Abstract
The environment encountered by Mycobacterium tuberculosis during infection is genotoxic. Most bacteria tolerate DNA damage by engaging specialized DNA polymerases that catalyze translesion synthesis (TLS) across sites of damage. M. tuberculosis possesses two putative members of the DinB class of Y-family DNA polymerases, DinB1 (Rv1537) and DinB2 (Rv3056); however, their role in damage tolerance, mutagenesis, and survival is unknown. Here, both dinB1 and dinB2 are shown to be expressed in vitro in a growth phase-dependent manner, with dinB2 levels 12- to 40-fold higher than those of dinB1. Yeast two-hybrid analyses revealed that DinB1, but not DinB2, interacts with the β-clamp, consistent with its canonical C-terminal β-binding motif. However, knockout of dinB1, dinB2, or both had no effect on the susceptibility of M. tuberculosis to compounds that form N2-dG adducts and alkylating agents. Similarly, deletion of these genes individually or in combination did not affect the rate of spontaneous mutation to rifampin resistance or the spectrum of resistance-conferring rpoB mutations and had no impact on growth or survival in human or mouse macrophages or in mice. Moreover, neither gene conferred a mutator phenotype when expressed ectopically in Mycobacterium smegmatis. The lack of the effect of altering the complements or expression levels of dinB1 and/or dinB2 under conditions predicted to be phenotypically revealing suggests that the DinB homologs from M. tuberculosis do not behave like their counterparts from other organisms.
The emergence and global spread of multi- and extensively drug-resistant strains of Mycobacterium tuberculosis have further complicated the already daunting challenge of controlling tuberculosis (TB) (15). The mechanisms that underlie the evolution of drug resistance in M. tuberculosis by chromosomal mutagenesis and their association with the conditions that tubercle bacilli encounter during the course of infection are poorly understood (6). It has been postulated that hypoxia, low pH, nutrient deprivation, and nitrosative and oxidative stress impose environmental and host immune-mediated DNA-damaging insults on infecting bacilli (64). In addition, the observed importance of excision repair pathways for the growth and survival of M. tuberculosis in murine models of infection (13, 55) and the upregulation of M. tuberculosis genes involved in DNA repair and modification in pulmonary TB in humans provide compelling evidence that the in vivo environment is DNA damaging (51).
Damage tolerance constitutes an integral component of an organism's response to genotoxic stress, preventing collapse of the replication fork at persisting, replication-blocking lesions through the engagement of specialized DNA polymerases that are able to catalyze translesion synthesis (TLS) across the sites of damage (19, 21, 60). Most TLS polymerases belong to the Y family, which comprises a wide range of structurally related proteins present in bacteria, archaea, and eukaryotes (44). Of these, the DinB subfamily of Y family polymerases, whose founder member is Escherichia coli Pol IV (63), is conserved among all domains of life (44). The association of Y family polymerases with inducible mutagenesis has implicated these enzymes in the adaptation of bacteria to environmental stress (17, 20, 39, 54, 58, 59, 66). Their key properties are exemplified in E. coli Pol IV: the polymerase catalyzes efficient and accurate TLS across certain N2-dG adducts (27, 28, 34, 40, 45, 67) and has been implicated in the tolerance of alkylation damage (4); furthermore, overexpression of Pol IV significantly increases mutation rates in E. coli (reviewed in references 21 and 26), and dinB is the only SOS-regulated gene required at induced levels for stress-induced mutagenesis in this organism (20). Furthermore, overproduction of E. coli Pol IV inhibits replication fork progression through replacement of the replicative polymerase to form an alternate replisome in which Pol IV modulates the rate of unwinding of the DnaB helicase (25) and also reduces colony-forming ability (61).
The M. tuberculosis genome encodes two Y family polymerase homologs belonging to the DinB subfamily, designated herein as DinB1 (DinX, encoded by Rv1537) and DinB2 (DinP, encoded by Rv3056), as well as a third, distantly related homolog encoded by Rv3394c (see Fig. S1 in the supplemental material) (9). On the basis of sequence similarity with their counterparts from E. coli (63) and Pseudomonas aeruginosa (54), including the complete conservation of key acidic residues essential for catalysis, DinB1 and DinB2 may be functional DNA polymerases (see Fig. S1). In contrast, Rv3394c lacks these residues and as such is unlikely to have polymerase activity (see Fig. S1). Unlike most Y family polymerase-encoding genes investigated with other bacteria (17, 26, 54, 58), dinB1 and dinB2 expression in M. tuberculosis is not dependent on RecA, the SOS response, or the presence of DNA damage (5, 7, 52). That these genes are regulated by other mechanisms and so may serve distinct roles in DNA metabolism in M. tuberculosis is suggested by the observation that dinB1 is differentially expressed in pulmonary TB (51) and is a member of the SigH regulon (30), whereas expression of dinB2 is induced following exposure to novobiocin (5).
In this study, we adopted a genetic approach to investigate the function of dinB1 and dinB2 in M. tuberculosis. Mutants with altered complements or expression levels of dinB1 and/or dinB2 were analyzed in vitro and in vivo under conditions predicted to be phenotypically revealing based on DinB function established with other model organisms. The lack of discernible phenotypes in any of the assays employed suggests that the DinB homologs from M. tuberculosis do not behave like their counterparts from other organisms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains, plasmids, and oligonucleotides used in this study are described in Tables S1 and S2 in the supplemental material. M. tuberculosis strains were grown in Middlebrook 7H9 media (Difco) supplemented with 0.2% glycerol, Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment (Merck), and 0.05% Tween 80 or on Middlebrook 7H10 media supplemented with 0.5% glycerol and Middlebrook OADC enrichment. Mycobacterium smegmatis strains were grown in 7H9 media supplemented with 0.2% glycerol, 0.085% NaCl, 0.2% glucose, and 0.05% Tween 80 or on 7H10 media containing 0.5% glycerol, 0.085% NaCl, and 0.2% glucose. Hygromycin (Hyg) and kanamycin (Km) were used at final concentrations of 50 and 25 μg/ml, and histidine was used at 100 μg/ml.
