Abstract
Eukaryotic mRNA translation begins with recruitment of the 40S ribosome complex to the mRNA 5′ end through the eIF4F initiation complex binding to the 5′ m7G-mRNA cap. Spliced leader (SL) RNA trans splicing adds a trimethylguanosine (TMG) cap and a sequence, the SL, to the 5′ end of mRNAs. Efficient translation of TMG-capped mRNAs in nematodes requires the SL sequence. Here we define a core set of nucleotides and a stem-loop within the 22-nucleotide nematode SL that stimulate translation of mRNAs with a TMG cap. The structure and core nucleotides are conserved in other nematode SLs and correspond to regions of SL1 required for early Caenorhabditis elegans development. These SL elements do not facilitate translation of m7G-capped RNAs in nematodes or TMG-capped mRNAs in mammalian or plant translation systems. Similar stem-loop structures in phylogenetically diverse SLs are predicted. We show that the nematode eukaryotic translation initiation factor 4E/G (eIF4E/G) complex enables efficient translation of the TMG-SL RNAs in diverse in vitro translation systems. TMG-capped mRNA translation is determined by eIF4E/G interaction with the cap and the SL RNA, although the SL does not increase the affinity of eIF4E/G for capped RNA. These results suggest that the mRNA 5′ untranslated region (UTR) can play a positive and novel role in translation initiation through interaction with the eIF4E/G complex in nematodes and raise the issue of whether eIF4E/G-RNA interactions play a role in the translation of other eukaryotic mRNAs.
Cap-dependent translation initiation in eukaryotes is a complex process involving many factors and serves as the primary mechanism for eukaryotic translation (37, 44). The first step in the initiation process, recruitment of the m7G (7-methylguanosine)-capped mRNA to the ribosome, is widely considered the rate-limiting step. It begins with recognition of and binding to the m7G cap at the 5′ end of the mRNA by the eukaryotic translation initiation factor 4F (eIF4F) complex, which contains three proteins: eIF4E (a cap-binding protein), eIF4G (a scaffold protein with RNA binding sites), and eIF4A (an RNA helicase). eIF4G's interaction with eIF3, itself a multisubunit complex that interacts with the 40S ribosome, facilitates the actual recruitment of capped RNA to the ribosome. With the help of several other initiation factors, the small ribosomal subunit scans the mRNA from 5′ to 3′ until a translation initiation codon (AUG) in appropriate context is identified and an 80S ribosomal complex is formed, after which the first peptide bond is formed, thus ending the initiation process (37, 44). The AUG context can play an important role in the efficiency of translation initiation (23, 44). The length, structure, and presence of AUGs or open reading frames in the mRNA 5′ untranslated region (UTR) can negatively affect cap-dependent translation and ribosomal scanning. In general, long and highly structured 5′ UTRs, as well as upstream AUGs leading to short open reading frames, can impede ribosome scanning and lead to reduced translation (23, 44). In addition, 5′ UTRs less than 10 nucleotides (nt) in length are thought to be too short to enable preinitiation complex assembly and scanning (24). Thus, several attributes of the mRNA 5′ UTR are known to negatively affect translation initiation, whereas only the AUG context and the absence of negative elements are known to have a positive effect on translation initiation (44).
Two of the important mRNA features associated with cap-dependent translation, the cap and the 5′ UTR, are significantly altered by an RNA processing event known as spliced leader (SL) trans splicing (3, 8, 17, 26, 36, 47). This takes place in members of a diverse group of eukaryotic organisms, including some protozoa, sponges, cnidarians, chaetognaths, flatworms, nematodes, rotifers, crustaceans, and tunicates (17, 28, 39, 55, 56). In SL trans splicing, a separately transcribed small exon (16 to 51 nucleotides [nt]) with its own cap gets added to the 5′ end of pre-mRNAs. This produces mature mRNAs with a unique cap and a conserved sequence in the 5′ UTR. In metazoa, the m7G cap is replaced with a trimethylguanosine (TMG) cap (m2,2,7GpppN) (27, 30, 46, 49). In nematodes, ∼70% of all mRNAs are trans spliced and therefore have a TMG cap and an SL (2). In general, eukaryotic eIF4E proteins do not effectively recognize the TMG cap (35). This raises the issues of how the translation machinery in trans-splicing metazoa effectively recognizes TMG-capped trans-spliced mRNAs, what role the SL sequence plays in translation initiation, and how the conserved translation initiation machinery has adapted to effectively translate trans-spliced mRNAs.
Previous work has shown that efficient translation of TMG-capped messages in nematodes requires the SL sequence (22 nt) immediately downstream of the cap (5, 25, 29). In the current studies, we sought to understand the manner in which the SL enhanced the translation of TMG-capped mRNAs. Using a cell-free nematode in vitro translation system, we carried out mutational analyses that define the specific sequences in the SL that are required and sufficient for efficient translation of TMG-capped mRNAs. These analyses led to the discovery of a small, discrete stem-loop immediately adjacent to the TMG cap in trans-spliced messages required for efficient translation. Notably, the sequences involved in the base pairing of the stem are highly conserved in alternative SL sequences found in nematodes. We further show that the nematode eIF4E/G complex plays a major role in facilitating the SL enhancement of TMG-capped mRNA that likely occurs after the initial cap-binding step. The results demonstrate the importance of specific enhancing elements in the 5′ UTR and adaptation in the eIF4F complex necessary for optimal cap-dependent translation.
MATERIALS AND METHODS
Cell-free translation.
Whole-cell extracts were prepared from Ascaris embryos as previously described, except in some cases in which they were not dialyzed (25). Translation assays were carried out without nuclease treatment of the extracts under competitive translation conditions that mimicked translation in vivo (5, 25). Translation was carried out with the addition of reporter luciferase RNAs to the reaction mixture at 1 to 2.5 μg/ml, and translation was conducted for 40 to 60 min at 30°C. Luciferase assays were carried out as described previously (5, 25).
Extracts were depleted of the eIF4F complex essentially as described previously (10, 11). In brief, 200 μl of Ascaris embryo extract was incubated with 250 μl of m7GTP-Sepharose 4B beads (GE Healthcare, Piscataway, NJ) equilibrated in translation dialysis buffer (20 mM Tris-HCl [pH 7.8], 50 mM KCl, 0.2 mM EDTA, 1 mM dithiothreitol [DTT], and 20% glycerol) at 4°C for 15 min with slow rotation. The samples were centrifuged at 800 × g for 2 min at 4°C, and the supernatant (depleted extract) was collected and immediately frozen. To reconstitute the depleted extract with recombinant proteins, either Ascaris eIF4G-4E3 complex at ∼12.5 ng/μl or wheat germ eIF4F, -4A, and -4B complex at 175 ng/μl was mixed with an equal volume of depleted extract so that a 10-μl reconstituted reaction mixture contained 25 ng luciferase mRNA, ∼3 ng of Ascaris protein, or 43.75 ng of wheat germ protein or an equivalent volume of buffer, 2.5 μl of depleted extract, and 1× translation buffer (25). As a control for 4G and 4E activity, 4E binding peptide (4E-BP) (RIIYDRKFLMECRNSPV) was added to reconstitution reaction mixtures at a final concentration of 45.4 μM. In those cases, the protein was preincubated with peptide at 30°C for 10 min. Translation assays were then carried out as previously described (25).
Wheat germ translation assays supplemented with recombinant proteins were carried out in a manner similar to that described for Ascaris translation assays. Briefly, a 20-μl reaction mixture contained 125 mM KOAc, 2.6 mM MgOAc, 0.5 mM GTP, 0.5 mM ATP, 10 mM creatine phosphate, 50 μg/ml creatine kinase, 2 mM DTT, and 50 μM amino acid mix with 0.625 ng/μl of luciferase RNA, either 62.5 ng/μl of recombinant Ascaris eIF4G-4E3 or 42 ng/μl of recombinant wheat germ 4G-4E or the equivalent volume of buffer, and 2.5 μl of prepared wheat germ extract. Prior to addition of the extract, all other components were preincubated at 30°C for 5 min and returned to ice. Upon addition of the extract, reactions were incubated at 25°C for 2 h. Each reaction mixture was diluted at 1:25 into cold 1× passive lysis preparation, and luciferase activity measurements were obtained as previously described (18).
eIF4E-3 and eIF4G expression.
