Abstract
We have previously described a novel Escherichia coli detoxification system for the removal of toxic and mutagenic N-hydroxylated nucleobases and related compounds that requires the molybdenum cofactor. Two subpathways (ycbX and yiiM) were identified, each employing a novel molybdo activity capable of inactivating N-hydroxylated compounds by reduction to the corresponding amine. In the present study, we identify the cysJ gene product as one additional component of this system. While the CysJ protein has been identified as the NADPH:flavin oxidoreductase component of the CysJI sulfite reductase complex (CysJ8I4), we show that the role of CysJ in base analog detoxification is unique and independent of CysI and sulfite reductase. We further show that CysJ functions as a specific partner of the YcbX molybdoenzyme. We postulate that the function of CysJ in this pathway is to provide, via its NADPH:flavin reductase activity, the reducing equivalents needed for the detoxification reaction at the YcbX molybdocenter. In support of the proposed interaction of the CysJ and YcbX proteins, we show that an apparent CysJ-YcbX “hybrid” protein from two Vibrio species is capable of compensating for a double cysJ ycbX defect in E. coli.
Mutagenic base analogs are chemically modified nucleobases that can be incorporated in the cellular metabolism through purine or pyrimidine salvage pathways. Once converted to the deoxynucleoside triphosphate (dNTP) level, they may participate in DNA replication in an error-prone manner because of their ambivalent base-pairing capacity (11). Such synthetic base analogs are often used as a sensitive tool for studying DNA replication fidelity, DNA repair, or the metabolism of nucleic acid precursors. Mutagenic base analogs such as 8-oxoguanine or 3-methyladenine can also be formed in vivo as a consequence of normal cellular metabolism or produced by chemical and physical factors, such as alkylating agents or ionizing radiation.
An important group of mutagenic and cytotoxic analogs are the N-hydroxylated nucleobases (or ribosides) such as 6-N-hydroxylaminopurine (HAP), 2-amino-HAP, or N4-hydroxycytidine (15). Specifically, HAP was found to be a very strong mutagen in bacteria and fungi, as well as mammalian cells (2, 20, 27). Some data have suggested that HAP may also be formed in vivo under oxidative stress (30) or as a by-product of certain purine salvage/interconversion pathways (5, 22).
The genetic control of HAP-induced mutagenesis has been studied in some detail in the yeast Saccharomyces cerevisiae and in the bacterium Escherichia coli. In S. cerevisiae, resistance to HAP depends primarily on genes involved in adjusting and regulating the DNA or RNA precursor pools (HAM1 [ITP/XTPase], AAH1 [adenine aminohydrolase], and ADE genes involved in de novo AMP biosynthesis) (34).
In E. coli, the major pathway that protects cells against HAP and related N-hydroxylated compounds is controlled by the moa, moe, and mog genes, which are required for biosynthesis of molybdenum cofactor (MoCo) (18, 19). MoCo is an essential cofactor for a varied group of oxidoreductases that are widely distributed from bacteria to humans. Chemically, MoCo is a pterin derivative (molybdopterin) that coordinates a molybdenum atom that serves as a catalytic redox center (for reviews, see references 23, 28, and 29). Based on catalytic details and sequence homology, molybdopterin-containing enzymes have been divided in four families: the xanthine oxidase family, the sulfite oxidase family, the dimethyl sulfoxide (DMSO) reductase family, and the aldehyde ferredoxin oxidoreductase family (14, 16). However, our previous studies on the MoCo-dependent resistance to HAP showed that none of the known or putative E. coli members of these families are responsible for the major HAP resistance mechanism (19). Instead, we discovered that HAP resistance is dependent on two newly described proteins, YcbX and YiiM, that are characterized by a so-called MOSC domain (molybdenum cofactor sulfurase C-terminal domain) (1, 17). This domain was first described as part of eukaryotic MoCo sulfurases (MOSs) (1), and it most likely represents a novel class of MoCo-binding domain, as indicated by studies on two mammalian MOSC-containing proteins (mARC1 and mARC2) discovered in mitochondria (12, 13).
Our studies in E. coli showed that cell-free bacterial extracts were capable of converting HAP to adenine by an N-reductive reaction (17). Importantly, this conversion was entirely dependent on the presence of MoCo and the YcbX or YiiM proteins (17). Consequently, we suggested that this reduction of HAP to adenine forms the basis of the in vivo MoCo-dependent detoxification in E. coli (17). Interestingly, the mammalian MOSC-containing proteins mARC1 and mARC2 were shown to mediate the reduction of the N-hydroxylated prodrug benzamidoxime to its active amino form benzamidine (12, 13). Thus, the reduction of N-hydroxylated compounds may be a defining feature for the broadly distributed MOSC proteins (1).
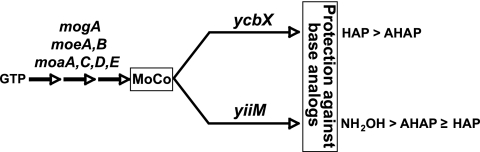
Our previous analyses also revealed that the E. coli ycbX and yiiM genes define two independent subpathways within the MoCo-dependent system (17). This is illustrated in the overall scheme shown in Fig. 1. MoCo is synthesized in a series of steps from GTP by-products of the moa, moe, and mog operons. MoCo is then used as a cofactor for the YcbX and YiiM proteins, which reduce the N-hydroxylated compound to the corresponding amino form. The ycbX and yiiM pathways are genetically distinct as determined by epistasis experiments (17). They also differ by their substrate specificity patterns: YcbX protects most strongly against HAP, whereas YiiM has its largest effects toward hydroxylamine (NH2OH) (17).
FIG. 1.
