculated from the results for 3 to 6 parallel cultures.
E. coli murQ mutant strain TJ2 (MC4100 murQ::Kmr) was a kind gift from Christoph Mayer, University of Konstanz (14). Mutant TJ2 and the complemented strains were cultivated in shaking flasks at 37°C in minimal medium A [10.5 g liter−1 K2HPO4, 4.5 g liter−1 KH2PO4, 1 g liter−1 (NH4)2SO4, 0.5 g liter−1 sodium citrate, 0.1 g liter−1 Mg2SO4] supplemented with 0.2% N-acetylmuramic acid (Sigma) as described by Jaeger et al. (14).
Plasmid construction.
Molecular manipulations were performed by using standard methods. Tool enzymes were used according to the manufacturers' instructions. PCR fragments were cloned in the T-vector pMD18-T (Takara) and confirmed by sequencing. The primers used are listed in Table 1.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Derivation and/or relevant characteristicsa | Source or reference(s) |
|---|---|---|
| Synechocystis sp. strains | ||
| PCC 6803 | Wild type | J. Zhao |
| wt-1188 | Kmr, Kmr cassette integrated into the neutral platform in slr0168 | 19, 34 |
| DRHB199b | Kmr, sll0861::C.K, generated by transformation with pHB1099 | This study |
| DRHB200 | Kmr, sll0862::C.K, generated by transformation with pHB200 | This study |
| DRHB199 DRHB3054 | Kmr Spr, omega-P6803rbcL-mlla-1 integrated into the neutral platform in the genome of the sll0861::C.K mutant, generated by transformation of DRHB199 with pHB3054 | |
| DRHB199 DRHB3055 | Kmr Emr, sll0860-sll0861-C.CE2 integrated into the neutral platform in the genome of the sll0861::C.K mutant, generated by transformation of DRHB199 with pHB3055 | This study |
| DRHB199 DRHB3157 | Kmr Spr, omega-P6803rbcL-murQ integrated into the neutral platform in the genome of the sll0861::C.K mutant, generated by transformation of DRHB199 with pHB3157 | This study |
| Anabaena sp. strains | ||
| PCC 7120 | Wild type | FACHBc |
| DRHB322-2 | Nmr, alr2432::C.K, generated by homologous double crossover of pHB322-2 with Anabaena chromosome | This study |
| DRHB322-2(pHB2949) | Nmr Spr, pHB2949 carrying omega-P7120rbcL-mlla-1 introduced into the alr2432::C.K mutant | This study |
| DRHB322-2(pHB3158) | Nmr Spr, pHB3158 carrying omega-P7120rbcL-murQ introduced into the alr2432::C.K mutant | This study |
| Microcystis aeruginosa PCC 7806 | Wild type | FACHBc |
| Escherichia coli TJ2 | murQ::Kmr | 14 |
| Plasmidsd | ||
| pET21b | Apr, expression vector | Novagen |
| pHB187 | Apr, PCR fragment bearing sll0861-sll0862 (Synechocystis PCC 6803 chromosomal bp 1161737 to 1165229), generated with primers sll0861-F and sll0861-R, cloned into the EcoRI site of pRL500 | This study |
| pHB199 | Apr, C.K cloned into KpnI-cut and T4 DNA polymerase-blunted pHB187, with sll0861 disrupted | This study |
| pHB200 | Apr, C.K cloned into NheI-cut and T4 DNA polymerase-blunted pHB187, with sll0862 disrupted | This study |
| pHB313 | Apr, PCR fragment bearing alr2432 (Anabaena PCC 7120 chromosomal bp 2923983 to 2925710), generated with primers alr2432-F and alr2432-R, cloned into pMD18-T | This study |
| pHB322-1 | Kmr Apr, C.K inserted into HpaI-cut and T4 DNA polymerase-blunted pHB313, with alr2432 disrupted | This study |
| pHB322-2 | Cmr Kmr, alr2432::C.K fragment excised with SphI and SalI from pHB322-1, cloned into SphI/SalI-cut pRL271 | This study |
| pHB1347 | Kmr Spr, plasmid containing omega-P7120rbcL in pHB912 | 32 |
| pHB2739 | Apr, PCR fragment containing P6803rbcL (Synechocystis PCC 6803 chromosomal bp 2477976 to 2478410), generated with primers 6803rbcL-F and 6803rbcL-R, cloned into pMD18-T | This study |