Ascaris eIF4E-3, which is the dominant isoform of eIF4E in early embryos and recognizes both monomethyl- and trimethylguanosine caps, has previously been described (25). Fragments of Ascaris eIF4G were generated using degenerate PCR and then 5′ and 3′ rapid amplification of cDNA ends (RACE) (GenBank accession no. GQ373389). By the use of the sequence from these portions of the eIF4G mRNA, the full-length Ascaris eIF4G open reading frame (see Fig. S1 in the supplemental material) was amplified by PCR (using GACGACGACAAGATCAGGTTCAAACTTACTATGTCCAGTAG and CGCGGGCGGCCGTTCTTAATTGAGCAACTCATAGAGATTTTC primers). Purified PCR products were treated with T4 DNA polymerase and simultaneously annealed into pET30a with an LIC Duet miniadaptor (Novagen, Madison, WI) to form a plasmid that would coexpress both proteins. The final construct, as confirmed by sequencing, consisted of eIF4G in the orf1 position with an N-terminal 6× His tag followed by the miniadaptor and then eIF4E-3 in the orf2 position. The plasmid, called 4G-Mini-4E3-pET30, was transformed into the Rosetta 2 DE3 expression strain (Novagen). A 1-liter volume of culture at an optical density at 600 nm (OD600) of 0.4 in LB-kanomycin (30 μg/ml) was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to achieve a final concentration of 0.2 mM and was incubated with shaking at 16°C for 24 h. Cells were harvested and stored at −80°C. The cell pellet was resuspended in 20 ml of lysis buffer (20 mM HEPES-KOH [pH 7.5], 300 mM urea, 200 mM NaCl, 100 mM KCl, 10% glycerol, 10 mM imidazole, 1 mM DTT, 2 Complete Mini EDTA-free protease inhibitor tablets [Roche, Indianapolis, IN], and 1 μg/ml lysozyme) and incubated on ice for 30 min. The lysate was then sonicated on ice for 10 min at power setting 7.5 using 10-s pulses with 20-s rests (Misonix Sonicator 3000; Fisher Scientific). The sonicated sample was clarified by centrifugation at 16,000 × g for 20 min at 4°C. The resulting supernatant was incubated with 1 ml nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen) that had been equilibrated with His wash buffer (20 mM HEPES-KOH [pH 7.5], 300 mM urea, 200 mM NaCl, 100 mM KCl, and 10% glycerol) for 1 h at 4°C on a nutator. Protein was eluted from the Ni-NTA beads by using a stepwise gradient of imidazole concentrations in His wash buffer. Eluates (1 ml) were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the three eluates containing the eIF4E/G complex were collected. The sample buffer was exchanged using buffer-20 (20 mM HEPES-KOH [pH 7.6], 20 mM KCl, 0.1 mM EDTA, 1 mM DTT, and 10% glycerol) and an Amicon Ultra-4 centrifugal filter device (Millipore) (100-K cutoff). SDS-PAGE analysis revealed the two protein bands of interest (eIF4G at ∼170 kDa and eIF4E-3 at ∼26 kDa), constituting ∼40% of the total protein in the sample.
RNA preparation.
Reporter RNAs (Renilla or Gaussia luciferase with an 85-nt adenylate tail) were transcribed from PCR templates by the use of T7 RNA polymerase and an Ambion Megascript kit and purified as previously described (5, 25). Cap priming was carried out at a ratio of 8:1 for cap:GTP by the use of m7GpppG or m2,2,7GpppG prepared as described previously (5, 25). Transcription reaction mixtures were treated with DNase I and extracted with TRIzol (Invitrogen, Carlsbad, CA), and the RNAs were precipitated twice, once with isopropanol and then with ammonium acetate-ethanol. Precipitated RNAs were further washed with 70% ethanol, dissolved in water, quantitated spectrophotometrically, and examined by agarose-formaldehyde denaturing gel electrophoresis. The 5′ UTR sequences used are illustrated in the figures for each experiment (shown as DNA sequences), and the 3′ UTRs were as previously described (5, 25).
SL mutations.
Blocks of random mutations in the RNAs were introduced using primers that incorporated N into PCR templates at specific positions, resulting in transcribed RNAs with all four nucleotides represented at the N position. Specific nucleotide substitutions were prepared using PCR primers to generate mutant PCR templates, with the corresponding mutant RNAs transcribed in vitro as described above.
RNA stability and 5′ monophosphate RNA analysis.
32P-labeled transcripts were synthesized using Promega's Riboprobe in vitro transcription system (Madison, WI) as described previously (5, 25). For stability analyses, uniformly labeled RNAs were added to the cell-free translation system, purified at different time points, and electrophoretically separated under denaturing conditions, and the amount of RNA remaining was determined by phosphorimager analysis using a Molecular Dynamics STORM 860 phosphorimager and ImageQuant software. Monophosphate RNAs (5′) were produced using RNA priming with a GMP:GTP ratio of 40:1. Dcp2 decapping of an mRNA in the extract would generate an RNA with a 5′ monophosphate (pGNx... versus TMGpppGNx.....). Monophosphate RNA (5′) was used to mimic decapped RNA in stability assays. For analysis of the amount of 5′ monosphosphate RNAs produced during translation in the cell-free system, RNAs were isolated at various time points and treated with Terminator 5′ phosphate-dependent exonuclease (Epicentre, Madison, WI) and the RNAs were analyzed and quantified by phosphorimager analysis of denaturing gel electrophoresis separations. Terminator is a 5′ → 3′ exonuclease (Epicentre) that is highly specific for RNAs with a 5′ monophosphate and does not degrade capped, 5′ triphosphate, or diphosphate RNA (52).
SPR.
The binding of proteins to short RNAs was analyzed using a BIACORE 3000 surface plasmon resonance (SPR) instrument (Biacore, Uppsala, Sweden) and Sensor Chip SA (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The chip was conditioned by three injections of 100 μl of 1 M NaCl-50 mM NaOH at a flow rate of 100 μl/minute. HBS-P buffer (GE Healthcare Bio-Sciences AB) (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 0.005% Surfactant P-20) was used as a running buffer at a flow rate of 20 μl/min. Immobilization of ligand RNA was a two-step process. First, 40 μl of 2 μM biotinylated oligo(dT)23 was immobilized on all four channels (saturated response). Then, 30 μl of 2 μM RNA [consisting of 41-nt RNA with 20 additional As for a 3′ poly(A) tail] was bound to each channel through dT/A base pairing. The responses for each RNA immobilization were observed to be relatively similar for all channels (1,700 to 1,900 relative units). A 100-μl volume of purified recombinant Ascaris 4E-3/4G complex at various concentrations (1.73 nM, 3.45 nM, 6.9 nM, 13.8 nM, and 27.6 nM) was injected on all four channels, and response was measured. The chip was regenerated using 40 μl of 6 M guanidine-HCl (pH 2.2). The data were analyzed using Scrubber-2 software (University of Utah, Salt Lake City, UT), and the bulk refractive index was floated to fit the data.
Sucrose gradient analysis of translation reactions.