Genetic framework for the major molybdenum cofactor (MoCo)-dependent pathways of detoxification of N-hydroxylated base analogs in E. coli (17). moaA to mogA indicate the series of genes required for MoCo biosynthesis (19, 28), while ycbX and yiiM represent the two independent subpathways identified within the MoCo-dependent pathway (17). Specifically, ycbX and yiiM produce apoenzymes that react with MoCo to form the active YcbX and YiiM proteins. The diagram also indicates the differential specificity of the two subpathways toward the model N-hydroxylated compounds used in our studies: 6-N-hydroxylaminopurine (HAP), 2-amino-HAP (AHAP), and hydroxylamine (NH2OH). For simplicity, the diagram does not distinguish between the MPT and MGD forms of MoCo (19). As shown elsewhere (19), YcbX and YiiM likely employ the MPT form. One additional, minor pathway for HAP detoxification dependent on biotin sulfoxide reductase (an MGD-requiring enzyme) is observable only in the double ycbX yiiM-deficient background and is likewise not shown here (see reference 17 for details).
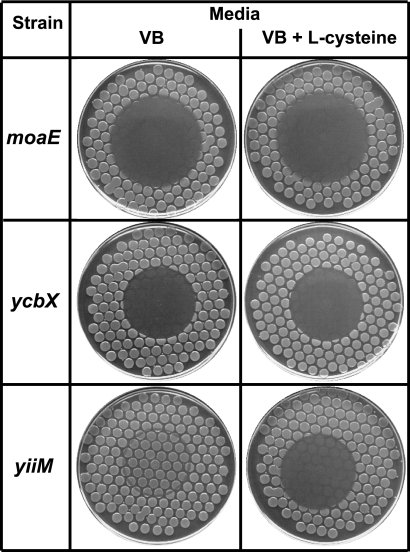
Prior to the establishment of this scheme of YcbX and YiiM as molybdoproteins, we had entertained certain alternative possibilities for the precise function of the ycbX and yiiM open reading frames (ORFs), including a possible role in MoCo sulfuration (which is a required modification of MoCo in certain molybdoenzymes, such as xanthine oxidase) (23, 29). This sulfuration model was ultimately eliminated (17), but certain experiments related to this hypothesis yielded interesting further clues regarding the detailed mechanisms of HAP resistance. These observations included an unexpected HAP-sensitive phenotype for cysJ mutants as well as a noted sensitization of wild-type strains to HAP by l-cysteine. In the present work, we describe these experiments and show the cysJ gene to be an essential component of the ycbX branch of HAP resistance. In a related mechanism, the observed sensitization of wild-type strains by l-cysteine results from the suppression, by l-cysteine, of the cys regulon. Overall, our experiments suggest that CysJ is a specific protein partner of YcbX and that CysJ mediates the N-reductive reaction through its NADPH:flavin oxidoreductase activity. This activity provides reducing equivalents to its partner YcbX, which ultimately performs the reduction of HAP to nontoxic adenine at its molybdocenter.
MATERIALS AND METHODS
Media and chemicals.
Bacteria were cultivated in LB broth (24) or minimal Vogel-Bonner (VB) medium (35) containing 0.2% glucose as a carbon source. To support growth of cysteine auxotrophs, VB medium was supplemented with either 0.33 mM l-cysteine or 0.8 mM sodium thiosulfate. Solid media contained 1.5% agar. For selection of antibiotic-resistant clones, medium was supplemented with 35 μg/ml of kanamycin, 20 μg/ml of chloramphenicol, 15 μg/ml of tetracycline, or 100 μg/ml of ampicillin. For selection of cysB(Con) constitutive mutants, VB medium was supplemented with 10 mM 1,2,4-triazole (VB-TRZ plates) (32). HAP (in the form of free base) was purchased from Midwest Research Institute (Kansas City, MO). All other chemicals were from Sigma-Aldrich.
Bacterial strains.
The E. coli strains used in this study are listed in Table 1 along with their source or derivation. All mutagenesis and base analog sensitivity tests were performed using strain NR10836 and its mutant derivatives. The deletions ΔcysJ::tet, ΔcysB::kan, ΔcysI::kan, and ΔcysM::kan were generated in strain BW25113/pKD46 by the PCR-based gene replacement method of Datsenko and Wanner (8) using the Kanr module of plasmid pKD13 (8) or the tetA tetR tetracycline resistance (Tetr) module of transposon Tn10 as a template. Primers for the PCRs were as follows (uppercase letters indicate the sequences of Tn10 or pKD13): for ΔcysJ::tet, cysJ-p1T (5′-tta gta gac atc tcg ctg ata acg gcg ctc tac gcg cag ctc act taa aaA AGA GGG TCA TTA TAT TTC G-3′) and cysJ-p4T (5′-tta ctg gaa cat aac gac gca tga cga cac agg tcc cac ctt ccg cgt tgA CTC GAC ATC TTG GTT ACC G-3′); for ΔcysB::kan, cysB-p1 (5′-aaa cga tgt tct gat ggc gtc taa gtg gat ggt tta aca tga aat tac aaG TGT AGG CTG GAG CTG CTT CG-3′) and cysB-p4 (5′ ttc cgg cac ctt cgc tac ata aaa ggt gcc gaa aat aac gca aga aat taA TTC CGG GGA TCC GTC GAC C-3′); for ΔcysI::kan, cysI-p1 (5′-ggc tga tgg tta atc cca caa atc acg cgc cgg atc gag cac cgg gcg aaG TGT AGG CTG GAG CTG CTT CG-3′) and cysI-p4 (5′-tga gcg aaa aac atc cag ggc ctt tag tgg tcg aag gaa aac tga cag acA TTC CGG GGA TCC GTC GAC C-3′); and for ΔcysM::kan, cysM-p1 (5′-agc gca tca ggc agt ttt gcg ttt gtc atc agt ctc cga tgc tat taa tcG TGT AGG CTG GAG CTG CTT CG-3′) and cysM-p4 (5′-acg tat gga tag aga tcg tga gta cat tag aac aaa caa tag gca ata cgA TTC CGG GGA TCC GTC GAC C-3′). When necessary, the resulting deletion-insertions were transferred into strain NR10836 by P1 transduction using P1virA (Table 1).