Yeast two-hybrid analysis.
Interaction with the replicative processivity factor (the dnaN-encoded β-clamp) was assessed using the Clontech Matchmaker yeast two-hybrid (Y2H) system. The open reading frames for dinB1, dinB2, dnaE1, dnaE2, and dnaN were PCR amplified and cloned in pGADT7 and pGBKT7 to produce GAL4 DNA activation domain (AD) and binding domain (BD) fusions, respectively (see Table S2 in the supplemental material), which were used in interaction assays according to the manufacturer's instructions.
Construction of mutant strains.
The dinB1::hyg and ΔdinB2 mutants of M. tuberculosis H37Rv were prepared by two-step allelic exchange (47) using the suicide plasmids pDINB1KO and pDINB2KO (see Table S1 in the supplemental material). The dinB1::hyg ΔdinB2 double mutant was generated from dinB1::hyg using pDINB2KO. Genetically complemented derivatives of the single mutants were constructed by integration of pMDINB1 and pMDINB2 at the attB site in the dinB1::hyg and ΔdinB2 mutant strains, respectively. To construct a suicide vector for introducing a frameshift mutation at codon 205 in M. smegmatis hisD, a 1,132-bp PstI fragment was amplified using primers HISF and HISR and cloned in pGEM3Z(+)f. The frameshift mutation was introduced by the Megaprimer method (57) using the mutagenic oligonucleotide HISM to form pGHISD5T, which was used to produce pHISD5T (see Tables S1 and S2 in the supplemental material).
Gene expression analysis.
Bacteria were grown in 7H9 media, and aliquots sampled during different phases of growth. RNA was isolated as described previously (16, 42), and real-time, quantitative reverse transcription-PCR (qRT-PCR) carried out with the primer pairs detailed in Table S2 in the supplemental material, using the LightCycler FastStart DNA Master SYBR green I kit in the Roche LightCycler and standard curves based on 10-fold serial dilutions of genomic DNA (29). Primers for detection of dinB1 transcript flanked the site used for insertional inactivation with the hyg marker. Amplification conditions were thus adjusted to ensure that only wild-type dinB1 transcript was detected. Absolute numbers of transcripts were normalized to the number of sigA transcripts in the same sample.
Sensitivity testing of M. tuberculosis strains.
MICs were determined by broth microdilution (14). Sensitivity testing was also performed by spotting 5- or 10-μl aliquots of 3- or 10-fold serial dilutions, respectively, of parallel cultures of the wild-type and mutant strains, grown to an optical density at 600 nm (OD600) of ∼0.5 to 0.7, on Middlebrook 7H10 agar plates supplemented with compound and scoring growth after 2 or 3 weeks.
Mutagenesis assays.
Rates of spontaneous mutation of M. tuberculosis to rifampin (Rif) resistance were determined by Luria-Delbrück fluctuation analysis (48, 53) using a modification of the method described for use with M. smegmatis (36). For each strain, 36 3-ml cultures containing an initial inoculum of ∼1 or 2 × 103 CFU/ml were set up in 6-well tissue culture plates (Greiner Bio-One) which were incubated in a humidified incubator at 37°C for 3 weeks with daily agitation. Aliquots from 6 cultures were serially diluted and plated on solid media to enumerate the total colony count (nt). Each of the remaining 30 cultures was spread in its entirety onto a 7H10 agar plate containing Rif at 2 μg/ml. For analysis of mutational spectra, a representative Rif-resistant colony was picked from each culture and a 449-bp fragment from the rpoB gene, spanning the rifampin resistance-determining region (RRDR), was amplified using the primers RpoBF2 and RpoBR2, and the RRDR was then sequenced using an internal primer, RpoBS (see Table S2 in the supplemental material). The rates of spontaneous mutation of wild-type M. smegmatis to Rif resistance or reversion of the hisD auxotrophic mutant were determined by fluctuation analysis (36). For reversion assays, 2-ml cultures of the auxotrophic mutant HISD5T, carrying pOLYGaa, pGaaDinB1, or pGaaDinB2 seeded at 102 to 103 CFU/ml, were grown in media supplemented with histidine and Hyg to stationary phase (5 to 7 days) before plating on histidine-free 7H10 agar to determine the number of revertants per culture. nt values were determined from three parallel cultures plated on histidine-supplemented media, and mutation rates determined as previously described (36).
M. tuberculosis growth in macrophages.
Human peripheral blood-derived monocytes (PBMCs) and peritoneal macrophages from 10- to 12-week old female B6D2F1 mice were isolated as described previously (18, 37). Infections were carried out using a multiplicity of infection (MOI) of 1 bacillus per 2 monocytes, and bacterial CFU assessed daily for 4 days, as described previously (37, 49). Where indicated, human PBMCs and murine macrophages were activated by the addition of lipopolysaccharide (LPS) or gamma interferon (IFN-γ) to a final concentration of 1 μg/ml or 100 U/ml, respectively, 2 h after infection.
Aerosol infection of mice.
Eight- to 10-week-old female B6D2F1 mice from Charles River Laboratories (Wilmington, MA) were infected with M. tuberculosis strains through the respiratory route (38). Approximately 200 to 1,000 organisms were implanted in the lungs of each mouse, as confirmed by plating lung homogenates 3 h after infection. Bacterial loads (numbers of CFU) in lungs and spleens of infected mice were assessed over a period of 77 to 350 days.
Statistics.
The independent Student t test or paired t test was applied to compare means from two different groups. A value of P of <0.05 was considered significant.
RESULTS
dinB1 and dinB2 are expressed in M. tuberculosis.
Transcript levels of dinB1 and dinB2 were determined by qRT-PCR during logarithmic- and stationary-phase growth of M. tuberculosis in vitro (Table 1). For comparison, genes encoding the replicative polymerase DnaE1 and the SOS-inducible polymerase DnaE2 were included in the analysis. Transcript was detected for all genes assessed, albeit at levels that differed significantly between genes and as a function of growth phase. Expression of dnaE1 was highest in early logarithmic phase and declined markedly in stationary phase, whereas dinB1 and dinB2 showed little change between phases. Notably, dinB2 was expressed at a level 12- to 40-fold higher than that of dinB1 (Table 1).