| pHB2759 | Apr Spr, omega cassette excised with BamHI from pRL57, blunted with T4 DNA polymerase, cloned into SalI-cut and T4 DNA polymerase-blunted pHB2739 | This study |
| pHB2909a | Apr, PCR fragment containing mlla-1 (Microcystis PCC 7806 chromosomal bp 65241 to 66280), generated with primers 7806-F and 7806-R, cloned into pMD18-T, oriented against PlacZ | This study |
| pHB2909b | Apr, PCR fragment containing mlla-1 (Microcystis PCC 7806 chromosomal bp 65241 to 66280), generated with primers 7806-F and 7806-R, cloned into pMD18-T, oriented against like PlacZ | This study |
| pHB2912 | Apr Spr, omega-P6803rbcL excised with PstI/XbaI from pHB2759, blunted with T4 DNA polymerase, inserted into SalI-cut and T4 DNA polymerase-blunted pHB2909a, oriented like mlla-1 | This study |
| pHB2913 | Apr Spr, omega-P7120rbcL excised with NheI from pHB1347, blunted with T4 DNA polymerase, inserted into SalI-cut and T4 DNA polymerase-blunted pHB2909a, oriented like mlla-1 | This study |
| pHB2949 | Apr Spr Kmr, omega-P7120rbcL-mlla-1 excised with PvuII from pHB2913, inserted into EcoRI-cut and T4 DNA polymerase-blunted pRL25C | This study |
| pHB2982 | Apr, PCR fragment containing sll0861 (Synechocystis PCC 6803 chromosomal bp 1163322 to 1164363), generated with primers 6803sll0861-F and 6803sll0861-R, cloned into pMD18-T, oriented like PlacZ | This study |
| pHB3012 | Apr, PCR fragment bearing sll0860-sll0861 (Synechocystis PCC 6803 chromosomal bp 1163322 to 1165090), generated with primers 6803sll0860-F and 6803sll0861-R, cloned into pMD18-T | This study |
| pHB3032 | Apr Cmr, C.CE2 excised with SalI from pRL598, blunted with T4 DNA polymerase, inserted into BamHI-cut and T4 DNA polymerase-blunted pHB3012 | This study |
| pHB3054 | Apr Spr, omega-P6803rbcL-mlla-1 excised with PvuII from pHB2912, cloned into EcoRI-cut and T4 DNA polymerase-blunted pKW1188, replacing the Kmr fragment | This study |
| pHB3055 | Apr Cmr, sll0860-sll0861-C.CE2 excised with SacI/PstI from pHB3032, cloned into EcoRI-cut and T4 DNA polymerase-blunted pKW1188, replacing the Kmr fragment | This study |
| pHB3146 | Apr, PCR fragment bearing E. coli murQ, generated by PCR with primers murQ-F and murQ-R, cloned into pMD18-T | This study |
| pHB3147 | Apr Spr, omega-P6803rbcL excised with PstI/XbaI from pHB2759, blunted with T4 DNA polymerase, inserted into SalI-cut and T4 DNA polymerase-blunted pHB3146, oriented like murQ | This study |
| pHB3148 | Apr Spr, omega-P7120rbcL excised with NheI from pHB1347, blunted with T4 DNA polymerase, inserted into SalI-cut and T4 DNA polymerase-blunted pHB3146, oriented like murQ | This study |
| pHB3157 | Apr Spr, omega-P6803rbcL-murQ excised with PvuII from pHB3147, cloned into EcoRI-cut and T4 DNA polymerase-blunted pKW1188, replacing the Kmr fragment | This study |
| pHB3158 | Apr Spr Kmr, omega-P7120rbcL-murQ excised with PvuII from pHB3148, inserted into EcoRI-cut and T4 DNA polymerase-blunted pRL25C | This study |
| pHB3908 | Apr, fragment bearing an E. coli ribosome-binding site and partial sll0861 sequence was excised from pHB1330 with BglII/BalI and used to replace the BalI/BamHI fragment in pHB2982, resulting in a clone with sll0861 expressed based on PlacZ and E. coli ribosome-binding site (designated sll0861′) | This study |
| pKW1188 | Apr Kmr, plasmid bearing a neutral integrative platform for Synechocystis PCC 6803 | 34 |
| pMD18-T | Apr, T-cloning vector | Takara, Dalian, People's Republic of China |