TABLE 1.
E. coli strains used in this study
| Strain | Genotype | Reference(s) or derivation |
|---|---|---|
| BW25113/pKD46 | lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78/pKD46 | 8 |
| CAG12028 | rph-1 zcj-233::Tn10 | 25, 33 |
| NR10836 | ara thi Δ(pro-lac) F′CC106 | 18 |
| NR15870 | NR10836 but ΔyiiM::kan | 17 |
| NR15871 | NR10836 but ΔyiiM | 17 |
| NR15873 | NR10836 but ycbX::mini-Tn10cam | 17 |
| NR15996 | NR10836 but ΔmoaE::kan | 19 |
| NR16043 | NR10836 but ΔcysJ::tet | This study |
| NR16069 | NR10836 but ΔcysI::kan | This study |
| NR16085 | NR15871 but ΔcysJ::tet | NR15871 × P1/NR16043 |
| NR16087 | NR15873 but ΔcysJ::tet | NR15873 × P1/NR16043 |
| NR16098 | NR10836 but ΔcysB::kan | This study |
| NR16189 | BW25113/pKD46 but zcj-233::Tn10 | BW25113/pKD46 × P1/CAG12028 |
| NR16191 | NR16189 but ΔcysM::kan | This study |
| NR16193 | NR16191 but cysB(Con) (T149P) | This study |
| NR16195 | NR10836 but cysB(Con) (T149P) zcj-233::Tn10 | NR10836 × P1/NR16193 |
| NR16262 | NR10836 but ΔycbX | 17 |
| NR16523 | NR10836 but ΔmoaE | NR15996 but kanamycin sensitivea |
| NR16743 | NR16262 but ΔcysJ::tet | NR16262 × P1/NR16043 |
| NR16757 | NR16523 but ΔcysJ::tet | NR16523 × P1/NR16043 |
| NR16918 | NR16262 but ΔycbX::VCA0924 | This study |
| NR16919 | NR16262 but ΔycbX::VPA0411 | This study |
| NR16920 | NR16918 but ΔcysJ::tet | NR16918 × P1/NR16043 |
| NR16921 | NR16919 but ΔcysJ::tet | NR16919 × P1/NR16043 |
The Kanr module was eliminated from moaE using plasmid pCP20 as described by Datsenko and Wanner (8).
Construction of a cysB(Con) (constitutive) mutant.
A method for isolating Salmonella enterica serovar Typhimurium cysB(Con) mutants was developed by Sledziewska and Hulanicka (32) using resistance to the agent 1,2,4-triazole in a cysM bacterial background, and specific amino acid changes in S. Typhimurium CysB leading to constitutive cysB(Con) phenotype (Thr149 → Pro and Thr149 → Met) were described by Colyer and Kredich (6). Based on the 95% identity of E. coli CysB and S. Typhimurium CysB (26), we assumed that the T149P (or T149M) change in E. coli CysB protein would also produce a constitutive phenotype. We designed a 70-mer oligonucleotide corresponding to the E. coli cysB gene but specifying the T149P substitution: cysB-T149P (5′-aac atc act aaa tct tca tac aga tgc agc gct tc TGG ggc gat agc gaa atc agc att gcc ttt aga ga-3′) (uppercase letters indicate the residues changing the wild-type Thr149 codon [ACA] to Pro [CCA]). To create the desired mutation, we used the gene replacement method of Datsenko and Wanner (8). Strain NR16191 (a cysM zcj-233::Tn10 derivative of BW25113/pKD46) was transformed with 200 ng of the cysB-T149P oligonucleotide by electroporation. Cells were plated on triazole-containing medium (VB-TRZ [see “Media and chemicals” above]), yielding a few thousand Trzr clones per transformation. Several of the Trzr transformants were then colony purified on VB-TRZ plates, and the presence of the T149P substitution in selected clones (strain NR16193) was confirmed by DNA sequencing using the PCR amplification primers cysB-diaA (5′-AAC GAT GTT CTG ATG GCG TC-3′) and cysB-diaB (5′-TCG CTA CAT AAA AGG TGC CG-3′). Sequencing was done in both directions and using the same primers, cysB-diaA and cysB-diaB. Strain NR16193 contains the zcj-233::Tn10 insertion (25, 31) located near cysB (cysB, 28.7 min [3]; zcj-233::Tn10, 29.5 min [31]). Using this linkage, the cysB(Con) was transferred from NR16193 to NR10836 by P1 transduction using double selection to triazole and tetracycline resistance on VB-TRZ plates supplemented with 15 μg/ml of tetracycline.
Chromosomal replacement of the E. coli ycbX gene with Vibrio cholerae VCA0924 and Vibrio parahaemolyticus VPA0411 ORFs.