TABLE 1.
Normalized levels of polymerase gene transcripts in M. tuberculosis H37Rv during logarithmic- and stationary-phase growth in liquid culture
| OD600 | Expression level of transcript relative to sigA (×103)a |
|||
|---|---|---|---|---|
| dnaE1 | dnaE2 | dinB1 | dinB2 | |
| 0.3 | 4.0 ± 1.3 | 0.29 ± 0.14 | 0.8 ± 0.4 | 9.8 ± 1.7 |
| 0.7 | 1.3 ± 0.6 | 0.04 ± 0.01 | 0.2 ± 0.2 | 8.9 ± 1.2 |
| 1.5 | 0.8 ± 0.2 | 0.01 ± 0.007 | 0.3 ± 0.1 | 7.9 ± 0.6 |
According to this representation, a value of 10 corresponds to 1% of the level observed for sigA.
DinB1 but not DinB2 interacts with the mycobacterial β-clamp.
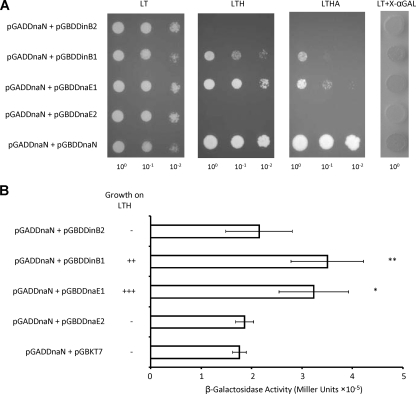
DNA polymerases that bind the β-clamp associate with this protein via a short pentapeptide motif with the consensus sequence QL[SD]LF (11). In other organisms, interactions of Y family polymerases with the β-clamp play a central role in regulating their activity and determining access to the DNA (1-3, 8, 17, 22, 28, 35, 62, 63, 65), suggesting that an analogous situation may apply in mycobacteria. A consensus β-binding motif is readily identified in M. tuberculosis DinB1 (QESLF), but not DinB2, which lacks the C-terminal region in which this motif is located in other homologs (see Fig. S1 in the supplemental material). To confirm the assignment, the β-binding potential of DinB1 and DinB2 was assessed in Y2H analyses measuring interaction by growth on selective media (Fig. 1A) and LacZ activity (Fig. 1B). As controls, we tested the ability of DnaN to interact with itself; with the replicative polymerase DnaE1, which contains a consensus β-binding motif; or with DnaE2, which does not (6). A strong DnaN-DnaN interaction was observed, in accordance with the homodimeric structure of the β-clamp. The DnaN-polymerase interactions were consistent with bioinformatics-based predictions: whereas DnaE1 and DinB1 interacted with DnaN, DnaE2 and DinB2 did not (Fig. 1A and B). The ability of DinB1 and DinB2 to interact with themselves, with each other, and with DnaE1 or DnaE2 was also assessed, but no positive interactions were identified (data not shown).
FIG. 1.
Y2H analysis of polymerase interactions with the β-clamp. (A) Growth on selective media. Potential β-interacting partners, cloned as GAL4 BD fusions, were tested for interaction with AD-DnaN by cotransformation and scoring for growth on selection (LT), medium stringency (LTH), and high stringency (LTHA) media. Individual colonies of each strain were resuspended in sterile water, and the cell density was adjusted to an OD600 of 1. Tenfold serial dilutions were then plated on media of differing stringencies. LT, Leu-Trp; LTH, Leu-Trp-His; LTHA, Leu-Trp-His-Ade dropout-supplemented media. Screening for autoactivation of the HISD reporter was carried out by plating cotransformants on dropout-supplemented media containing 3-amino-1,2,4-triazole (3-AT) at 0.5 to 25 mM. A 3-AT concentration of 1 mM was found to inhibit all background growth, and all further interactions were assessed at this concentration. (B) LacZ activity. Positive interactions were confirmed by cotransformation of the corresponding vectors into strain Y187, and LacZ assays were carried out according to the manufacturer's instructions. *, P < 0.01; **, P < 0.005. + and − denote growth or lack of growth of AH109 recombinants on LTH dropout-supplemented media.
Effect of DinB homolog deficiency on susceptibility of M. tuberculosis to genotoxic agents and novobiocin.
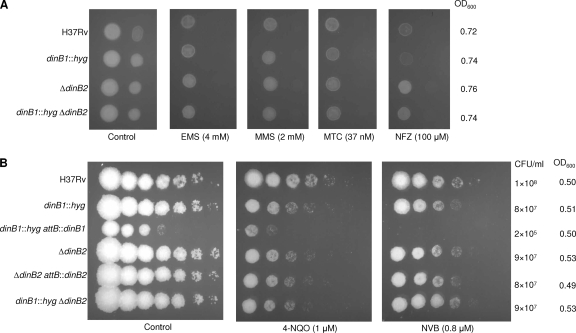
Recent work has uncovered a role for DinB in error-free TLS across deoxyguanine lesions carrying adducts at the N2 position that are generated by treatment with agents such as benzo[a]pyrene, acrolein, nitrofurazone (NFZ), or 4-nitroquinoline-1-oxide (4-NQO) (27, 34, 40, 43, 54, 56, 67). Loss of dinB also sensitized E. coli to the cytotoxic effects of alkylating agents (4). To determine whether the M. tuberculosis dinB homologs may function in tolerance of damage induced by such agents, we constructed derivatives of M. tuberculosis H37Rv carrying mutant dinB1 and/or dinB2 alleles (see Fig. S2A in the supplemental material). The dinB1::hyg and ΔdinB2 single mutants were genetically complemented by integration, at the attB site, of a copy of the corresponding wild-type gene expressed under the control of its own promoter. RT-PCR analysis confirmed that the mutations eliminated the dinB1 and dinB2 transcript detectable in the wild type and that expression was restored in the complemented strains (see Fig. S2B).