| pRL25C | Kmr, pDU1-based E. coli-Anabaena shuttle vector | 36 |
| pRL57 | Spr, cloning vector with the spectinomycin resistance cassette omega | 4 |
| pRL271 | Cmr, sacB-bearing positive selection vector | 4 |
| pRL446 | Kmr, cloning vector with the C.K cassette | 9; GenBank accession no EU346690.1 |
| pRL500 | Apr, positive-selection vector | 9 |
| pRL598 | Cmr Emr, cloning vector with the C.CE2 cassette | 9 |
| Primers (5′-3′) | ||
| 6803rbcL-F | CCGATGAAGTGGTGGAGCA | |
| 6803rbcL-R | GGTCAGTCCTCCATAAACATTG | |
| 6803sll0860-F | CTAGTTCATTTCTCCACCGG | |
| 6803sll0861-F | CCCGATCATCTTTCTCCCATCC | |
| 6803sll0861-R | TGGCGAAGACAATGGGGACT | |
| 7806-F | TCTTACGAAAAGCTCAATTAAACCG | |
| 7806-R | AAGATAGCACCAGCGACGAGG | |
| alr2432-F | TCCCAAGATGATGCTGTCCGT | |
| alr2432-R | CCGACAACCGTAGGAGGTAA | |
| murQ-F | CGTAAATAGTAAGGTCACCACCG | |
| murQ-R | CGACACGGGTAAGAATGGTG | |
| sll0861-F | TTGAATTCCACTGTCCAACGACCATAGAC | |
| sll0861-R | ACGAATTCAAGATACGGAAGTAGTGCTG |
Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Km, kanamycin; Nm, neomycin; Sp, spectinomycin.
DRHB indicates a product of double homologous recombination between a pHB plasmid and the Synechocystis sp. or Anabaena sp. genome.
FACHB, Freshwater Algal Culture Collection of the Institute of Hydrobiology.
The templates used for PCRs were genomic DNA of Anabaena PCC 7120, E. coli DH5α, and Synechocystis PCC 6803; PCR clones were confirmed by sequencing.
(i) Plasmids used for gene disruption.
To inactivate sll0861 or sll0862 in Synechocystis PCC 6803, pHB187 carrying sll0861::C.K and pHB200 carrying sll0862::C.K were constructed. To inactivate alr2432 in Anabaena PCC 7120, pHB322-2 carrying alr2432::C.K and sacB was constructed. C.K is a kanamycin resistance cassette (GenBank accession no. EU346690.1) (9).
(ii) Plasmids used for complementation.
For sll0861, pHB2982 carrying PlacZ-sll0861 and pHB3908 carrying PlacZ-sll0861′ were constructed to complement E. coli TJ2, and pHB3055 carrying sll0860-sll0861-C.CE2 was constructed to complement the Synechocystis sll0861::C.K mutant. sll0861′ was sll0861 fused with an E. coli ribosome-binding site. C.CE2 is a chloramphenicol-erythromycin resistance cassette (9).
For mlla-1, the sll0861 homolog in Microcystis PCC 7806 (NCBI GenBank accession no. AM778954.1), pHB2909b carrying PlacZ-mlla-1 was constructed to complement E. coli TJ2, pHB2949 carrying omega-P7120rbcL-mlla-1 was constructed to complement the Anabaena alr2432::C.K mutant, and pHB3054 carrying omega-P6803rbcL-mlla-1 was constructed to complement the Synechocystis sll0861::C.K mutant.
For E. coli murQ, pHB3157 carrying omega-P6803rbcL-murQ was constructed to complement the Synechocystis sll0861::C.K mutant, and pHB3158 carrying omega-P7120rbcL-murQ was constructed to complement the Anabaena alr2432::C.K mutant.
Details concerning construction of these plasmids are shown in Table 1.
Generation of cyanobacterial mutants.
Transformation of Synechocystis PCC 6803 was performed as described by Williams (34). Conjugative transfer of plasmids into Anabaena PCC 7120 was performed as described by Elhai and Wolk (9). Synechocystis mutants were generated by transformation with corresponding plasmids. Anabaena mutants were generated using a two-step protocol involving sacB-based positive selection of double-crossover mutants (6). Complete segregation of mutants was confirmed by PCR. Details concerning mutant generation are shown in Table 1.
Western blot detection.