The ycbX gene replacement procedure was based on the assumption that VCA0924 and VPA0411 ORFs may complement the HAP sensitivity of the E. coli ycbX mutant. We employed the PCR-based gene replacement method of Datsenko and Wanner (8) using the Vibrio VCA0924 and VPA0411 open reading frames as “HAP resistance cassettes”. Primers for amplification/replacement were designed to replace, precisely, the ycbX coding sequence with the VCA0924 or VPA0411 gene, without affecting the ycbX gene surrounding regions such as the ycbX promoter. A 2,089-bp PCR product containing the 1,989-bp VCA0924 ORF was amplified from genomic DNA of V. cholerae (ATCC, catalog number 17802D) using primers VCA0924-S (5′-gcg cgt cat gtc ata aag ttg agg gct tat ttt cat ttg agg acc gca cc ATG TTC TAT GAT GTT GCG CA-3′) and VCA0924-E (5′-agg ctg aaa ccg cag gtt aat gtt gac agc ttc agc ctc gaa cag gca gt CTA AAA CTC GAC ATC CAG ATC-3′), and a 1,918-bp PCR product containing the 1,818-bp VPA0411 ORF was amplified from genomic DNA of V. parahaemolyticus (ATCC, catalog number 39315D) using primers VPA0411-S (5′-gcg cgt cat gtc ata aag ttg agg gct tat ttt cat ttg agg acc gca cc ATG TCG CAA CCC GTT TTA TC-3′) and VPA0411-E (5′-agg ctg aaa ccg cag gtt aat gtt gac agc ttc agc ctc gaa cag gca gt TTA TTC CGT TAC CGT GAC ATC-3′) (lowercase letters indicate E. coli sequences, uppercase letters indicate Vibrio sequences, and the start and stop codons of the Vibrio ORFs are in bold). These PCR products were used to transform strain NR16262 (ΔycbX) carrying plasmid pKD46, using conditions described by Datsenko and Wanner (8). HAP-resistant recombinants were selected on minimal VB medium containing 10 μg/ml of HAP (under these conditions, ycbX mutants do not form colonies, whereas YcbX+ strains are recovered with 100% efficiency). To verify correct gene replacement events, genomic DNAs of several HAP-resistant clones were amplified with primers ycbX-seqA (5′-AAC CCT GCG TTG TTA TTG CG-3′) and ycbX-seqB (5′-TGA GGA TTT CGA GCA TAA CC-3′), corresponding to the left and right flanking sequences of the ycbX gene, and the product was sequenced in both directions using the same primers, ycbX-seqA and ycbX-seqB.
Cloning of VCA0924 and VPA0411 into pBluescript II SK(+).
The 1,089-bp PCR product containing the VCA0924 ORF or the 1,918-bp VPA0411 PCR product described above were blunt ended with T4 DNA polymerase and inserted into the SmaI site of pBluescript II SK(+) (Stratagene). Clones containing the VCA0924 or VPA0411 ORF (pBluescript-VCA0924 and pBluescript-VPA0411) in the same orientation as the lac promoter were used for the complementation experiments.
Spot test for HAP sensitivity.
Stationary-phase E. coli cultures grown in LB were diluted 30-fold in 10 ml of 0.9% NaCl in petri dishes and transferred to VB minimal plates using a multiprong replicator device (approximately 0.1 ml total per plate). After the spots had dried, a few microliters of a 20-μg/μl solution of HAP in DMSO was spotted onto the center of the plate. The plates were incubated overnight at 37° C and inspected the next day for zones of inhibition.
Liquid incubation tests for inhibition by HAP.
The test was performed as described earlier (17). Briefly, a single colony of each strain to be tested was inoculated into liquid LB and grown overnight at 37° C. 80 μl of each culture was inoculated into 40 ml of VB medium supplemented with 1 μg/ml of thiamine and 0.8 mM sodium thiosulfate, and incubated to reach mid-log-phase (OD600 = 0.2, ∼ 108 cells/ml). At this point, for each strain and each concentration of HAP to be tested, three 1-ml aliquots were transferred into glass culture tubes, and an appropriate amount of HAP (solution in DMSO) was added to each tube. Cultures were incubated for 2 h at 37°C on a rotating drum. The cultures were diluted 104-fold in 0.9% NaCl, and appropriate volumes of the dilution were plated on LB plates in triplicate to count surviving cells. Plates were incubated for 20 to 24 h at 37°C. The fraction of surviving cells was determined by dividing the average number of cells in HAP-exposed cultures by the average number in the nonexposed cultures. For each strain, this test was repeated two or three times, and average survival values were used to obtain the survival curves.
RESULTS
CysJ, but not the sulfite reductase CysJI, is involved in HAP resistance.
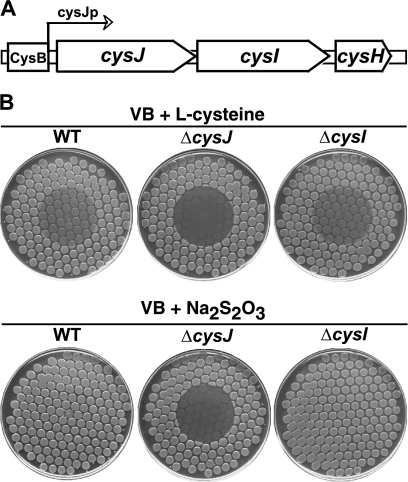
In Fig. 2B we show that cysJ cysteine auxotrophs are hypersensitive to HAP when tested in our standard plate assay. In this test, cells from an overnight culture are transferred, using a multiprong replicator device, to a minimal medium plate, followed by placement of a small drop of HAP on the center of the plate; upon overnight incubation, HAP-sensitive strains display a circle of inactivation or growth inhibition (17, 18). This sensitivity is most easily seen in the bottom row of Fig. 2B, where sodium thiosulfate was added as a nutritional supplement (cys mutants require an appropriate sulfur source such as l-cysteine or thiosulfate for growth on minimal medium). The HAP sensitivity of the cysJ mutant was not due to its defect in cysteine biosynthesis per se, because no other cysteine auxotrophs in our collection (cysD, cysN, cysH, and cysG) were found to be HAP sensitive (data not shown). Also, addition of cysteine did not alter the sensitivity of the strain (Fig. 2B), although addition of cysteine does sensitize the wild-type strain. (This related effect is discussed in the next section.) The main established function of CysJ is that of an essential component of the enzyme sulfite reductase. Sulfite reductase, a complex of CysJ and CysI proteins (CysJ8I4), reduces sulfite to sulfide, which then serves as the sulfur donor for l-cysteine biosynthesis (21). In the complex, CysJ is an NADPH:flavin oxidoreductase that passages electrons from NADPH through its flavin module to CysI serohemoprotein, which uses them to achieve the six-electron reduction of sulfite (21). cysI and cysJ comprise, along with cysH, an operon (Fig. 2A). The cysH gene encodes 3′-phosphoadenylylsulfate reductase, an enzyme required for the production of sulfite from sulfate (21). Interestingly, we found that deletion of cysI (or cysH) did not cause any HAP sensitivity (Fig. 2B). This result indicates that the flavin reductase CysJ per se, and not the sulfite reductase CysJI, is involved in the HAP tolerance mechanism.