The mutant strains grew normally when cultured in liquid media (data not shown). They were tested for sensitivity to various agents in liquid culture by MIC determination. Similar MICs were observed for NFZ (75 to 150 μM) and 4-NQO (18 to 37 μM) for all strains tested. Similarly, no differences in susceptibility to the alkylating agents, ethyl methane sulfonate (EMS) and methyl methane sulfonate (MMS) were observed, with all strains showing MICs of 10 mM and 5 mM, respectively, for these compounds. The strains were also equally susceptible to mitomycin C (MTC; MIC of 0.07 to 0.15 μM). Since treatment of M. tuberculosis with novobiocin (NVB) results in induction of dinB2 expression (5), we also assessed the effect of loss of dinB1 and/or dinB2 function on sensitivity to this drug, but no differences were observed, with all strains showing an MIC for NVB of 0.5 to 1 μM.
Susceptibility was further assessed by spotting serial dilutions of cultures of wild-type and mutant strains sampled at the same stage of growth (OD600 value and number of CFU/ml) on plates containing the compounds (Fig. 2). In this assay, all strains showed equivalent susceptibility to EMS, MMS, and MTC. The dinB1::hyg mutant showed a slight increase in NFZ sensitivity; however, this effect was not observed in the double dinB1::hyg ΔdinB2 mutant (Fig. 2A) and thus may not be significant in the dinB1::hyg mutant. A slight increase in sensitivity to 4-NQO was also observed for the ΔdinB2 and ΔdinB1::hyg ΔdinB2 mutants in some assays. However, this defect was not reversed in the ΔdinB2 attB::dinB2 mutant in which dinB2 expression was restored (Fig. 2B) and therefore is unlikely to be associated with the loss of dinB2 function. Although it grew as well as the other strains in liquid culture, as determined by the OD600, the complemented dinB1 mutant surprisingly displayed poorer growth on solid media than either its parental dinB1::hyg or wild-type strains (Fig. 2B). The possibility that the growth defect of the dinB1::hyg attB::dinB1 mutant strain was due to aberrant expression of the complementing dinB1 gene was investigated by qRT-PCR analysis. As expected, no dinB1 transcript was detected with the dinB1::hyg mutant. However, transcript was detected with the complemented derivative at levels comparable to those of the wild type (2.3 ± 0.3% versus 2.1 ± 1.1% of the level of expression of sigA in H37Rv and the dinB1::hyg attB::dinB1 mutant, respectively, P = 0.7), suggesting that the growth defect of the dinB1::hyg attB::dinB1 mutant on solid media was not due to aberrant expression of dinB1 integrated at the attB locus.
FIG. 2.
Effect of DinB1 and/or DinB2 deficiency on susceptibility of M. tuberculosis to genotoxic agents and novobiocin. Ten or five microliters of neat culture (∼106 CFU) and 10-fold (A) or 3-fold (B) serial dilutions thereof were spotted on plates containing compounds at the indicated concentrations. Plates were incubated for 2 or 3 weeks before scoring growth.
Role of DinB homologs in mutagenesis.
The contribution of the DinB homologs to the fidelity of replication in M. tuberculosis was evaluated by determining the rates of spontaneous mutation to Rif resistance in the mutant strains. All strains showed similar mutation rates (2.3 × 10−9 mutations per cell per generation for the dinB1::hyg, ΔdinB2, and ΔdinB1::hyg ΔdinB2 mutants versus 2.9 × 10−9 for M. tuberculosis H37Rv). The spectrum of rpoB mutations was assessed by sequencing the RRDR in 23 or 24 Rif-resistant mutants isolated from different parallel cultures of each strain (Table 2). The spectrum was consistent with that reported for in vitro-selected Rif-resistant mutants of M. tuberculosis H37Rv (41), and the distributions were similar between strains. In all strains, the Ser531→Leu and His526→Arg mutations, which are the result of C→T transitions, were most common. The His526→Arg mutation, resulting from an A→G transition, was also observed with all strains, albeit at a slightly lower frequency in the dinB1::hyg mutant. In addition to these dominant mutations, C→G and A→C transversion mutations conferring a Rif-resistant phenotype were also observed with some strains.
TABLE 2.
Spectrum of rpoB mutations in spontaneous Rif-resistant mutants derived from M. tuberculosis mutant strains lacking dinB1 and/or dinB2
| Base change | Codon mutation | Substitution | Frequency of mutation (%)a |
|||
|---|---|---|---|---|---|---|
| H37Rv | dinB1::hyg mutant | ΔdinB2 mutant | ΔdinB1::hyg ΔdinB2 mutant | |||
| C→T | TCG→TTG | Ser531→Leu | 69.6 | 82.6 | 56.5 | 50 |
| A→G | CAC→CGC | His526→Arg | 21.7 | 8.7 | 17.4 | 20.8 |
| C→G | CAC→GAC | His526→Asp | 8.7 | 0 | 0 | 0 |
| C→T | CAC→TAC | His526→Tyr | 0 | 8.7 | 21.7 | 25 |
| A→C | CAC→CCC | His526→Pro | 0 | 0 | 0 | 4.2 |
| C→G | TCG→TGG | Ser522→Trp | 0 | 0 | 4.4 | 0 |
Twenty-three independent Rif-resistant mutants were analyzed for H37Rv and the dinB1::hyg and ΔdinB2 mutants, and 24 mutants were analyzed for the ΔdinB1::hyg ΔdinB2 mutant.