Synechocystis PCC 6803 grown with different light intensities was collected by centrifugation at 6,000 rpm, resuspended in 40 mM Tris-Cl (pH 8.0) with 1 mM phenylmethylsulfonyl fluoride (PMSF), and ruptured by ultrasonication on ice. The cell debris and unbroken cells were removed by centrifugation at 6,000 rpm and 4°C for 10 min. The supernatant was then centrifuged at 30,000 rpm at 4°C for 30 min to separate membrane proteins from soluble proteins. Equal amounts of membrane and soluble proteins were loaded, separated by 12% SDS-PAGE, transferred to nitrocellulose filters (Millipore), detected with anti-Sll0861 rabbit antiserum, and visualized with goat anti-rabbit alkaline phosphatase antibody (Invitrogen) with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Amresco, United States) as the substrates. SDS-PAGE and Western blotting were performed using standard methods.
RESULTS AND DISCUSSION
Role of sll0861 in low-light adaptation in Synechocystis PCC 6803.
Synechocystis PCC 6803 can grow heterotrophically in the dark with daily brief exposure to weak light (1). This phenomenon is called light-activated heterotrophic growth (LAHG). In an effort to identify genes required for LAHG, we obtained Synechocystis mutants whose growth rates were significantly reduced under such conditions (18). In one of the mutants, sll0861 was inactivated. We then generated mutants with a kanamycin resistance cassette (C.K) inserted into sll0861 or the downstream gene sll0862 (Table 1). The sll0861::C.K mutant showed greatly reduced growth under LAHG conditions, while the sll0862::C.K mutant had the wild-type phenotype (Table 2). The LAHG phenotype of the sll0861 mutant was fully complemented by a DNA fragment containing the sll0860-sll0861 sequence (Table 2). Because sll0861 may be cotranscribed with sll0860, sll0860 and the upstream sequence were included to promote expression of sll0861 in the complementation experiment.
TABLE 2.
Specific growth rates of Synechocystis PCC 6803 and mutants of this strain cultured under LAHG conditions
| Strain | Specific growth rate (day−1)a |
|---|---|
| Wild type | 0.435 ± 0.014 |
| sll0861::C.K mutant | 0.166 ± 0.007 |
| sll0862::C.K mutant | 0.441 ± 0.024 |
| sll0861::C.K mutant complemented with: | |
| sll0860-sll0861 | 0.458 ± 0.023 |
| mlla-1 | 0.447 ± 0.028 |
| E. coli murQ | 0.415 ± 0.006 |
The values are means ± standard deviations and were calculated using the results for three parallel cultures.
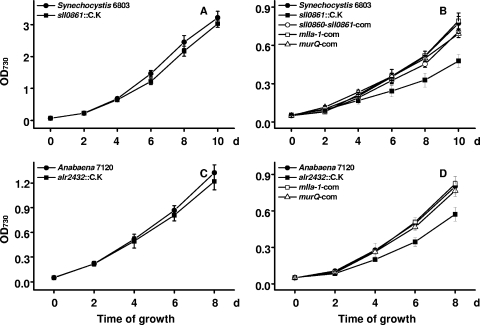
We wondered if sll0861 affects photoautotrophic growth under low-light conditions. The sll0861::C.K mutant had a growth rate almost identical to that of the wild type at a light intensity of 30 μmol photons m−2 s−1 (Fig. 2A). However, when cultured with 5 μmol photons m−2 s−1, the mutant showed slower growth than the wild type (Fig. 2B). When sll0860-sll0861 was added to the mutant, the level of growth was restored to the wild-type level (Fig. 2B). These results suggest that sll0861 probably plays a role in low-light adaptation in Synechocystis PCC 6803.
FIG. 2.
Autotrophic growth of the Synechocystis PCC 6803 (A and B) and Anabaena PCC 7120 (C and D) strains with light intensities of 30 μmol photons m−2 s−1 (A and C), 5 μmol photons m−2 s−1 (B), and 10 μmol photons m−2 s−1 (D). Cells were cultured in BG11 medium at 30°C, and the growth was determined by monitoring the optical density at 730 nm (OD730). sll0860-sll0861-com, complemented with sll0860-sll0861; mlla-1-com, complemented with omega-P6803rbcL-mlla-1 or omega-P7120rbcL-mlla-1; murQ-com, complemented with omega-P6803rbcL-murQ or omega-P7120rbcL-murQ.
Using Western blot analysis, we found similar levels of Sll0861 in Synechocystis PCC 6803 cultures exposed to light at intensities of 30, 5, and 0 μmol photons m−2 s−1 (see Fig. S1 in the supplemental material). Unlike Sll0886 (21), another protein involved in LAHG, Sll0861 was not associated with membranes.
sll0861 homologs in Anabaena and Microcystis.