Translation reactions were analyzed on sucrose gradients prepared in polypropylene tubes (Seton Scientific, Los Gatos, CA) (14 by 89 mm) by layering equal volumes of 5% sucrose on 25% sucrose (5% or 25% sucrose, 50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml cycloheximide). Gradients were formed using a Gradient Station apparatus (Biocomp, Fredericton, New Brunswick, Canada). Cell-free translation reactions were performed using micrococcal nuclease-treated Ascaris extract scaled up to 200 μl and preincubated with 0.5 mM cycloheximide for 5 min at 30°C. Translation reactions were initiated by the addition of 250 ng of universally 32P-labeled RNA. Following an incubation of 30 min at 30°C, reaction mixtures were diluted with an equal volume of cold homogenization buffer (100 mM Tris-HCl [pH 7.5], 100 mM NaCl, 40 mM MgCl2, 300 μg/ml cycloheximide, 100 μg/ml heparin) and 40 μl of 10× detergent solution (5% Trition X-100, 121 mM sodium deoxycholate). Samples were layered on top of the gradients and centrifuged at 36,000 rpm for 3 h at 4°C in a SW41 Ti rotor (Beckman Coulter, Inc., Fullerton, CA). Sucrose gradients were fractionated using a Gradient Station apparatus (Biocomp) and an FC203B fraction collector (Gilson, Inc., Middleton, WI). Fractions (350 μl) were collected, and radioactivity was detected using Cerenkov counting. During fractionation, the absorbance at 254 nm was recorded using a Pharmacia Biotech UV-MII lamp (1-A sensitivity) and RecIII recorder (GE Healthcare, Piscataway, NJ) (0.2 V, 20 mm/min).
RESULTS
SL sequence and translation of TMG-capped RNAs.
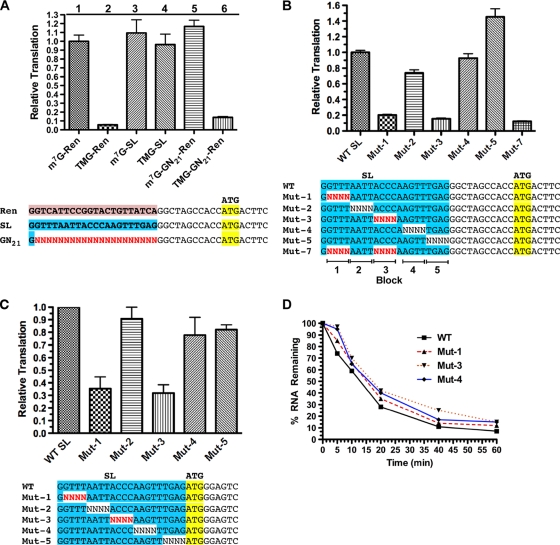
To examine the function of the SL sequence in translation, we used a nematode embryo cell-free translation system that is cap and poly(A)-tail dependent, exhibits cap-tail synergism, and reproduces characteristics of trans-spliced mRNA translation observed in vivo, including the ability of the SL sequence to stimulate translation of TMG-capped RNAs (5, 25). As previously observed, TMG-capped RNA translation is less efficient than m7G-capped RNA translation in the nematode cell-free system (see Fig. 1A; compare columns 1 and 2). Replacement of the first 22 nt of the test RNA with the nematode SL1 sequence significantly enhanced translation of the TMG-capped RNA but did not significantly stimulate translation of m7G-capped RNA (Fig. 1A; compare columns 1 and 3 to columns 2 and 4). Efficient translation of the TMG-capped RNA appears to be dependent on sequences within the spliced leader, since substitution of random sequences for the SL did not facilitate translation of the TMG-capped RNA (Fig. 1A; compare columns 5 and 6). Overall, these data show that the SL is needed for efficient translation of TMG-capped mRNAs.
FIG. 1.
Two regions within the SL are required for optimal translation of TMG-capped RNAs. (A) The spliced leader sequence is required for efficient translation of TMG-capped RNAs. Renilla luciferase reporter RNAs with a 3′ 85-nt polyadenylate tail were transcribed in vitro and translated in an Ascaris embryo cell-free translation system. RNA differences included the type of cap (m7G, m7GpppG; TMG, m2,2,7GpppG) and sequences in the 5′ UTR as illustrated. Translation levels were set at 1 for the m7GpppG-capped RNA with the Renilla 5′ UTR (m7G-Ren). Translation of this m7G-capped Renilla RNA resulted in ∼1.5 × 107 relative light units (RLUs)/ng of RNA in the extracts. (B) Random block mutations within the SL define two regions that contribute to translation of TMG-capped Renilla luciferase RNA. N, all four nucleotides represented at these positions in the DNA template and translated RNA. (C) Translation of Gaussia luciferase RNA with the SL and its mutants immediately adjacent to the AUG codon defines the same two regions within the SL important for translation of TMG-capped RNA. (D) Stability of RNAs with block mutations in the SL is not decreased during translation in the extracts. The percentages of 1,100-nt Renilla RNA remaining were calculated as a function of time and plotted. Note that RNAs with mutations in the SL and decreased translation (Mut-1 and Mut-3) did not exhibit reduced RNA stability during the translation period.
Key regions of the SL required for TMG cap translation.
To determine the specific sequences within the spliced leader that contribute to translation of TMG-capped mRNAs, we tested a series of random 4-nt substitutions across the entire SL in the in vitro translation system (Fig. 1B). Mutations in two regions of the SL, Mut-1 and Mut-3, led to significant reductions in the translation of the TMG-capped luciferase reporter (Fig. 1B). Similar results were obtained using a second luciferase reporter RNA (Gaussia luciferase) with the SL sequence placed immediately upstream of the reporter AUG (Fig. 1C). The configuration of the SL relative to the AUG in these two 5′ UTRs corresponds to the two major types of native trans-spliced 5′ UTRs on nematode mRNAs (25). We next determined whether the SL mutations in blocks 1 and 3 were additive by evaluating translation of a double mutant, Mut-7. The reduction in translation observed in Mut-7 was only slightly greater than that seen with either Mut-1 or Mut-3 (Fig. 1B). To determine whether the Mut-1 and Mut-3 mutations reduced luciferase expression by decreasing mRNA stability, we examined the decay of the reporter RNAs during translation. Mut-1 and Mut-3 RNAs were slightly more stable than RNA with the wild-type (WT) SL during translation (Fig. 1D). Similar results were obtained for the second Gaussia luciferase reporter RNA (data not shown). Therefore, mutations in the SL inhibit translation and not RNA stability.
To test the possibility that mutations in blocks 1 or 3 led to decapping of mutant versus wild-type SL RNA, we examined the rate of decay of 5′ monophosphate RNAs (the products of RNA decapping) and compared the amounts of decapped RNAs derived from wild-type or mutant SL mRNAs that accumulated during translation in the extracts. These analyses demonstrated that decapping of mRNAs with the wild-type SL was not different from decapping of mRNAs with the mutant SLs (see Fig. S2 in the supplemental material). We conclude from these data that SL nucleotides within blocks 1 and 3 promote translation of capped mRNAs.
Specific SL nucleotides are necessary and sufficient for translation of TMG-capped RNAs.
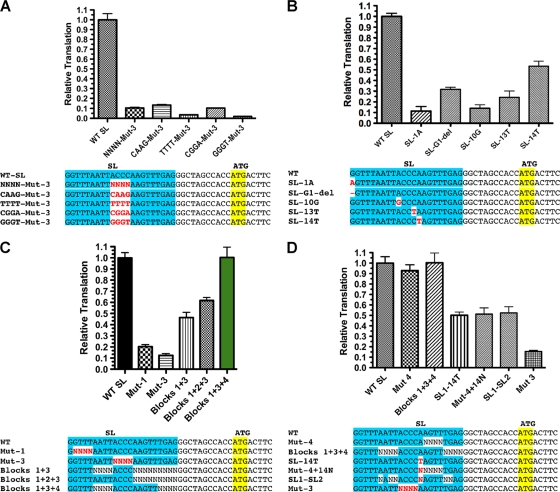
To determine the contribution of individual nucleotides within blocks 1 and 3, we examined random 2-nt mutations. All 2-nt sets within these regions contribute to the translation of TMG-capped mRNAs (see Fig. S3A and B in the supplemental material). We used blocks of random nucleotides in a nonbiased and rapid approach to define sequences that contribute to this translation effect. However, as each 4-nt block of substitutions represents 256 different RNA transcripts, we predicted that the observed reductions in translation are likely underestimates of the total effect of altering these nucleotides. We next examined several specific nucleotide mutations in block 3 for their effect on translation. Specific nucleotide substitutions led to a significant reduction in translation (Fig. 2A; GGGT-Mut-3 levels were only 3% of wild-type levels). Single nucleotide substitutions in block 3 and SL nt 14 (not previously tested as part of a Renilla block mutation) also greatly reduced translation (Fig. 2B). Finally, substitution of the first nucleotide of the SL (G → A) or deletion of this G led to a reduction in translation (Fig. 2B). Thus, specific nucleotides within the SL are required for translation of TMG-capped RNA in nematodes.