Elevated expression of dinB homologs has been shown to confer a mutator phenotype in other organisms (31, 54). We therefore investigated the effects of ectopic expression of M. tuberculosis dinB1 or dinB2 on mutagenesis in the nonpathogenic saprophyte M. smegmatis. The M. tuberculosis genes were cloned on an episomal vector under the control of the M. smegmatis acetamidase promoter (46). RT-PCR analysis confirmed expression of both genes, even in the absence of acetamide (see Fig. S3 in the supplemental material), allowing mutation rates to be assessed in the absence of inducer. However, ectopic expression of M. tuberculosis dinB1 or dinB2 had no effect on the rate of mutation of M. smegmatis to Rif resistance (Table 3). Since the rpoB reporter is restricted in terms of the spectrum of mutations it can detect (base substitutions), we developed a novel reporter for assessing frameshift mutagenesis at a homopolymeric run in mycobacteria by transferring a +1 chromosomal frameshift mutation into a 4T run located in the hisD gene of M. smegmatis (23). The mutation resulted in histidine auxotrophy through abrogation of the hisD function (E. E. Machowski, unpublished data). The M. tuberculosis dinB1 and dinB2 expression vectors were introduced into the auxotroph HIS5T, and the rates of reversion to histidine prototrophy by −1 frameshift mutagenesis (5T→4T) compared. However, no significant differences were observed between strains (Table 3).
TABLE 3.
Mutation rates for M. smegmatis strains overexpressing M. tuberculosis dinB1 and dinB2
| Strain | Mutation rate |
|
|---|---|---|
| Rif resistance (rpoB) (×10−8) | Histidine prototrophy [hisD(5T) reversion] (×10−8)a | |
| mc2155 | 4.9 | NA |
| HIS5T(pOLYGaa) | 3.2 | 4.8 |
| HIS5T(pGaaDinB1) | 2.6 | 6.3 |
| HIS5T(pGaaDinB2) | 8.4 | 8.7 |
NA, not applicable.
dinB1 and dinB2 are individually dispensable for growth of M. tuberculosis in macrophages and mice.
To investigate the role of the DinB homologs in virulence, intracellular growth was assessed in various macrophage infection models. No significant growth differences were observed between the wild type and the dinB1::hyg mutant in resting human PBMCs or in murine peritoneal macrophages or bone marrow-derived macrophages (see Fig. S4A in the supplemental material and data not shown). Moreover, growth of the dinB1::hyg and ΔdinB2 mutants in LPS-activated human PBMCs was not significantly different from that of the wild type (see Fig. S4B and C).
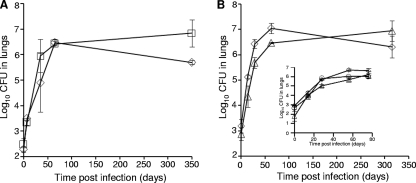
Groups of B6D2F1 mice were then aerosol infected with the single mutant or wild-type strains, and bacillary growth was monitored over time. The dinB1::hyg mutant, seeded at 2.5 ± 0.2 log10 CFU, displayed no attenuation of growth in the lungs of infected mice compared to the wild-type strain, seeded at 2.3 ± 0.2 log10 CFU. The lung bacillary loads of the mutant and wild-type strains were comparable; the 1.2 log10-higher number of CFU of the mutant over the wild type at 350 days postinfection was not statistically significant (P > 0.08) (Fig. 3A). The mutant was also fully proficient in hematogenous dissemination to the spleen, and no significant differences in survival of mice infected with the dinB1::hyg mutant were noted (data not shown). The ΔdinB2 mutant, inoculated at 2.9 ± 0.3 log10 CFU, displayed slightly reduced bacillary loads in the lung compared to the wild type (seeded at 3.2 ± 0.3 log10 CFU) during early infection but achieved comparable numbers of CFU during late infection (P > 0.1) (Fig. 3B). Similarly, dissemination of this mutant to the spleen was reduced during early infection, possibly due to the reduced lung bacillary loads, but this effect was lost over time with no difference in the number of spleen CFU between the wild type and the ΔdinB2 mutant recorded at 315 days postinfection (data not shown). To further analyze the virulence of the ΔdinB2 mutant during early infection, bacillary loads in the lungs of mice infected with the ΔdinB2 mutant and wild-type strains were assessed alongside the complemented mutant in a separate experiment. However, no significant differences were observed (Fig. 3B, inset).
FIG. 3.
Growth and survival of DinB1 and DinB2 mutants of M. tuberculosis H37Rv in infected mice. Growth of the dinB1::hyg (A) and ΔdinB2 (B) mutants with the parental wild type was monitored over a period of 350 and 315 days postinfection, respectively. The inset in panel B shows growth and survival of the ΔdinB2 mutant, its complemented derivative, and the parental wild type over a period of 77 days postinfection. Three mice were sacrificed at each time point for each strain. ⋄, wild type; □, dinB1::hyg mutant; ▵, ΔdinB2 mutant; ○, ΔdinB2 attB::dinB2 mutant.
DISCUSSION
Insight into the expression, activity, and regulation of specialized polymerases belonging to the DinB subfamily of Y family polymerases has been gained from studies of E. coli (reviewed in reference 26), P. aeruginosa (54), and Bacillus subtilis (17). Important similarities and differences have been identified between organisms in terms of the number of DinB homologs and the complement of other DNA polymerases that are present, the mechanisms regulating their expression (SOS regulated or not), interaction with the β-clamp and the functional significance thereof, and the effects of loss-of-function mutations or DinB overproduction on susceptibility to DNA damage and/or mutagenesis. Therefore, defining a “canonical” role for DinB in bacterial DNA metabolism has proven to be elusive, although some characteristic functions have emerged, such as error-free TLS across N2-dG lesions.
In this study, we investigated the role of the two DinB homologs in M. tuberculosis. We identified marked differences between DinB1 and DinB2 at the level of expression and interaction with the β-clamp and also between them and homologs in other bacteria, in terms of protection against the cytotoxic effects of DNA damage and mutator activity. Sequence analysis suggests that DinB1 and DinB2 may be functional DNA polymerases (see Fig. S1 in the supplemental material). Attempts were made to confirm this prediction by overexpression, purification, and enzymatic characterization of these proteins as E. coli recombinants. However, these efforts met with limited success (data not shown), and as a result, the DNA polymerase activities of DinB1 and DinB2 have yet to be established.