Homologs of sll0861 have been found in most cyanobacterial species (see Fig. S2 in the supplemental material). We used the homologous genes of the filamentous N2-fixing cyanobacterium Anabaena PCC 7120 and the bloom-forming cyanobacterium Microcystis PCC 7806 to investigate the roles of these genes in low-light adaptation. Unlike Synechocystis PCC 6803, Anabaena PCC 7120 and Microcystis PCC 7806 are not able to grow heterotrophically. Anabaena PCC 7120 can produce specialized cells, which are called heterocysts, upon nitrogen stepdown to fix dinitrogen (35).
We inactivated the homologous alr2432 gene in Anabaena PCC 7120 by inserting the C.K cassette. The alr2432::C.K mutant grew like the wild type when it was cultured with 30 μmol photons m−2 s−1 (Fig. 2C), but with 10 μmol photons m−2 s− its growth was slower than that of the wild type (Fig. 2D). Because illumination is required for initiation of heterocyst differentiation in Anabaena (5), we used induction of heterocyst differentiation as an alternative criterion to evaluate the role of alr2432 in low-light adaptation. With 5 μmol photons m−2 s−1, heterocyst differentiation was significantly slower in the mutant (Table 3). Apparently, alr2432 plays a role in low-light adaptation similar to that of sll0861.
TABLE 3.
Heterocyst frequencies of Anabaena PCC 7120 and mutants of this strain with 5 μmol photons m−2 s−1
| Time after nitrogen stepdown (h) | Heterocyst frequency (‰)a |
|||
|---|---|---|---|---|
| Wild type | alr2432::C.K mutant |
alr2432::C.K mutant complemented with: |
||
| mlla-1 | E. coli murQ | |||
| 24 | 47.7 ± 11.3 | 6.0 ± 0.7 | 54.3 ± 2.8 | 35.9 ± 6.2 |
| 48 | 65.2 ± 10.0 | 25.9 ± 6.7 | 69.0 ± 4.8 | 52.1 ± 5.2 |
| 72 | 74.5 ± 15.9 | 46.0 ± 17.2 | 74.0 ± 6.4 | 62.2 ± 4.9 |
The heterocyst frequency was calculated by determining the number of heterocysts in 1,000 cells. The values (means ± standard deviations) were calculated by using the results for 6 cultures (2 × 3) of the wild-type or alr2432::C.K strain or for 3 cultures of each complemented strain.
We attempted to inactivate the homologous gene in Microcystis PCC 7806 but obtained no transformant. We then used the homologous gene from Microcystis PCC 7806, mlla-1 (NCBI GenBank accession no. AM778954.1), to complement the Synechocystis PCC 6803 sll0861::C.K and Anabaena PCC 7120 alr2432::C.K mutants. mlla-1 was expressed from PrbcL (the promoter of the ribulose 1,5-bisphosphate carboxylase/oxygenase [Rubisco] large subunit-encoding gene) of Synechocystis PCC 6803 or Anabaena PCC 7120. As shown in Fig. 2B, expression of mlla-1 in the Synechocystis mutant fully restored growth with 5 μmol photons m−2 s−1. Also, the levels of growth and heterocyst differentiation of the Anabaena mutant under low-light conditions were restored to the wild-type levels by complementation with mlla-1 (Fig. 2D and Table 3). Based on these results, we concluded that sll0861 in Synechocystis and homologs of this gene in Anabaena and Microcystis should have the same function in low-light adaptation.
sll0861 and homologs of this gene function like murQ (N-acetylmuramic acid 6-phosphate etherase) genes.