FIG. 2.
Sequences necessary and sufficient for optimal translation of TMG-capped RNAs. (A) Fixed nucleotide substitutions within block 3 further reduced translation of TMG-capped RNAs. For the comparisons, WT SL translation was set to 1. (B) Single nucleotide substitutions significantly reduce translation of TMG-capped RNAs. (C) Sequences within blocks 1, 3, and 4 are necessary and sufficient for optimal translation of TMG-capped RNAs. (D) Nucleotide 14 is the key residue in block 4 required for optimal translation of TMG-capped RNAs. Experiments were carried out as described for Fig. 1.
We next determined which SL sequences were sufficient for optimal translation of a TMG-capped RNA by comparing the translation of RNAs with fixed regions of SL sequence combined with other randomized regions (Fig. 2C). Although mutations in blocks 1 and 3 of the SL led to the most dramatic reductions in translation, these two regions alone were not able to fully rescue translation of TMG-capped RNA. However, blocks 1, 3, and 4 that included the native nucleotide 14 were able to translate TMG-capped RNA as efficiently as the wild-type SL sequence. The key block 4 residue was the A at nucleotide 14 (Fig. 2D and data not shown). Thus, the core SL sequence sufficient for optimal translation when the TMG cap is present is GGUUUNNNNACCCANNNNNNNN.
Core SL sequences contribute to the RNA secondary structure required for translation of TMG-capped RNA.
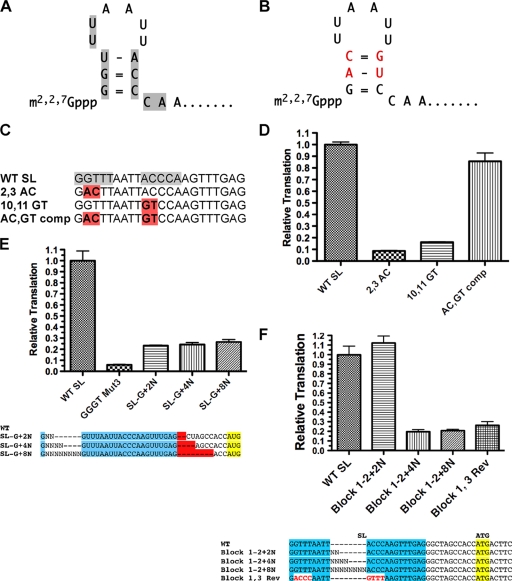
Examination of the SL sequence led to the identification of a potential stem-loop structure in the 5′ end of the SL (Fig. 3A). To determine whether the stem contributed to facilitating translation of TMG-capped RNAs, we tested RNAs in which each side of the stem was altered to disrupt potential base pairing along with a double mutant that restored the stem base pairing (Fig. 3B). The mutations that disrupted the putative stem led to significant reductions in translation, but when the two mutations were combined to restore base pairing, translation was also restored (Fig. 3D). As is consistent with these observations, selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (33, 51) demonstrated that some of the core SL sequences were constrained and that mutations in block 3 (ACCC → GGGU) led to increased flexibility in blocks 1 and 3 (see Fig. S4 in the supplemental material). Together, these data demonstrate that a small stem-loop at the 5′ end of the SL plays an important role in translation of TMG-capped RNAs.
FIG. 3.
Conserved SL sequences contribute to a 5′ stem-loop, and spacing of sequences within the SL are required for efficient translation of TMG-capped RNA. (A) Predicted 5′ stem-loop in the nematode SL1. Gray shading highlights the SL residues that contribute to the efficient translation of TMG-capped mRNAs. (B) Mutations in both stems of the SL that together would retain base pairing in the stem. Red-shaded residue sequences represent the mutated residues. (C) Designations and alignment of SL mutations that alter and reconstitute base pairing in the SL stem. Gray shading shows key sequences in the SL required for translation of TMG-capped RNA, and red shading denotes the nucleotide mutations. (D) Translation of RNAs with SL mutations that disrupt and then reconstitute the 5′ stem of the SL. (E) Block 1 sequences must be within 2 nt of the TMG to enable efficient translation of TMG-capped RNA. GGGT Mut3 is represented as described for Fig. 2C. Insertion mutations (e.g., +2N) were designed and compared with compensating deletions (e.g., −2N) following the SL to maintain a constant length in the 5′ UTR for translation comparisons. The plotted values were obtained by dividing the translation level of the “SL-G+XN” value by the “−XN” value (see Fig. S3 in the supplemental material for the illustration of the raw data). (F) The spacing of sequences between blocks 2 and 3 in the loop of the SL is important for efficient translation of TMG-capped RNA. Experiments were carried out as described for Fig. 1.
The position and spacing of the SL determine its activity.
We next examined whether spacing between the stem and block 1 nucleotides is important. Separation of the TMG cap and block 1 by as little as 2 random nucleotides dramatically reduced translation (Fig. 3E). We next examined the importance of spacing between the two key blocks of residues (blocks 1 and 3) by evaluating the effect of increasing the distance between these elements. Spacing of more than 2 nt significantly reduced the overall translation (Fig. 3F). We conclude that spacing between the TMG cap, block 1, and block 3 is important to translation, which in turn suggests that the loop, its length, and its location with respect to the cap are important for translation.
The nematode eIF4E/G complex, but not the wheat germ eIF4F complex, enables translation of TMG-SL RNAs.
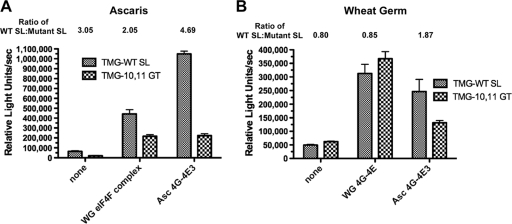
To investigate specific nematode proteins that might be involved in this translation effect, we first asked whether components of the nematode eIF4F complex were able to facilitate the enhanced translation of TMG-SL mRNAs. We depleted the eIF4F complex (typically eIF4E, eIF4G, and eIF4A) from Ascaris extracts by the use of m7GTP-Sepharose affinity chromatography (10, 11). Western blot analysis of the depleted extract demonstrated that the majority of eIF4G and the three Ascaris eIF4E isoforms (eIF4E-1, eIF4E-3, and eIF4E-4) were depleted from the extracts (see Fig. S5 in the supplemental material). m7GTP-Sepharose depletion of the extracts reduced translation competence to 1% of the nondepleted extract levels (data not shown). Reconstitution of the depleted extract with purified recombinant Ascaris eIF4E-3 and eIF4G, the two major components of eIF4F complex, increased translation 25-fold (Fig. 4A). Notably, eIF4E/G rescue of translation led to the translation profile observed with nondepleted extracts: significant translation of TMG-SL mRNA but limited translation of TMG-capped RNAs with SL mutations (Fig. 4A). 4E binding proteins and peptides derived from these proteins, namely, 4E binding peptide (4E-BP), compete with eIF4G for binding to eIF4E. Preincubation of Ascaris eIF4E/G complex with a 4E-BP peptide (RIIYDRKFLMECRNSPV) prior to their addition to the extract resulted in ∼95% loss of reconstituted translation, indicating that both the complex and interaction between the two proteins were required for reconstitution (data not shown). Reconstitution of the m7GTP-Sepharose-depleted Ascaris extract with wheat germ eIF4F complex (eIF4E, eIF4G, and eIF4A) did not result in the same effect for TMG-capped mRNAs (compare the ratio of WT SL/mutant SL for wheat germ [2.05] to that determined for Ascaris extracts [4.69]). These data suggest that the Ascaris eIF4E-3/G complex specifically contributed to the translation of TMG-SL mRNAs. We also added the Ascaris eIF4E-3/G complex to wheat germ extracts, for which levels of TMG-mediated translation of WT and mutant SL mRNAs are similar (Fig. 4B and Fig. 5A). However, the addition of Ascaris eIF4E-3/G complex increased translation of the TMG-capped WT SL mRNA nearly 2-fold (Fig. 4B). Together, these results suggest that the nematode eIF4E-3/G complex is one of the major determinants for the translational enhancement of TMG-capped mRNA by the SL stem-loop.