The level of sigA-normalized dinB2 transcript detected in cultures of M. tuberculosis grown in vitro was significantly higher than that of dinB1 and also higher than that of dnaE1, which encodes the replicative polymerase. Moreover, unlike dnaE1, which was expressed at lower levels in stationary- versus logarithmic-phase cells, the levels of dinB1 and dinB2 transcript did not change significantly as a function of growth phase. The comparatively high level of dinB2 expression in M. tuberculosis is therefore reminiscent of the situation in E. coli under non-SOS-inducing conditions, in which DinB is present at a level more than 10-fold higher than that of the Pol III holoenzyme (32).
One feature of M. tuberculosis DinB2 that differentiates it from DinB1 and the DinB homologs from E. coli and B. subtilis is an inability to bind to the β-clamp, which may impair or even preclude putative TLS and replication slow-down functions (10, 25). Thus, overexpression of dinB2 would not be expected to affect the growth or colony-forming ability of M. tuberculosis, whereas overexpression of dinB1, which encodes a protein with β-binding activity, might. However, aberrant dinB1 expression was ruled out as an explanation for the impaired colony-forming ability of the dinB1::hyg attB::dinB1 mutant, a phenotype which has yet to be resolved. Therefore, the functional implications of the differential β-binding activity of DinB1 and DinB2 remain unclear and await further information on their DNA polymerase activity, the importance (or otherwise) of β-binding for polymerase function in mycobacteria and association between β-binding by a specialized DNA polymerase, replication fork slowdown, and the effects thereof on mycobacterial growth (25, 61).
The loss of dinB1 and/or dinB2 function had little or no effect on the sensitivity of M. tuberculosis to N2-dG adduct-forming compounds (27, 54) or to alkylating agents (4), in contrast to that of other organisms. While evidence suggests that the in vivo environments encountered by M. tuberculosis are genotoxic, the types of DNA lesions generated in these environments have not been defined (6, 13, 55). The lesion N2-(1-carboxyethyl)-2′-dG (N2-CEdG) has been detected with human melanoma cells, and its frequency shown to increase in a dose-responsive manner by treatment with methylglyoxal (MG) or glucose (67). Interestingly, transcriptional profiling of M. tuberculosis in pulmonary TB has shown that the tissue environment of M. tuberculosis at sites of infection is rich in MG (51), which is present at elevated levels in macrophages during mycobacterial infection (50). The upregulation of dinB1 observed during pulmonary TB (51) suggests a plausible link between the production of host and/or M. tuberculosis-derived MG during infection, the induction of N2-CEdG lesions in M. tuberculosis, and dinB1 function. However, the dispensability of DinB1 (and DinB2) for the growth of M. tuberculosis in vivo suggests that M. tuberculosis possesses another mechanism(s) for dealing with the cytotoxic effects of lesions, such as N2-CEdG, that may obscure the effects of DinB homolog deficiency. One possibility is nucleotide excision repair (NER), which provides the major mechanism for processing NFZ-induced damage in E. coli (45). NER has been implicated in the virulence of M. tuberculosis in mice (13) through the processing of DNA damage caused by nitrosative stress (12). Given the comparatively low rate of DNA replication in M. tuberculosis (24), NER may play an even more dominant role than damage tolerance in enabling this organism to contend with such lesions.
Consistent with the findings for E. coli (33), the loss of dinB1 and/or dinB2 function had no effect on the replication fidelity of M. tuberculosis in vitro, as evidenced by the rate of spontaneous mutation to Rif resistance. Minor differences in the spectrum of rpoB mutations were observed between strains, but in all cases, Rif resistance arose predominantly from C→T mutations. Instead, our results suggest the value of developing a mutational reporter suitable for readout of the G→T mutations that might be expected to increase in frequency in strains defective in error-free bypass of lesions such as N2-CEdG (67). However, in a further deviation from findings for other organisms (31, 54), ectopic expression of the M. tuberculosis dinB homologs had no discernible effect on rates of (base substitution) mutation to Rif resistance or −1 frameshift mutagenesis at a homopolymeric (5T) run in M. smegmatis. Again, it is possible that the reporters employed are not suitable for detecting mutations that the DinB homologs may be particularly prone to inducing, such as −1 frameshifts within homopolymeric runs of deoxyguanylic acids (31, 54) or alternatively that the M. tuberculosis dinB1 and dinB2 expression levels were not sufficiently high to confer an effect. To test these possibilities, a broader range of mutational reporters and further expression vectors are under development in our laboratory.
In conclusion, the physiological roles of DinB1 and DinB2 in DNA metabolism in M. tuberculosis have proven remarkably refractory to elucidation using models that were predicted to be phenotypically revealing on the basis of insights from other organisms. Together, our findings suggest that the DinB homologs from M. tuberculosis differ significantly from their counterparts from other bacteria.
Supplementary Material
Acknowledgments
This work was funded by grants from the Wellcome Trust (Collaborative Research Initiative grant 065578 to N.G.S. and V.M.), the Medical Research Council of South Africa (to V. M.), the National Research Foundation (to V. M.), the National Institutes of Health (R01 AI 54338 to G.K.), the Howard Hughes Medical Institute (International Research Scholar's grant to V.M.), and the Welch Foundation (to J.C.S.). B.D.K. was supported by a traineeship from the Columbia University-Southern Africa Fogarty AIDS International Research and Training Program (NIH/FIC D43 TW00231 [to Q. Abdool Karim]). N.S. was supported by a postdoctoral fellowship from the Heiser Program for Research in Leprosy and Tuberculosis.
We thank Stewart Cole for providing BACC12, Tanya Parish for providing pAGAN11, Dorothy Fallows for constructively reviewing the manuscript, members of the Mizrahi, Kaplan, and Sacchettini laboratories for advice and assistance, and anonymous reviewers for helpful suggestions.