FIG. 2.
(A) Organization of the E. coli cysJIH operon. The binding element for the CysB positive regulator of the operon is boxed. CysJp indicates the promoter of the operon. (B) HAP sensitivity of the wild-type (WT) strain (NR10836) and its cysJ (NR16043) and cysI (NR16069) derivatives on two different types of media. Spot tests are shown to reveal the HAP-induced inactivation of each of the strains. Cells were plated using a multiprong replicator device to minimal medium plates supplemented either with 0.33 mM l-cysteine or with 0.8 mM sodium thiosulfate as indicated, and 50 μg of HAP was spotted onto the center of each plate. The plates are shown as they appear after 24 h of incubation. In these assays, the edges of the inhibition zones are typically sharp and well defined. The reason for the sharpness is not fully understood but presumably reflects the precise mode of killing by HAP (not yet fully established) along with certain regulatory systems that appear to be triggered by exposure to HAP (unpublished data).
HAP sensitization by l-cysteine is due to CysB-mediated repression of cysJ.
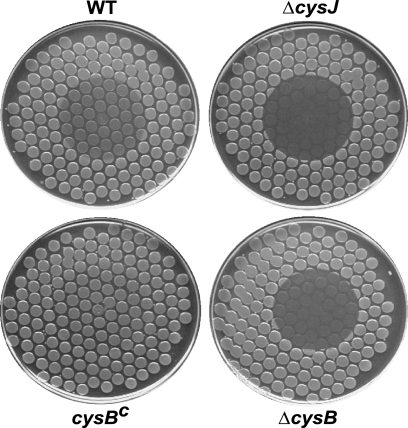
Expression of the genes of the cys regulon, including that of the cysJIH operon (Fig. 2A), is positively controlled by CysB protein (CysB4), a member of the LysR family of transcriptional regulators (21). CysB, in turn, is activated by N-acetylserine derived from O-acetylserine, a direct l-cysteine precursor (21). l-Cysteine exerts negative feedback on the production of O- and N-acetylserine, reducing the amount of active CysB and, in this manner, downregulating the cys regulon, including cysJIH (21). Indeed, as shown in Salmonella, addition of l-cysteine (0.2 mM) to the growth medium led to a complete lack of detectable sulfite reductase (CysJI) activity in cell extracts, while the activity was unaffected in cells grown on media containing sulfate (0.2 mM) or thiosulfate (0.25 mM) (9). In view of the sensitivity of the cysJ mutant to HAP, the inhibitory effect of l-cysteine on cysJ expression may provide a plausible explanation for the l-cysteine-mediated sensitization of wild-type strains, as seen in Fig. 2B. We further explored this mechanism by investigating the HAP sensitivity of a constitutive cysB [cysB(Con)] mutant. Such a mutant carries an altered CysB protein that promotes transcription of the cys genes even in the absence of its activator N-acetyl-l-serine (21). Using such a mutant, two predictions can be made: (i) a cysB-deficient mutant (in which transcription of cysJ is virtually absent) will be HAP sensitive, possibly as sensitive as the ΔcysJ mutant, and (ii) a cysB(Con) mutant (in which transcription of cysJ is largely constitutive) will not be sensitized by addition of l-cysteine. We constructed both ΔcysB and cysB(Con) mutants (see Materials and Methods) and tested their HAP sensitivities in the presence of l-cysteine. As shown in Fig. 3, the cysB deletion markedly sensitized the strain to HAP, to nearly the level observed in the cysJ strain, while the cysB(Con) mutant was markedly more HAP resistant than the wild-type strain.
FIG. 3.
HAP sensitivity of cysB strains. Spot test results are shown for a ΔcysB strain and a cysB-constitutive (cysBc) strain (NR16098 and NR16195, respectively), along with wild-type (NR10836) and ΔcysJ (NR16043) controls. Cells were plated using a multiprong replicator device to minimal medium plates supplemented with 0.33 mM l-cysteine, and 50 μg of HAP was spotted onto the center of each plate.
The action of CysJ in protection against HAP is MoCo and YcbX dependent.
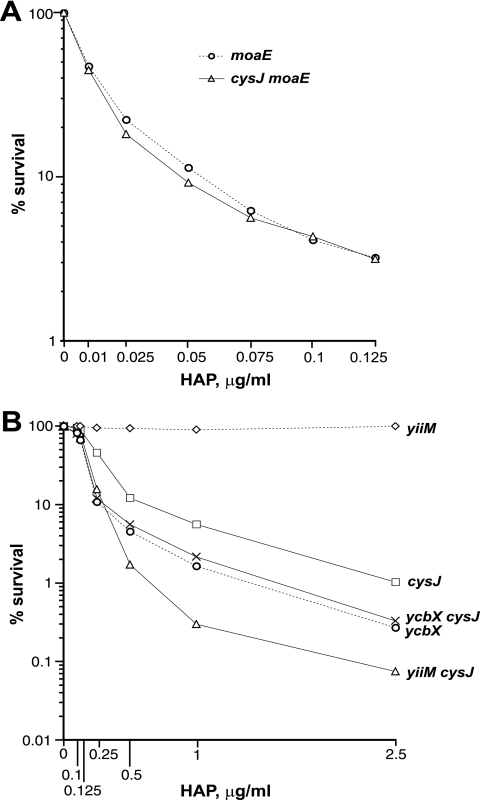
Referring back to Fig. 1, HAP detoxification in E. coli requires MoCo and proceeds in two subpathways dependent on the ycbX and yiiM genes. To investigate the genetic relationship between cysJ and these established pathways, we tested the HAP sensitivities of cysJ strains in combination with moaE (MoCo deficiency), and ycbX or yiiM defects. In Fig. 4, we present the results of liquid incubation tests that measure the inactivation (killing) of the strain after a 2-h incubation in the presence of HAP. It can be seen that inactivation of the cysJ moaE double mutant is essentially indistinguishable from that of the single moaE mutant (Fig. 4A), indicating that CysJ operates within the MoCo-dependent pathway. Likewise, the cysJ ycbX double mutant was equally as sensitive as the single ycbX mutant (Fig. 4B), indicating that CysJ operates in the ycbX subpathway. In contrast, the cysJ yiiM double mutant was significantly more sensitive than either single cysJ or yiiM mutant (Fig. 4B), indicating that CysJ is apparently not required for the yiiM subpathway. Also, spot tests (not shown) revealed that the single cysJ defect mirrored the single ycbX defect in its limited sensitivity to 2-amino-HAP (17). Finally, we found that the sensitizing effect of l-cysteine, mediated by suppression of CysJ, disappeared in moaE and ycbX mutants but not in the yiiM mutant (Fig. 5). The combined observations are fully consistent with CysJ protein participating, specifically, in the ycbX pathway of HAP resistance.