FIG. 4.
The Ascaris eIF4E/G complex enabled efficient translation of TMG-capped RNA in the context of the WT spliced leader, but wheat germ eIF4F did not. (A) Depletion and reconstitution of Ascaris extracts with eIF4E/G. Note that reconstitution of eIF4F-depleted Ascaris extracts with Ascaris eIF4G/E recapitulated the “SL effect” and that efficient translation of a TMG-capped RNA required an intact SL. Note also that translation in nondepleted extracts resulted in a value of ∼7 × 106 relative light units/s. (B) Addition of eIF4E/G to wheat germ extracts. Note that in wheat germ translation extracts (B), translation of TMG capped mRNAs was very inefficient (<5% of m7G capped RNAs; see Fig. 5). WG, wheat germ; Asc, Ascaris. WG eIF4F complex included wheat germ eIF4E, eIF4G, eIF4A, and eIF4B.
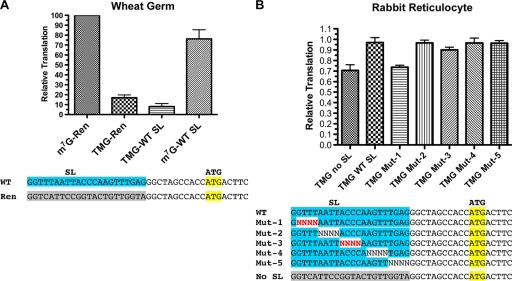
FIG. 5.
SL-enhanced translation of TMG-capped RNAs was nematode specific. (A) The nematode spliced leader did not enhance translation of a TMG-capped Renilla reporter RNA in a wheat germ cell-free system. (B) The nematode spliced leader and mutations within the SL did not significantly enhance or affect translation of a TMG-capped Renilla reporter RNA in a heterologous rabbit reticulocyte translation system. Experiments were carried out as described for Fig. 1. Red-shaded characters represent random nucleotides whose presence led to a reduction in nematode translation of TMG-capped RNAs.
The contribution of these nematode initiation factors to the translation of TMG-capped RNAs suggests that the SL affects translation initiation. To confirm this, we examined the kinetics of translation and the formation of 80S ribosomes on TMG-capped RNAs. Time course translation experiments revealed that the difference between WT and mutant SL is a difference in the rate of translation (see Fig. S6A and B in the supplemental material), which indicates that the initiation processes for these two messages are different (34, 40). We also observed that more 80S ribosomes accumulate on the wild-type SL mRNA (see Fig. S6B in the supplemental material). Overall, these data confirm that the SL's enhancement of translation takes place at the initiation step. Unfortunately, using sucrose gradients with Ascaris extract, we were unable to resolve or trap 43S ribosome-loaded mRNAs, so we could not determine whether translation is affected before or after 43S complex loading.
SL does not affect the affinity of eIF4E/G for capped RNA.
One hypothesis addressing how the nematode eIF4E/G complex works with the SL is that the SL stem-loop enhances eIF4E/G affinity for TMG-capped RNA. In order to examine this hypothesis, we used surface plasmon resonance (SPR) (32, 41) and different TMG-capped, polyadenylated small RNAs annealed to 5′ biotin-oligo(dT) immobilized on a streptavidin chip (see Materials and Methods). The recombinant Ascaris eIF4E-3/G complex showed the same relative affinity for the WT SL, the 10,11 GT Mut 3 SL, and the stem-loop compensatory AC,GT Mut SL RNAs (Table 1; also see Fig. S7A in the supplemental material). These binding data are in agreement with translation experiments demonstrating that the trimethylguanosine cap analog, which directly blocks capped-RNA recruitment to eIF4F (24), does not differentially inhibit TMG-capped messages with or without the SL (see Fig. S7B in the supplemental material). UV cross-linking experiments showed that recombinant Ascaris eIF4E-3 alone did not have an increased affinity for TMG-capped RNAs in the presence of the SL stem-loop (see Fig. S7C in the supplemental material). Similar experiments using filter binding and other SPR experiments confirmed these results (data not shown). Overall, these data suggest that the SL enhancement of translation does not involve the cap-binding affinity of the eIF4E/G complex for the RNA. In the wheat germ experiments represented by Fig. 4B, the Ascaris eIF4E-3/G complex provided a nearly 2-fold boost to TMG-WT SL mRNA translation. Thus, it remains possible that additional factors may be required to promote cap-binding or translation initiation.
TABLE 1.
Surface plasmon resonance data for Ascarid eIF4E-3/G complex with m2,2,7G-capped wild-type SL, stem mutant SL, and stem compensatory mutant SLa
| RNA | ka | kd | Rmax | Residual SD | KD (nM) |
|---|---|---|---|---|---|
| TMG-SL WT | 3.27 × 105 | 1.09 × 10−3 | 3,812 | 55.9 | 3.34 |
| TMG-10,11 (GT) | 2.88 × 105 | 1.32 × 10−3 | 3,408 | 54.1 | 4.57 |
| TMG-AC,GT | 2.248 × 105 | 1.22 × 10−3 | 3,788 | 56.0 | 5.45 |
TMG-SL WT, m2,2,7G-capped wild-type SL; TMG-10,11 (GT), stem mutant SL; TMG-AC,GT, stem compensatory mutant SL; ka, rate constant association; kd, rate constant dissociation; Rmax, theoretical response maximum for channel; Residual SD, residual standard deviation; KD, dissociation constant.
Specificity of TMG-SL translation enhancement.
The depletion and reconstitution of Ascaris extracts and addition of Ascaris eIF4E/G to wheat germ extracts suggest that this nematode complex is a key determinant in the translation enhancement of TMG-capped SL mRNAs and that the enhanced translation would be observed only in nematode extracts. We therefore asked whether the spliced leader sequence specifically enhanced translation of TMG-capped mRNAs in heterologous translation systems. mRNAs in eukaryotes typically have a m7G cap, and eIF4E, the cap-binding protein required for cap-dependent translation initiation, has a low affinity for the TMG cap (35, 43, 54). Translation in wheat germ extracts is dependent on the presence of cap; however, translation in rabbit reticulocytes is quite promiscuous, enabling some translation of TMG-capped RNAs (6). As shown in Fig. 5, the SL sequence does not play the same role in enhancing translation in other eukaryotic translation systems. Thus, the effect of the SL on TMG-capped translation is specific to the nematode translation system and is enabled by nematode trans-acting factors, including eIF4E/G.
Nematode SL core sequences are conserved.