Footnotes
Published ahead of print on 5 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Becherel, O. J., R. P. Fuchs, and J. Wagner. 2002. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair (Amst) 1:703-708. [DOI] [PubMed] [Google Scholar]
- 2.Bertram, J. G., L. B. Bloom, M. O'Donnell, and M. F. Goodman. 2004. Increased dNTP binding affinity reveals a nonprocessive role for Escherichia coli beta clamp with DNA polymerase IV. J. Biol. Chem. 279:33047-33050. [DOI] [PubMed] [Google Scholar]
- 3.Beuning, P. J., D. Sawicka, D. Barsky, and G. C. Walker. 2006. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol. Microbiol. 59:460-474. [DOI] [PubMed] [Google Scholar]
- 4.Bjedov, I., C. N. Dasgupta, D. Slade, S. Le Blastier, M. Selva, and I. Matic. 2007. Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics 176:1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boshoff, H. I., T. G. Myers, B. R. Copp, M. R. McNeil, M. A. Wilson, and C. E. Barry, 3rd. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279:40174-40184. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff, H. I., M. B. Reed, C. E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, P. C., F. Movahedzadeh, and E. O. Davis. 2001. Identification of some DNA damage-inducible genes of Mycobacterium tuberculosis: apparent lack of correlation with LexA binding. J. Bacteriol. 183:4459-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunting, K. A., S. M. Roe, and L. H. Pearl. 2003. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 22:5883-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Courcelle, J. 2009. Shifting replication between IInd, IIIrd, and IVth gears. Proc. Natl. Acad. Sci. U. S. A. 106:6027-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. U. S. A. 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin, K. H., S. Ehrt, J. C. Gutierrez-Ramos, N. Weich, and C. F. Nathan. 2003. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302:1963-1966. [DOI] [PubMed] [Google Scholar]
- 13.Darwin, K. H., and C. F. Nathan. 2005. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect. Immun. 73:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domenech, P., M. B. Reed, and C. E. Barry III. 2005. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73:3492-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donald, P. R., and P. D. van Helden. 2009. The global burden of tuberculosis-combating drug resistance in difficult times. N. Engl. J. Med. 360:2393-2395. [DOI] [PubMed] [Google Scholar]
- 16.Downing, K. J., J. C. Betts, D. I. Young, R. A. McAdam, F. Kelly, M. Young, and V. Mizrahi. 2004. Global expression profiling of strains harbouring null mutations reveals that the five rpf-like genes of Mycobacterium tuberculosis show functional redundancy. Tuberculosis (Edinb.) 84:167-179. [DOI] [PubMed] [Google Scholar]
- 17.Duigou, S., S. D. Ehrlich, P. Noirot, and M. F. Noirot-Gros. 2004. Distinctive genetic features exhibited by the Y-family DNA polymerases in Bacillus subtilis. Mol. Microbiol. 54:439-451. [DOI] [PubMed] [Google Scholar]
- 18.Freeman, S., F. A. Post, L. G. Bekker, R. Harbacheuski, L. M. Steyn, B. Ryffel, N. D. Connell, B. N. Kreiswirth, and G. Kaplan. 2006. Mycobacterium tuberculosis H37Ra and H37Rv differential growth and cytokine/chemokine induction in murine macrophages in vitro. J. Interferon Cytokine Res. 26:27-33. [DOI] [PubMed] [Google Scholar]
- 19.Friedberg, E. C., R. Wagner, and M. Radman. 2002. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296:1627-1630. [DOI] [PubMed] [Google Scholar]
- 20.Galhardo, R. S., R. Do, M. Yamada, E. C. Friedberg, P. J. Hastings, T. Nohmi, and S. M. Rosenberg. 2009. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182:55-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman, M. F. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71:17-50. [DOI] [PubMed] [Google Scholar]
- 22.Heltzel, J. M., S. K. Scouten Ponticelli, L. H. Sanders, J. M. Duzen, V. Cody, J. Pace, E. H. Snell, and M. D. Sutton. 2009. Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli. J. Mol. Biol. 387:74-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinshelwood, S., and N. G. Stoker. 1992. Cloning of mycobacterial histidine synthesis genes by complementation of a Mycobacterium smegmatis auxotroph. Mol. Microbiol. 6:2887-2895. [DOI] [PubMed] [Google Scholar]
- 24.Hiriyanna, K. T., and T. Ramakrishnan. 1986. Deoxyribonucleic acid replication time in Mycobacterium tuberculosis H37 Rv. Arch. Microbiol. 144:105-109. [DOI] [PubMed] [Google Scholar]
- 25.Indiani, C., L. D. Langston, O. Yurieva, M. F. Goodman, and M. O'Donnell. 2009. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc. Natl. Acad. Sci. U. S. A. 106:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarosz, D. F., P. J. Beuning, S. E. Cohen, and G. C. Walker. 2007. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 15:70-77. [DOI] [PubMed] [Google Scholar]
- 27.Jarosz, D. F., V. G. Godoy, J. C. Delaney, J. M. Essigmann, and G. C. Walker. 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439:225-228. [DOI] [PubMed] [Google Scholar]
- 28.Jarosz, D. F., V. G. Godoy, and G. C. Walker. 2007. Proficient and accurate bypass of persistent DNA lesions by DinB DNA polymerases. Cell Cycle 6:817-822. [DOI] [PubMed] [Google Scholar]
- 29.Kana, B. D., B. G. Gordhan, K. J. Downing, N. Sung, G. Vostroktunova, E. E. Machowski, L. Tsenova, M. Young, A. Kaprelyants, G. Kaplan, and V. Mizrahi. 2008. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol. Microbiol. 67:672-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. U. S. A. 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S. R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. U. S. A. 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, S. R., K. Matsui, M. Yamada, P. Gruz, and T. Nohmi. 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet Genomics 266:207-215. [DOI] [PubMed] [Google Scholar]