FIG. 4.
CysJ operates within the MoCo-dependent pathway of base analog detoxification (A) and within the ycbX-dependent pathway (B). Shown are results of survival tests after a 2-h exposure to the indicated concentrations of HAP in liquid medium. Strains used were NR16523 (moaE), NR16757 (cysJ moaE), NR15871 (yiiM), NR15873 (ycbX), NR16043 (cysJ), NR16087 (cysJ ycbX), and NR16085 (cysJ yiiM). See Materials and Methods for further details.
FIG. 5.
Lack of HAP sensitization by l-cysteine in moaE (NR15996) and ycbX (NR16262) mutants. In contrast, clear sensitization is observed in a yiiM strain (NR15870). Cells were plated using a multiprong replicator device to minimal medium plates supplemented with 0.8 mM sodium thiosulfate (VB) or with 0.33 mM l-cysteine, and 50 μg of HAP was spotted onto the center of each plate.
A functional YcbX-CysJ complex?
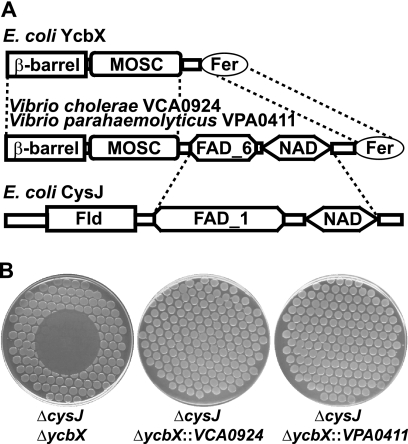
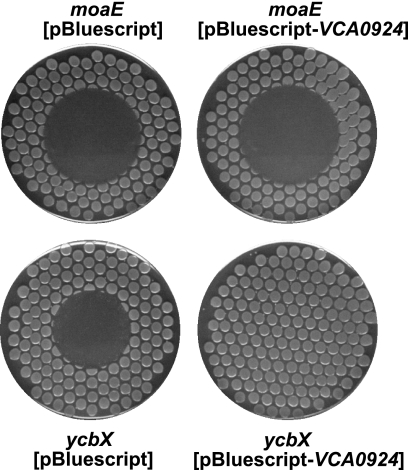
In database searches focusing on MOSC-containing proteins, we noted the presence in Vibrio species of apparent hybrid proteins combining elements of the CysJ and YcbX proteins in one polypeptide. As depicted in Fig. 6A, the YcbX protein contains three domains, the β-barrel, MOSC, and Fer (a ferredoxin-like domain) domains, while CysJ also contains three domains, the flavodoxin-like, FAD-binding, and NAD(P)(H)-binding domains. In the two Vibrio proteins, the FAD-NAD(P) unit of CysJ appears to be inserted between the MOSC and ferredoxin domains of YcbX (Fig. 6A). Assuming that the Vibrio proteins are also capable of detoxifying HAP, their existence is likely indicative of close interaction of the YcbX and CysJ units in the HAP detoxification reaction. We tested this idea by investigating whether the single Vibrio proteins might be able to complement the double ycbX cysJ deficiency. We amplified V. cholerae and V. parahaemolyticus ORFs VCA0924 and VPA0411, encoding the YcbX-CysJ-like “hybrid” proteins (Fig. 6A), and used them to replace the exact ycbX coding sequence on the E. coli chromosome (see Materials and Methods). With both constructs, full complementation of the ycbX and cysJ defects was achieved (Fig. 6B). The VCA0924 and VPA0411 genes were not able to suppress the HAP sensitivity of a MoCo-deficient (moaE) strain either as a single chromosomal copy (data not shown) or from a multicopy plasmid (Fig. 7), suggesting that VCA0924 and VPA0411 function in a MoCo-dependent manner.
FIG. 6.
(A) Domain architecture of the E. coli YcbX and CysJ proteins and the corresponding Vibrio proteins, VCA0924 and VPA0411, which may be regarded as hybrids of YcbX and CysJ. The presented domains as obtained by Pfam database searches (10; http://pfam.sanger.ac.uk/) are MOSC (MoCo sulfurase C-terminal domain, PF03473), β-barrel (MOSC N-terminal β-barrel domain, PF03476), Fer ([2Fe-2S] iron-sulfur cluster binding domain, PF00111), FAD_1 (oxidoreductase FAD-binding domain, PF00667), FAD_6 (oxidoreductase FAD-binding domain, PF00970), NAD (NAD-binding domain, PF00175), and FLD (flavodoxin, PF00258). (B) HAP sensitivities of strain NR16743 (ycbX cysJ) and its isogenic derivatives NR16920 and NR16921, which carry the VCA0924 and VPA0411 Vibrio genes, respectively, replacing the ycbX gene at its endogenous chromosomal location under expression from the ycbX promoter. See Materials and Methods for details. Cells were plated using a multiprong replicator device to minimal medium plates supplemented with 0.33 mM l-cysteine, and 100 μg of HAP was spotted onto the center of each plate.