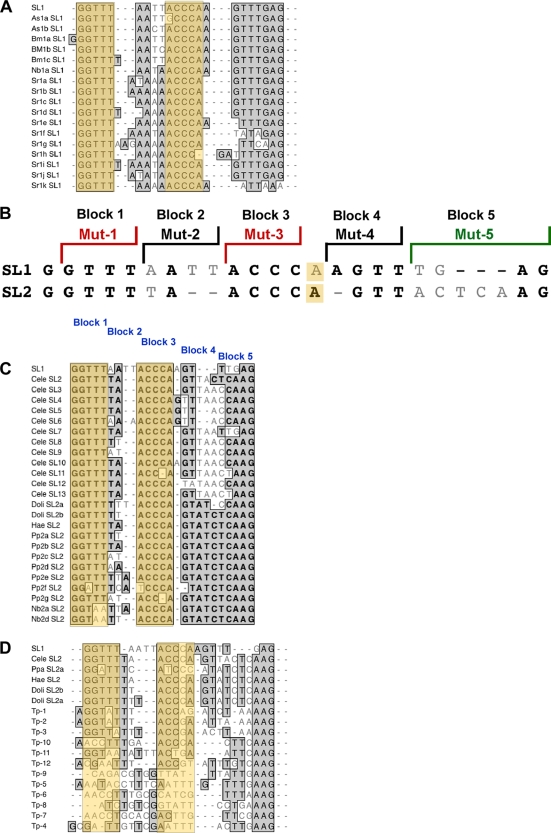
The SL1 sequence is highly conserved in rhabditid nematodes (Fig. 6A). A second spliced leader (SL2) and a number of related variants are present in Caenorhabditis elegans (19, 42). Alignment of Ascaris SL1, C. elegans SL2 (Fig. 6B), and several additional SL variants in nematodes (Fig. 6C) demonstrated that the key SL sequences are conserved (15). Notably, the conservation of the SL stem is supported by the existence of covariations in some of the SLs. For example, in Pp2f SL2, covariation in nt 3 and 9 maintains the stem structure. Similar covariations are also found in Bm1a SL1 and Ppa SL2a (Fig. 6A, C, and D).
FIG. 6.
Alignment and sequence conservation of nematode spliced leaders identified key SL regions associated with efficient translation of the TMG cap. (A) Rhabditid SL1 sequences. (B) Comparison of C. elegans SL1 and the primary SL2 sequence, illustrating the conserved sequences and regions mutated as shown in previous figures. (C) Rabditid SL2-like sequences. (D) Diversity of SL sequences in nematodes. Bold or shaded sequences represent identical residues, light gray residues represent variants, and dashes represent spacing to facilitate alignments. The orange shading depicts blocks 1 and 3 of the spliced leader. Nucleotide 14 is included here in block 3.
The major differences between SL1 and SL2 are represented by the nucleotides in blocks 2 and 5 (Fig. 6C). We examined several of these variant SLs and hybrid SLs in the Ascaris in vitro translation system (see Fig. S8A in the supplemental material). These analyses demonstrated that a hybrid SL that used the 5′ end of SL2 with the 3′ end of SL1 enabled efficient translation of the TMG cap (see 5′ SL2-SL1 3′ in Fig. S8A in the supplemental material) and that the 3′ end of the SL plays a minimal role in facilitating translation of the TMG cap (see Fig. S8A in the supplemental material). Overall, these data suggest that the residues (GGUUUNNNNACCCANNNNNNNN) are important for efficient translation of TMG-capped mRNAs not only in Ascaris species but also in many divergent nematode species.
In all spliced leaders identified, the three 3′-most nucleotides are invariably purines. We substituted three Ts for these purines but observed no effect on translation (see SL-TTT in Fig. S8A in the supplemental material). Recently, variant spliced leaders in what is considered a basal nematode lineage, Trichinella spiralis, were described (38). In contrast to the high conservation of SL1- and SL2-like sequences observed in other nematodes, Trichinella SLs are numerous and divergent. However, several of the spliced leaders (Tp1, Tp2, and Tp3) have portions of the conserved blocks that we have identified as important. These regions can contribute to the enhanced translation of TMG-capped mRNAs in Ascaris extracts (see Fig. S8B in the supplemental material).
DISCUSSION
Efficient translation of a TMG-capped mRNA in the Ascaris nematode requires the associated 22-nt trans-spliced SL1 sequence (5, 25, 29). We refer to this phenomenon as the “SL effect.” Here, we define the nucleotides within the highly conserved nematode SL1 spliced leader (nucleotides 1 to 5 and 10 to 14 [GGUUUNNNNACCCANNNNNNNN]) required for the SL effect (Fig. 1 and 2). The identified core nucleotides contribute to a functionally required stem-loop at the 5′ end of the SL (Fig. 3). The contribution of the SL to efficient translation is specific to both the TMG cap and nematodes (Fig. 1A and 5). Several lines of evidence demonstrate that the nematode eIF4E-3/G complex is a critical determinant for the SL effect. First, depletion of the complex from nematode extracts and its reconstitution with recombinant nematode eIF4E/G complex, but not the wheat germ complex, demonstrates the SL effect (Fig. 4A and B). Second, the SL effect is not observed in wheat germ translation extracts. Third, the addition of the nematode eIF4E/G complex, but not the wheat germ complex, to wheat germ extracts leads to the SL effect in wheat germ extracts. Overall, these data demonstrate that the nematode eIF4E/G complex works with the SL stem-loop and associated sequences to facilitate nematode translation of TMG-capped mRNAs.
Nematode SL stem-loop required for efficient translation of TMG-capped RNA.
Spacing of nucleotides within SL1 is important, with a minimal distance between the TMG cap and block 1 required (Fig. 3E and F). The length of the loop plays a role in translation, as insertion of more than 2 nt into the loop abrogates the enhancement of translation (Fig. 3F). The loop can be reduced by 2 nt, as illustrated by SL2, which is translated efficiently. Mutations on both sides of the stem that restore base pairing indicate that the stem is required for enhancing translation. The stem mutations introduced in this study were all transitions and thus maintained some character of the bases. While the structure is required, there also may be some sequence requirements. In addition, nucleotides within the loop and immediately 3′ of the stem also facilitate translation of TMG-capped RNAs (Fig. 3). The second guanine nucleotide of the TMG cap is also required for efficient TMG-capped RNA translation (Fig. 2B). A 3-bp stem would be predicted to be quite weak, so we hypothesize that the trimethylated guanosine cap may physically interact with the cytosine or adenosine immediately 3′ of the stem or the RNA structure to stabilize the stem and/or contribute to a novel structure. Overall, it is likely that the stem-loop structure, a higher-order structure, or its stability is dependent on some attribute specific to the TMG cap because m7G-capped RNA translation is not enhanced by the SL. Interestingly, high-throughput RNA sequencing of Ascaris mRNAs demonstrated that the majority of the mRNA sequences that extend 5′ to the SL stop predominantly at the C that is immediately downstream of the stem (data not shown). These data further suggest that a structural element is present at the 5′ end of the SL.
Mechanisms through which the eIF4E/4G complex might facilitate translation initiation.
Recognition of the mRNA cap by eIF4E (and recruitment of the ribosome through eIF4G in the eIF4F complex) is a rate-limiting step of translation (13, 37, 50). In contrast with most eukaryotic eIF4Es, Ascaris eIF4E-3 and several C. elegans eIF4Es recognize the TMG cap in addition to the m7G cap (20, 22, 25). Addition of Ascaris eIF4E-3 directly to translation extracts or m7GTP-Sepharose-depleted extracts did not significantly enhance translation or show the “SL effect.” We previously reported (using UV cross-linking) that (i) recombinant nematode eIF4E bound only to capped RNA and (ii) binding to the TMG cap appeared greater when the SL was present (25). However, efforts to further characterize and quantify the potential interaction between eIF4E-3 and TMG-SL RNA by the use of filter binding assays, surface plasmon resonance, and other approaches did not demonstrate that the core residues, GGUUUNNNNACCCANNNNNNNN, play a large role in enhancing cap binding (see Fig. S7B in the supplemental material). Overall, these data suggest that while nematode eIF4E may recognize the TMG cap, the SL and nematode eIF4G may be necessary to either stabilize the complex or enhance recruitment of the 43S complex for translation initiation. Therefore, it would be predicted that in the absence of the SL or with mutations in the SL, ribosome recruitment would be less efficient. The kinetics of translation and the formation of 80S ribosomes on wild-type and mutant SL mRNAs demonstrate that the SL does indeed enhance translation initiation (see Fig. S6 in the supplemental material). Experiments using cap analogs as competitive inhibitors of translation suggest the SL may contribute to translation of TMG-capped RNAs after the initial cap-binding step (see Fig. S7A in the supplemental material).