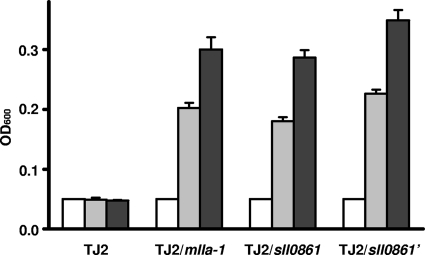
sll0861 is predicted to encode a sugar isomerase (SIS) domain (2) protein with similarity to the glucokinase regulatory protein (GKRP) in humans (E value, 4 × 10−10). Also, this protein is similar to the N-acetylmuramic acid 6-phosphate etherase (MurQ) in E. coli (E value, 3 × 10−65). We first compared the glucose kinase activities in the mutant and wild type and found no difference. Thus, we tested if sll0861 and its homologs can complement E. coli murQ::Kmr mutant TJ2. Unlike the wild type, the TJ2 mutant could not grow on N-acetylmuramic acid (MurNAc) as the sole source of carbon and energy (14). We expressed sll0861 and mlla-1 in E. coli TJ2 from the lacZ promoter. In one of the constructs, pHB3908 (Table 1), a typical E. coli ribosome-binding site (RBS) was used to promote translation of sll0861 (designated sll0861′). sll0861, sll0861′, and mlla-1 all enabled E. coli TJ2 to grow in liquid medium with MurNAc as the sole carbon source (Fig. 3). The use of an E. coli ribosome-binding site did not have an apparent effect on growth. Tests on plates also clearly showed that there was complementation of TJ2 by the cyanobacterial genes (data not shown).
FIG. 3.
Growth of E. coli murQ mutant TJ2 and this mutant containing pHB2982 (TJ2/sll0861), pHB2993 (TJ2/mlla-1), or pHB3908 (TJ2/sll0861′) in minimal medium A supplemented with MurNAc. Growth in liquid medium at 37°C was monitored by determining the turbidity (optical density at 600 nm [OD600]) at zero time (open bars), 48 h (light gray bars), and 96 h (dark gray bars).
On the other hand, we complemented the cyanobacterial mutants with murQ from E. coli. murQ was expressed from the rbcL promoter of Synechocystis PCC 6803 and Anabaena PCC 7120 in the sll0861::C.K and alr2432::C.K mutants, respectively. The E. coli murQ gene restored the autotrophic growth of both mutants to wild-type levels (Fig. 2B and D) under low-light conditions. Also, it restored heterocyst differentiation (Table 3) in the Anabaena PCC 7120 alr2432::C.K mutant at an intensity of 5 μmol photons m−2 s−1 and LAHG (Table 2) of Synechocystis PCC 6803 sll0861::C.K. From the results described above, we concluded that sll0861 and its homologs have a function similar to that of murQ in E. coli and that the encoded N-acetylmuramic acid 6-phosphate etherase is involved in PG recycling in cyanobacteria.
Predicted MurQ and other enzymes involved in PG recycling in Prochlorococcus ecotypes.
Kettler et al. (17) classified 12 strains of P. marinus in six high-light-adapted ecotypes (MED4, MIT 9215, MIT 9301, MIT 9312, MIT 9515, and AS9601) and six low-light-adapted ecotypes (MIT 9303, MIT 9313, MIT 9211, SS120, NATL2A, and NATL1A). Homologs of murQ were found in the six low-light-adapted ecotypes but in none of the high-light-adapted ecotypes (Table 4).
TABLE 4.
Predicted genes involved in recycling of PG amino sugars in light-adapted ecotypes of P. marinusa
| Strain | Ecotypeb | Homolog ofc: |
||||
|---|---|---|---|---|---|---|
| nagZ | nagK | anmK | murQ | nagA | ||
| AS9601 | HL | − | − | − | − | − |
| MED4 | HL | − | − | − | − | − |
| MIT9215 | HL | − | − | − | − | − |
| MIT9301 | HL | − | − | − | − | − |
| MIT9312 | HL | − | − | − | − | − |
| MIT9515 | HL | − | − | − | − | − |
| MIT9211 | LL | + | − | + | + | + |
| MIT9303 | LL | + | + | + | + | + |
| MIT9313 | LL | + | + | + | + | + |
| NATL1A | LL | + | − | + | + | + |
| NATL2A | LL | + | − | + | + | + |
| SS120 | LL | + | − | + | + | + |
The genome information was obtained from the study of Kettler et al. (17).
HL, high-light-adapted ecotype; LL, low-light-adapted ecotype.
+, yes; −, no.
Consistently, there are no homologs of nagZ, nagK, anmK, and nagA, the other four genes involved in reutilization of PG amino sugars, in the six high-light-adapted ecotypes (Table 4). There are homologs of nagZ, anmK, and nagA in all six low-light-adapted ecotypes, while there are nagK homologs only in two of the low-light-adapted ecotypes, MIT 9303 and MIT 9313 (Table 4). The other four low-light-adapted ecotypes may reutilize MurNAc but not GlcNAc.
Hypothesis that MurQ in cyanobacteria may promote low-light adaptation through peptidoglycan recycling.