FIG. 7.
Lack of suppression of the HAP sensitivity of a moaE mutant (NR16523) by a multicopy vector expressing the Vibrio VCA0924 gene. Note that the same plasmid fully complements the HAP sensitivity of a ycbX strain (NR16262) or a ycbX cysJ strain (not shown). Cells were plated using a multiprong replicator device to minimal medium plates supplemented with ampicillin, and 50 μg of HAP was spotted onto the center of each plate.
DISCUSSION
In this study we have demonstrated that a defect in the E. coli cysJ gene confers HAP sensitivity (Fig. 2). cysJ encodes the NADPH:flavin oxidoreductase component of the sulfite reductase complex (CysJ8I4), involved in the de novo biosynthesis of l-cysteine (21). However, a cysI mutant was not sensitive to HAP, indicating that CysJ itself, and not sulfite reductase, is required for HAP resistance. We further showed that cysJ operates within the MoCo-dependent detoxification pathway, specifically in the ycbX subpathway, and that CysJ and YcbX may be closely interacting partners in this pathway.
YcbX and YiiM were initially described as hypothetical proteins belonging to the MOSC superfamily (1). Based on primary structures, members of the MOSC superfamily were classified into five families (1): the MOS family (eukaryotic MoCo sulfurases), the FLJ22390 family (eukaryotic proteins), the PA3022 family (prokaryotic proteins, including YcbX, VCA0924, and VPA0411 [Fig. 6A]), the YiiM family (prokaryotic proteins), and the YuaD family (prokaryotic proteins). Except for MoCo sulfurase (MOS), all other members of the MOSC superfamily were hypothetical proteins without a known cellular role (1). Subsequently, our work showed that the YcbX and YiiM proteins are required for the MoCo-dependent conversion (reduction) of HAP to adenine in cell extracts and that they are likely MoCo-containing molybdoenzymes that can perform this reaction (17). In other relevant studies, two mammalian MOSC-containing proteins of the FLJ22390 family (mARC1 and mARC2) were characterized as MoCo-binding catalytic components of a mitochondrial benzamidoxime-reducing complex. These mARC proteins were found to be capable, in conjunction with cytochrome (cyt) b5 and NADH:cyt b5 reductase, of reducing the prodrug benzamidoxime and related N-hydroxylated compounds to the corresponding amine (12, 13). Thus, it is likely that the oxidoreductase function is a general characteristic of the MOSC protein superfamily, whereas MoCo sulfuration function may be limited to the MOS family proteins.
The reduction of HAP to adenine in the E. coli system (17) as well as the reduction of the N-hydroxylated agents by the mammalian system (12, 13) each requires a source of reducing equivalents. In the mammalian system these equivalents likely arise from NADH and are transferred via NADH:cyt b5 reductase and cyt b5 to the mARC molybdenum center (12, 13). Similarly, our data for E. coli defining CysJ as a critical component in HAP detoxification suggest that the function of CysJ flavin reductase is to provide electrons for the reduction of HAP. We propose that CysJ obtains electrons from NADPH and transfers them via its FAD and flavodoxin domains to the YcbX protein, presumably to its ferredoxin domain (Fig. 6A). Electrons then travel to the molybdenum center in the MOSC domain, where they are available for reduction of HAP to adenine. Such an electron transfer chain would benefit from a functional interaction between the CysJ and YcbX proteins. Our discovery of a functional “CysJ-YcbX hybrid” protein in Vibrio species and its ability to complement the ycbX cysJ double deficiency (Fig. 6) fully support this possibility. Indeed, a large-scale analysis of protein complexes of E. coli also reported a physical association between YcbX and CysJ proteins (4). Studies with purified YcbX and CysJ proteins in vitro in our laboratory are also in support of this conclusion (unpublished data).
It is of interest to note that while there is high similarity in the primary structures of MOSC domains when comparing YcbX and the VCA0924/VPA0411 Vibrio proteins (1), there is no significant corresponding similarity in the FAD domains of CysJ and Vibrio VCA0924/VPA0411 (see Fig. 6A and the Pfam database [10; http://pfam.sanger.ac.uk/]), indicating that the nature of the electron-providing component does not play a critical role in the specificity of MOSC-containing enzymes. In contrast, there is substantial divergence between the MOSC domains of YcbX and YiiM (1), a divergence that presumably underlies the different substrate specificities of the ycbX and yiiM pathways (17) (Fig. 1).
As CysJ does not appear to operate in the YiiM branch of the HAP detoxification scheme (Fig. 1 and 4), it is an interesting question what the electron-donating component for the yiiM pathway of base analog detoxification is. One other E. coli flavin reductase, Fre (33), may represent a candidate for this role. The one established role for Fre is in the activation of the tyrosyl radical of the enzyme ribonucleotide reductase (RNR) (7). In preliminary experiments we found fre mutants to grow slightly less vigorously than the corresponding wild-type strains, perhaps due to this particular defect. Nevertheless, the mutant appeared to be modestly sensitive to HAP, similar to the case for a yiiM mutant, consistent with the suggestion that it may operate within this pathway. The absence of a ferredoxin domain in YiiM (it contains only a short α-helical domain in addition to its MOSC domain) (1) suggests that electron transfer might proceed differently in this case, which is further consistent with the operation of a different electron donor.
Acknowledgments
We thank Matthew Longley and Giang Nguyen of the NIEHS for their critical reading of the manuscript for this paper.