The data presented here suggest that the stem-loop structure does not increase eIF4E/4G's affinity for the capped messages (Fig. 4C and D). One possible model to explain the SL requirement for TMG-capped RNA translation is that the nematode eIF4G has adapted to interact, directly or indirectly, with the SL sequence to enhance recruitment of the 43S complex to the mRNA, form a stable 48S complex on the mRNA, enhance its scanning, or join to the 60S subunit. eIF4G is a known RNA binding protein, and interactions between eIF4E and eIF4G upon binding to the cap could enable conformation changes and interaction with the SL to facilitate translation (1, 14, 18, 21, 40, 53). A great deal of sequence and functional diversity between eIF4Gs exists (11, 13). Overall, Ascaris eIF4G shows only 15% identity and 12% similarity to wheat germ eIF4G and 21% identity and 14% similarity to human eIF4GI (see Fig. S1 in the supplemental material). While there is higher similarity in the core, central domain, it is difficult to identify regions of Ascaris eIF4G that might be unique and contribute to the SL effect. Experiments are under way to evaluate RNA binding of Ascaris eIF4G to the RNA stem-loop on trans-spliced mRNAs.
The specific spacing requirements necessary for efficient translation, including the distance from the TMG cap to the stem and size of the loop, suggest that a specific binding site is likely to be used by proteins. Our efforts to identify a trans-acting protein that binds TMG-SL RNA compared to m7G-SL RNA or the TMG cap alone have not yet identified any auxiliary proteins (data not shown). We were unable to detect recruitment or scanning of the 43S complex by the use of a variety of approaches and inhibitors in sucrose gradient analysis of translation extracts. Thus, we currently cannot determine whether the SL and eIF4E-3/G enhance recruitment of the 43S complex, increase ribosome scanning, or enhance formation of the 80S ribosome. Additional studies will be required to further define the detailed mechanism and contribution of nematode eIF4E/G in the enhancement of translation of TMG-capped mRNA by the SL.
Conservation of key SL1 nucleotides in other nematode spliced leaders.
C. elegans has a second spliced leader, SL2 (with a number of variants) (19, 42). Orthologous SL2 sequences are also present in several other nematodes (Fig. 6C). Alignment of these SL2-like sequences illustrates that the most highly conserved regions consist of the sequences defined here (GGUUUNNNNACCCANNNNNNNN) that exert the greatest effect in enhancing translation of TMG-capped RNA. We show here that the C. elegans SL2 sequence is efficiently translated in the Ascaris cell-free system (see Fig. S8 in the supplemental material).
Sequence alignments demonstrate that the 3′ end of the nematode SL exhibits considerable sequence variation (Fig. 6), and both mutation studies and translation assays of alternative and hybrid SL sequences on reporter RNAs show that these sequences do not play a significant role in translation (Fig. 1; see also Fig. S8 in the supplemental material). Although the final three nucleotides in all nematode SLs are purines, replacement of these purines with pyrimidines had little effect on translation. Many SLs are trans-spliced immediately upstream of the AUG mRNA coding region (25). In this context, these purines would contribute key nucleotides to the −1 to −3 in the context of eukaryotic initiation, where purines are known to play an important role in translation.
Recently, a number of highly divergent SLs in species of Trichinella, which is considered a basal member of the nematode lineage, were described (38). Interestingly, several key residues in blocks 1 and 3 of SL1 and SL2 in the rhabditid lineage are conserved in a few of the variable Trichinella spliced leaders. While the tested Trichinella spliced leaders supported TMG translation in the Ascaris cell-free system, they were at best only ∼50% as efficient (see Fig. S4 in the supplemental material). If the lineage is basal, the selection and conservation of these core sequences in blocks 1 and 3 may have given rise to SL1 and SL2 in some lineages. Alternatively, the sequence divergence in Trichinella SLs may indicate that this nematode lineage is highly derived.
The highly conserved rhabditid nematode SL1 sequence (see Fig. 6A) is a core, internal promoter element for transcription of SL RNA genes but is not a key element for the trans-splicing reaction (9, 16, 31). Here we show that the SL1 sequence conservation is also related to an important role in contributing to the overall translation of TMG-capped mRNAs. Therefore, at least two functional constraints maintain SL1 sequence conservation.
Role of key SL residues in vivo.
Maroney et al. first demonstrated that a 10-nt substitution (nt 7 to 16) within the SL sequence led to a decrease in translation in an Ascaris cell-free system (29). Ferguson and Rothman demonstrated that some sequences within the SL1 were critical for C. elegans embryonic survival, but their potential role in posttranscriptional gene expression was not defined (9). Interestingly, they showed that alterations within the core sequences identified here or altered spacing between these blocks was not able to rescue embryonic lethality. These alterations did not affect SL RNA expression or the trans-splicing reaction and therefore likely altered the postsplicing function of SL1. Nucleotide substitutions downstream of the ACCCA were still able to rescue embryonic lethality, which is consistent with our translation data. Notably, the SL1 regions required for C. elegans embryonic development correspond to the regions we have identified here as necessary for optimal translation of TMG-capped RNA in our cell-free system. Here we reveal the underlying mechanism for the requirement of these SL regions.
Evolution of trans splicing and the SL effect.
Efficient translation of TMG-capped mRNA in vivo or in vitro requires the spliced leader. This suggests that optimal translation of TMG-capped mRNAs evolved in the context of the downstream SL and that the initial appearance of trans splicing likely would have produced mRNAs that were not efficiently translated (5, 25). trans-spliced mRNAs are not translated more efficiently and the RNAs are not more stable in nematode embryos compared to m7G-capped non-trans-spliced mRNAs (5, 25). Therefore, the introduction and maintenance of trans splicing were most likely not related to an advantage in translation or mRNA stability but to some other advantage provided by this process. A number of trans-spliced metazoan mRNAs (apparently not the majority) are derived from polycistronic transcripts, and thus trans splicing contributes to their resolution into monocistronic mRNAs. If polycistronic transcripts were initially present in nematode lineages, trans splicing must have evolved either before or simultaneously with these atypical eukaryotic transcription units or else some alternate mechanism of translation initiation was present (perhaps internal ribosome entry sites) to enable translation of the proteins from these polycistronic transcripts.
SL stem-loops.
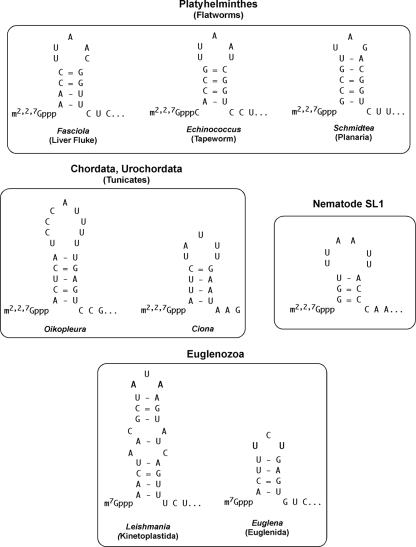
We examined other metazoan spliced leaders for the presence of a stem-loop adjacent to the cap. Indeed, many of the spliced leaders can be predicted to form a stem-loop adjacent to the cap (Fig. 7). Thus, the presence of an SL stem-loop adjacent to the cap may be a common feature of trans splicing.
FIG. 7.
Potential stem-loop structures adjacent to the cap in phylogenetically divergent spliced leaders. Note that while many SLs have been predicted to have a stem-loop adjacent to the cap, the sequences contributing to the stem are divergent. The nematode SL1 stem-loop is shown for comparison. Representatives of 3 separate classes of flatworms (Turbellaria, Trematoda, and Cestoda) and representatives of 2 different classes of the Euglenozoa (Kinetoplastida and Euglenida) are illustrated. Sequences are derived as follows: Schmidtea mediterranea (55), Fasciola hepatica (7), Echinococcus multilocularis (4), Oikopleura dioica (12), Ciona intestinalis (48), Leishmania spp. (34), and Euglena gracilis (45).
Conclusions.
We have defined elements within the nematode SL1 spliced leader that form a stem-loop and the eIF4F complex required for efficient translation of TMG-capped RNAs generated by trans splicing. While sequences within the 5′ UTR of eukaryotic mRNAs are known to have a modulating effect on translation (most often negative), our studies demonstrate the importance of specific elements in the 5′ UTR and adaptation in the eIF4F complex necessary for optimal cap-dependent translation.