Based on our experimental investigation of murQ from three cyanobacterial species and the bioinformatics analysis of light-adapted ecotypes of P. marinus, we propose that MurQ promotes low-light adaptation in cyanobacteria. However, we showed that the sll0861::C.K mutant of Synechocystis PCC 6803 did not differ in oxygen evolution or the transition state from the wild type (unpublished data); therefore, the role of murQ in low-light adaptation is different from that of rpaC (10, 22).
We hypothesize that the effect of MurQ on low-light adaptation is based on its role in PG recycling. PG is the major material in the Gram-negative cell wall, and it accounts for about 2% of the cell mass. The reutilization of PG degradation products should reduce the loss of fixed carbon in cyanobacteria. When there is sufficient light, the contribution of PG recycling to the increase in biomass can be neglected. However, at a low light intensity, this energy-saving strategy could have an effect on promoting cell propagation. In Anabaena, illumination in first several hours after nitrogen stepdown is required for initiation of heterocyst differentiation (5). No molecular mechanism for the role of illumination in heterocyst differentiation has been reported yet, but it is conceivable that photosynthesis can promote cell division and accumulation of 2-oxoglutarate, both of which are required for initiation of heterocyst differentiation (20, 25). Under light-limiting conditions, PG recycling may affect these processes by increasing the pool of carbon metabolites. Alternatively, MurQ may exert its effects on growth or cell differentiation by reducing the accumulation of MurNAc-6-P or upstream metabolites (Fig. 1). In the future, inactivation of other genes involved in PG recycling and analyses of metabolites in Synechocystis PCC 6803 and Anabaena PCC 7120 should provide further evidence to clarify these possibilities.
Supplementary Material
Acknowledgments
We thank Christoph Mayer, Fachbereich Biologie, University of Konstanz, for kindly providing E. coli strain TJ2 (MC4100 murQ::Kmr).
This study was supported by the National Natural Science Foundation of China (grant 30825003), the State Key Basic Research Development Program of China (grant 2008CB418001), and Key Project KZCX1-YW-14-1 of the Knowledge Innovation Program of the Chinese Academy of Sciences.
Footnotes
Published ahead of print on 5 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, S., and L. McIntosh. 1991. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803, a blue-light-requiring process. J. Bacteriol. 173:2761-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A. 1999. The SIS domain: a phosphosugar-binding domain. Trends Biochem. Sci. 24:94-95. [DOI] [PubMed] [Google Scholar]
- 3.Bibby, T., I. Mary, J. Nield, F. Partensky, and J. Barber. 2003. Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424:1051-1054. [DOI] [PubMed] [Google Scholar]
- 4.Black, T., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 5.Bradley, S., and N. G. Carr. 1976. Heterocyst development in Anabaena cylindrica: the necessity of light as the initial trigger and sequential stages of commitment. J. Gen. Microbiol. 101:291-297. [DOI] [PubMed] [Google Scholar]
- 6.Cai, Y., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castenholz, R. W. 2001. Phylum BX. Cyanobacteria. Oxygenic photosynthetic bacteria, p. 474-487. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, NY. [Google Scholar]
- 8.Cheng, Q., H. Li, K. Merdek, and J. Park. 2000. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 182:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 10.Emlyn-Jones, D., M. K. Ashby, and C. W. Mullineaux. 1999. A gene required for the regulation of photosynthetic light harvesting in the cyanobacterium Synechocystis 6803. Mol. Microbiol. 33:1050-1058. [DOI] [PubMed] [Google Scholar]
- 11.Garczarek, L., W. R. Hess, J. Holtzendorff, G. W. van der Staay, and F. Partensky. 2000. Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote. Proc. Natl. Acad. Sci. U. S. A. 97:4098-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoiczyk, E., and A. Hansel. 2000. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J. Bacteriol. 182:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs, C., L. Huang, E. Bartowsky, S. Normark, and J. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaeger, T., M. Arsic, and C. Mayer. 2005. Scission of the lactyl ether bond of N-acetylmuramic acid by Escherichia coli “etherase.” J. Biol. Chem. 280:30100-30106. [DOI] [PubMed] [Google Scholar]