This research was supported by funds allocated to project number Z01 ES050170 of the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Anantharaman, V., and L. Aravind. 2002. MOSC domains: ancient, predicted sulfur-carrier domains, present in diverse metal-sulfur cluster biosynthesis proteins including molybdenum cofactor sulfurases. FEMS Microbiol. Lett. 207:55-61. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, J. C. 1981. Induction of gene mutation in and cell transformation of mammalian cells by modified purines: 2-aminopurine and 6-N-hydroxylaminopurine. Proc. Natl. Acad. Sci. U. S. A. 78:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlyn, M. K. 1998. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 62:814-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butland, G., J. M. Peregrín-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 5.Clement, B., and T. Kunze. 1990. Hepatic microsomal N-hydroxylation of adenine to 6-N-hydroxylaminopurine. Biochem. Pharmacol. 39:925-933. [DOI] [PubMed] [Google Scholar]
- 6.Colyer, T. E., and N. M. Kredich. 1996. In vitro characterization of constitutive CysB proteins from Salmonella typhimurium. Mol. Microbiol. 21:247-256. [DOI] [PubMed] [Google Scholar]
- 7.Covès, J., V. Nivière, M. Eschenbrenner, and M. Fontecave. 1993. NADPH-sulfite reductase from Escherichia coli. A flavin reductase participating in the generation of the free radical of ribonucleotide reductase. J. Biol. Chem. 268:18604-18609. [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreyfuss, J., and K. J. Monty. 1963. Coincident repression of the reduction of 3′-phosphoadenosine 5′-phosphosulfate, sulfite, and thiosulfate in the cysteine pathway of Salmonella typhimurium. J. Biol. Chem. 238:3781-3783. [PubMed] [Google Scholar]
- 10.Finn, R. D., J. Mistry, B. Schuster-Böckler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freese, E. 1959. The specific mutagenic effect of base analogs on phage T4. J. Mol. Biol. 1:87-105. [Google Scholar]
- 12.Gruenewald, S., B. Wahl, F. Bittner, H. Hungeling, S. Kanzow, J. Kotthaus, U. Schwering, R. R. Mendel, and B. Clement. 2008. The fourth molybdenum containing enzyme mARC: cloning and involvement in the activation of N-hydroxylated prodrugs. J. Med. Chem. 51:8173-8177. [DOI] [PubMed] [Google Scholar]
- 13.Havemeyer, A., F. Bittner, S. Wollers, R. Mendel, T. Kunze, and B. Clement. 2006. Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 281:34796-34802. [DOI] [PubMed] [Google Scholar]
- 14.Hille, R. 2002. Molybdenum and tungsten in biology. Trends Biochem. Sci. 27:360-367. [DOI] [PubMed] [Google Scholar]
- 15.Khromov-Borisov, N. N. 1997. Naming the mutagenic nucleic acid base analogs: the Galatea syndrome. Mutat. Res. 379:95-103. [DOI] [PubMed] [Google Scholar]
- 16.Kisker, C., H. Schindelin, D. Baas, J. Retey, R. U. Meckenstock, and P. M. Kroneck. 1998. A structural comparison of molybdenum cofactor-containing enzymes. FEMS Microbiol. Rev. 22:503-521. [DOI] [PubMed] [Google Scholar]
- 17.Kozmin, S. G., P. Leroy, Y. I. Pavlov, and R. M. Schaaper. 2008. ycbX and yiiM, two novel determinants for resistance of E. coli to N-hydroxylated base analogs. Mol. Microbiol. 68:51-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozmin, S. G., Y. I. Pavlov, R. L. Dunn, and R. M. Schaaper. 2000. Hypersensitivity of Escherichia coli Δ(uvrB-bio) mutants to 6-hydroxylaminopurine and other base analogs is due to a defect in molybdenum cofactor biosynthesis. J. Bacteriol. 182:3361-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozmin, S. G., and R. M. Schaaper. 2007. Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in E. coli is independent of MobA function. Mutat. Res. 619:9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozmin, S. G., R. M. Schaaper, P. V. Shcherbakova, V. N. Kulikov, V. N. Noskov, M. L. Guetsova, V. V. Alenin, I. B. Rogozin, K. S. Makarova, and Y. I. Pavlov. 1998. Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat. Res. 402:41-50. [DOI] [PubMed] [Google Scholar]
- 21.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 22.Lieberman, I. 1956. Enzymatic synthesis of adenosine-5′-phosphate from inosine-5′-phosphate. J. Biol. Chem. 223:327-339. [PubMed] [Google Scholar]
- 23.Mendel, R. R., and F. Bittner. 2006. Cell biology of molybdenum. Biochim. Biophys. Acta 1763:621-635. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 25.Nichols, B. P., O. Shafiq, and V. Meiners. 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostrowski, J., G. Jagura-Burdzy, and N. M. Kredich. 1987. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 262:5999-6005. [PubMed] [Google Scholar]
- 27.Pavlov, Y. I., V. N. Noskov, E. K. Lange, E. V. Moiseeva, M. R. Pshenichnov, and N. N. Khromov-Borisov. 1991. The genetic activity of N6-hydroxyadenine and 2-amino-N6-hydroxyadenine in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae. Mutat. Res. 253:33-46. [DOI] [PubMed] [Google Scholar]
- 28.Rajagopalan, K. V. 1996. Biosynthesis of the molybdenum cofactor, p. 674-679. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 29.Schwarz, G., R. R. Mendel, and M. W. Ribbe. 2009. Molybdenum cofactors, enzymes and pathways. Nature 460:839-847. [DOI] [PubMed] [Google Scholar]
- 30.Simandan, T., J. Sun, and T. A. Dix. 1998. Oxidation of DNA bases, deoxyribonucleosides and homopolymers by peroxyl radicals. Biochem. J. 335:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sledziewska, E., and D. Hulanicka. 1978. Method of isolation of cysteine constitutive mutants of the cysteine regulon in Salmonella typhimurium. Mol. Gen. Genet. 165:289-293. [DOI] [PubMed] [Google Scholar]
- 33.Spyrou, G., E. Haggård-Ljungquist, M. Krook, H. Jörnvall, E. Nilsson, and P. Reichard. 1991. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J. Bacteriol. 173:3673-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stepchenkova, E. I., S. G. Kozmin, V. V. Alenin, and Y. I. Pavlov. 2005. Genome-wide screening for genes whose deletions confer sensitivity to mutagenic purine base analogs in yeast. BMC Genet. 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]