- 33.Kuban, W., P. Jonczyk, D. Gawel, K. Malanowska, R. M. Schaaper, and I. J. Fijalkowska. 2004. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J. Bacteriol. 186:4802-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumari, A., I. G. Minko, M. B. Harbut, S. E. Finkel, M. F. Goodman, and R. S. Lloyd. 2008. Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. J. Biol. Chem. 283:27433-27437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenne-Samuel, N., J. Wagner, H. Etienne, and R. P. Fuchs. 2002. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 3:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machowski, E. E., S. Barichievy, B. Springer, S. I. Durbach, and V. Mizrahi. 2007. In vitro analysis of rates and spectra of mutations in a polymorphic region of the Rv0746 PE_PGRS gene of Mycobacterium tuberculosis. J. Bacteriol. 189:2190-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manca, C., S. Paul, C. E. Barry III, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 67:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manca, C., L. Tsenova, A. Bergtold, S. Freeman, M. Tovey, J. M. Musser, C. E. Barry III, V. H. Freedman, and G. Kaplan. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl. Acad. Sci. U. S. A. 98:5752-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenzie, G. J., P. L. Lee, M. J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579. [DOI] [PubMed] [Google Scholar]
- 40.Minko, I. G., K. Yamanaka, I. D. Kozekov, A. Kozekova, C. Indiani, M. E. O'Donnell, Q. Jiang, M. F. Goodman, C. J. Rizzo, and R. S. Lloyd. 2008. Replication bypass of the acrolein-mediated deoxyguanine DNA-peptide cross-links by DNA polymerases of the DinB family. Chem. Res. Toxicol. 21:1983-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morlock, G. P., B. B. Plikaytis, and J. T. Crawford. 2000. Characterization of spontaneous, in vitro-selected, rifampin-resistant mutants of Mycobacterium tuberculosis strain H37Rv. Antimicrob. Agents Chemother. 44:3298-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mowa, M. B., D. F. Warner, G. Kaplan, B. D. Kana, and V. Mizrahi. 2009. Function and regulation of class I ribonucleotide reductase-encoding genes in mycobacteria. J. Bacteriol. 191:985-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Napolitano, R., R. Janel-Bintz, J. Wagner, and R. P. Fuchs. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 19:6259-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohmori, H., E. C. Friedberg, R. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. A. Kunkel, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate. 2001. The Y-family of DNA polymerases. Mol. Cell 8:7-8. [DOI] [PubMed] [Google Scholar]
- 45.Ona, K. R., C. T. Courcelle, and J. Courcelle. 2009. Nucleotide excision repair is a predominant mechanism for processing nitrofurazone-induced DNA damage in Escherichia coli. J. Bacteriol. 191:4959-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parish, T., E. Mahenthiralingam, P. Draper, E. O. Davis, and M. J. Colston. 1997. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143(7):2267-2276. [DOI] [PubMed] [Google Scholar]
- 47.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(8):1969-1975. [DOI] [PubMed] [Google Scholar]
- 48.Pope, C. F., D. M. O'Sullivan, T. D. McHugh, and S. H. Gillespie. 2008. A practical guide to measuring mutation rates in antibiotic resistance. Antimicrob. Agents Chemother. 52:1209-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Post, F. A., C. Manca, O. Neyrolles, B. Ryffel, D. B. Young, and G. Kaplan. 2001. Mycobacterium tuberculosis 19-kilodalton lipoprotein inhibits Mycobacterium smegmatis-induced cytokine production by human macrophages in vitro. Infect. Immun. 69:1433-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rachman, H., N. Kim, T. Ulrichs, S. Baumann, L. Pradl, A. Nasser Eddine, M. Bild, M. Rother, R. J. Kuban, J. S. Lee, R. Hurwitz, V. Brinkmann, G. A. Kosmiadi, and S. H. Kaufmann. 2006. Critical role of methylglyoxal and AGE in mycobacteria-induced macrophage apoptosis and activation. PLoS One 1:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rachman, H., M. Strong, T. Ulrichs, L. Grode, J. Schuchhardt, H. Mollenkopf, G. A. Kosmiadi, D. Eisenberg, and S. H. Kaufmann. 2006. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect. Immun. 74:1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rand, L., J. Hinds, B. Springer, P. Sander, R. S. Buxton, and E. O. Davis. 2003. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 50:1031-1042. [DOI] [PubMed] [Google Scholar]
- 53.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 20:4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders, L. H., A. Rockel, H. Lu, D. J. Wozniak, and M. D. Sutton. 2006. Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. J. Bacteriol. 188:8573-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen, X., J. M. Sayer, H. Kroth, I. Ponten, M. O'Donnell, R. Woodgate, D. M. Jerina, and M. F. Goodman. 2002. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9,10-epoxide A and G adducts. J. Biol. Chem. 277:5265-5274. [DOI] [PubMed] [Google Scholar]
- 57.Smith, A. M., and K. P. Klugman. 1997. “Megaprimer” method of PCR-based mutagenesis: the concentration of megaprimer is a critical factor. Biotechniques 22:438.-442. [DOI] [PubMed] [Google Scholar]
- 58.Sung, H. M., G. Yeamans, C. A. Ross, and R. E. Yasbin. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 185:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tegova, R., A. Tover, K. Tarassova, M. Tark, and M. Kivisaar. 2004. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 186:2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tippin, B., P. Pham, and M. F. Goodman. 2004. Error-prone replication for better or worse. Trends Microbiol. 12:288-295. [DOI] [PubMed] [Google Scholar]
- 61.Uchida, K., A. Furukohri, Y. Shinozaki, T. Mori, D. Ogawara, S. Kanaya, T. Nohmi, H. Maki, and M. Akiyama. 2008. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol. Microbiol. 70:608-622. [DOI] [PubMed] [Google Scholar]
- 62.Wagner, J., S. Fujii, P. Gruz, T. Nohmi, and R. P. Fuchs. 2000. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 1:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner, J., P. Gruz, S. R. Kim, M. Yamada, K. Matsui, R. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4:281-286. [DOI] [PubMed] [Google Scholar]
- 64.Warner, D. F., and V. Mizrahi. 2006. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin. Microbiol. Rev. 19:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang, W., and R. Woodgate. 2007. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 104:15591-15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. U. S. A. 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan, B., H. Cao, Y. Jiang, H. Hong, and Y. Wang. 2008. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:8679-8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.