- 15.Jürgens, U. J., and J. Weckesser. 1986. Polysaccharide covalently linked to the peptidoglycan of the cyanobacterium Synechocystis sp. strain PCC 6714. J. Bacteriol. 168:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jürgens, U. J., G. Drews, and J. Weckesser. 1983. Primary structure of the peptidoglycan from the unicellular cyanobacterium Synechocystis sp. strain PCC 6714. J. Bacteriol. 154:471-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kettler, G., A. Martiny, K. Huang, J. Zucker, M. Coleman, S. Rodrigue, F. Chen, A. Lapidus, S. Ferriera, J. Johnson, C. Steglich, G. Church, P. Richardson, and S. Chisholm. 2007. Patterns and implications of gene gain and loss in the evolution of. Prochlorococcus. PLoS Genet. 3:2515-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong, R., X. Xu, and Z. Hu. 2003. A TPR-family membrane protein gene is required for light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 219:75-79. [DOI] [PubMed] [Google Scholar]
- 19.Kunert, A., M. Hagemann, and N. Erdmann. 2000. Construction of promoter probe vectors for Synechocystis sp. PCC 6803 using the light-emitting reporter systems Gfp and LuxAB. J. Microbiol. Methods 41:185-194. [DOI] [PubMed] [Google Scholar]
- 20.Laurent, S., H. Chen, S. Bédu, F. Ziarelli, L. Peng, and C.-C. Zhang. 2005. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. U. S. A. 102:9907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, Z., X. Xu, and R. Kong. 2006. Studies on sll0886, a gene essential to light-activated heterotrophic growth in Synechocystis sp. PCC 6803. Acta Hydrobiol. Sin. 30:375-379. [Google Scholar]
- 22.Mullineaux, C., and D. Emlyn-Jones. 2005. State transitions: an example of acclimation to low-light stress. J. Exp. Bot. 56:389-393. [DOI] [PubMed] [Google Scholar]
- 23.Park, J. T., and T. Uehara. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plumbridge, J. A. 1991. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol. Microbiol. 5:2053-2062. [DOI] [PubMed] [Google Scholar]
- 25.Sakr, S., R. Jeanjean, C.-C. Zhang, and T. Arcondeguy. 2006. Inhibition of cell division suppresses heterocyst development in Anabaena sp. strain PCC 7120. J. Bacteriol. 188:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Templin, M., A. Ursinus, and J. Höltje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 18:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uehara, T., and J. Park. 2004. The N-acetyl-d-glucosamine kinase of Escherichia coli and its role in murein recycling. J. Bacteriol. 186:7273-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uehara, T., K. Suefuji, N. Valbuena, B. Meehan, M. Donegan, and J. Park. 2005. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 187:3643-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uehara, T., K. Suefuji, T. Jaeger, C. Mayer, and J. T. Park. 2006. MurQ etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. J. Bacteriol. 188:1660-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vötsch, W., and M. F. Templin. 2000. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J. Biol. Chem. 50:39032-39038. [DOI] [PubMed] [Google Scholar]
- 31.Walsby, A. E., and F. Jüttner. 2006. The uptake of amino acids by the cyanobacterium Planktothrix rubescens is stimulated by light at low irradiances. FEMS Microbiol. Ecol. 58:14-22. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Y., and X. Xu. 2005. Regulation by hetC of genes required for heterocyst differentiation and cell division in Anabaena sp. J. Bacteriol. 187:8489-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitton, B., and M. Potts. 2000. Introduction to the cyanobacteria, p. 1-11. In B. A. Whitton and M. Potts (ed.), Ecology of cyanobacteria: their diversity in time and space. Kluwer Academic, Dordrecht, Netherlands.
- 34.Williams, J. G. K. 1988. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167:766-778. [Google Scholar]
- 35.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic, Dordrecht, Netherlands.
- 36.Wolk, C. P., Y. Cai, L. Cardemil, E. Flores, B. Hohn, M. Murry, G. Schmetterer, B. Schrautemeier, and R. Wilson. 1988. Isolation and complementation of mutants of Anabaena sp. strain PCC 7120 unable to grow aerobically on dinitrogen. J. Bacteriol. 170:1239-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin, C., W. Li, Y. Du, R. Kong, and X. Xu. 2007. Identification of a gene, ccr-1 (sll1242), required for chill-light tolerance and growth at 15°C in Synechocystis sp. PCC 6803. Microbiology 153:1261-1267. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, W., Y. Du, I. Khudyakov, Q. Fan, H. Gao, D. Ning, C. P. Wolk, and X. Xu. 2007. A gene cluster that regulates both heterocyst differentiation and pattern formation in Anabaena sp. strain PCC 7120. Mol. Microbiol. 66:1429-1443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.