Supplementary Material
Acknowledgments
We are particularly grateful to Karen Browning for helpful discussions and for generously providing wheat germ translation extracts and eIF4F complex. We thank Tom Evans, Tom Blumenthal, Jeffrey Kieft, Tim Nilsen, and members of the Davis laboratory for their comments on the manuscript.
This work was supported by NIH grant AI49558 to R.E.D. and by grants from the Howard Hughes Medical Institute grant (55005604) and National Science Support Project 2008-2010 no. PBZ-MNiSW-07/1/2007 to E.D.
Footnotes
Published ahead of print on 12 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Berset, C., A. Zurbriggen, S. Djafarzadeh, M. Altmann, and H. Trachsel. 2003. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA 9:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumenthal, T. 25 June 2005, posting date. Trans-splicing and operons. In The C. elegans Research Community (ed.), WormBook. doi/ 10.1895/wormbook.1.5.1. http://www.wormbook.org/. [DOI] [PubMed]
- 3.Bonen, L. 1993. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 7:40-46. [DOI] [PubMed] [Google Scholar]
- 4.Brehm, K., K. Jensen, and M. Frosch. 2000. mRNA trans-splicing in the human parasitic cestode Echinococcus multilocularis. J. Biol. Chem. 275:38311-38318. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, G., L. Cohen, C. Mikhli, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R. E. Davis. 2007. In vivo translation and stability of trans-spliced mRNAs in nematode embryos. Mol. Biochem. Parasitol. 153:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darzynkiewicz, E., J. Stepinski, I. Ekiel, Y. Jin, D. Haber, T. Sijuwade, and S. M. Tahara. 1988. Beta-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Res. 16:8953-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, R. E., H. Singh, C. Botka, C. Hardwick, M. Ashraf el Meanawy, and J. Villanueva. 1994. RNA trans-splicing in Fasciola hepatica. Identification of a spliced leader (SL) RNA and SL sequences on mRNAs. J. Biol. Chem. 269:20026-20030. [PubMed] [Google Scholar]
- 8.Douris, V., M. J. Telford, and M. Averof. 25 November 2009, posting date. Evidence for multiple independent origins of trans-splicing in Metazoa. Mol. Biol. Evol. [Epub ahead of print.] [DOI] [PubMed]
- 9.Ferguson, K. C., and J. H. Rothman. 1999. Alterations in the conserved SL1 trans-spliced leader of Caenorhabditis elegans demonstrate flexibility in length and sequence requirements in vivo. Mol. Cell. Biol. 19:1892-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallie, D. R. 2007. Use of in vitro translation extract depleted in specific initiation factors for the investigation of translational regulation. Methods Enzymol. 429:35-51. [DOI] [PubMed] [Google Scholar]
- 11.Gallie, D. R., and K. S. Browning. 2001. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 276:36951-36960. [DOI] [PubMed] [Google Scholar]
- 12.Ganot, P., T. Kallesoe, R. Reinhardt, D. Chourrout, and E. M. Thompson. 2004. Spliced-leader RNA trans splicing in a chordate, Oikopleura dioica, with a compact genome. Mol. Cell. Biol. 24:7795-7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gingras, A.-C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 14.Gross, J. D., N. J. Moerke, T. von der Haar, A. A. Lugovskoy, A. B. Sachs, J. E. McCarthy, and G. Wagner. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115:739-750. [DOI] [PubMed] [Google Scholar]
- 15.Guiliano, D. B., and M. L. Blaxter. 2006. Operon conservation and the evolution of trans-splicing in the phylum Nematoda. PLoS Genetics 2:e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannon, G. J., P. A. Maroney, D. G. Ayers, J. D. Shambaugh, and T. W. Nilsen. 1990. Transcription of a nematode trans-spliced leader RNA requires internal elements for both initiation and 3′ end-formation. EMBO J. 9:1915-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hastings, K. E. 2005. SL trans-splicing: easy come or easy go? Trends Genet. 21:240-247. [DOI] [PubMed] [Google Scholar]
- 18.Hinton, T. M., M. J. Coldwell, G. A. Carpenter, S. J. Morley, and V. M. Pain. 2007. Functional analysis of individual binding activities of the scaffold protein eIF4G. J. Biol. Chem. 282:1695-1708. [DOI] [PubMed] [Google Scholar]
- 19.Huang, X. Y., and D. Hirsh. 1989. A second trans-spliced RNA leader sequence in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 86:8640-8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jankowska-Anyszka, M., B. J. Lamphear, E. J. Aamodt, T. Harrington, E. Darzynkiewicz, R. Stolarski, and R. E. Rhoads. 1998. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in C. elegans can distinguish between mono- and trimethylated mRNA cap structures. J. Biol. Chem. 273:10538-10542. [DOI] [PubMed] [Google Scholar]
- 21.Kaye, N. M., K. J. Emmett, W. C. Merrick, and E. Jankowsky. 2009. Intrinsic RNA binding by the eukaryotic initiation factor 4F depends on a minimal RNA length but not on the m7G cap. J. Biol. Chem. 284:17742-17750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keiper, B. D., B. J. Lamphear, A. M. Deshpande, M. Jankowska-Anyszka, E. J. Aamodt, T. Blumenthal, and R. E. Rhoads. 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 275:10590-10596. [DOI] [PubMed] [Google Scholar]
- 23.Kozak, M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13-37. [DOI] [PubMed] [Google Scholar]
- 24.Kozak, M. 1991. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1:111-115. [PMC free article] [PubMed] [Google Scholar]
- 25.Lall, S., C. C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R. E. Davis. 2004. Contribution of trans-splicing, 5′-leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J. Biol. Chem. 279:45573-45585. [DOI] [PubMed] [Google Scholar]
- 26.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liou, R. F., and T. Blumenthal. 1990. trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol. Cell. Biol. 10:1764-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marlétaz, F., A. Gilles, X. Caubit, Y. Perez, C. Dossat, S. Samain, G. Gyapay, P. Wincker, and Y. Le Parco. 2008. Chaetognath transcriptome reveals ancestral and unique features among bilaterians. Genome Biol. 9:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maroney, P. A., J. A. Denker, E. Darzynkiewicz, R. Laneve, and T. W. Nilsen. 1995. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA 1:714-723. [PMC free article] [PubMed] [Google Scholar]
- 30.Maroney, P. A., G. J. Hannon, and T. W. Nilsen. 1990. Transcription and cap trimethylation of a nematode spliced leader RNA in a cell-free system. Proc. Natl. Acad. Sci. U. S. A. 87:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maroney, P. A., G. J. Hannon, J. D. Shambaugh, and T. W. Nilsen. 1991. Intramolecular base pairing between the nematode spliced leader and its 5′ splice site is not essential for trans-splicing in vitro. EMBO J. 10:3869-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy, J. E., S. Marsden, and T. von der Haar. 2007. Biophysical studies of the translation initiation pathway with immobilized mRNA analogs. Methods Enzymol. 430:247-264. [DOI] [PubMed] [Google Scholar]
- 33.Merino, E. J., K. A. Wilkinson, J. L. Coughlan, and K. M. Weeks. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127:4223-4231. [DOI] [PubMed] [Google Scholar]
- 34.Miller, S. I., S. M. Landfear, and D. F. Wirth. 1986. Cloning and characterization of a Leishmania gene encoding a RNA spliced leader sequence. Nucleic Acids Res. 14:7341-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niedzwiecka, A., E. Darzynkiewicz, and R. Stolarski. 2004. Thermodynamics of mRNA 5′ cap binding by eukaryotic translation initiation factor eIF4E. Biochemistry 43:13305-13317. [DOI] [PubMed] [Google Scholar]
- 36.Nilsen, T. W. 1997. Trans-splicing, p. 310-334. In A. R. Krainer (ed.), Frontiers in molecular biology: eukaryotic mRNA processing. IRL Press, Oxford, United Kingdom.
- 37.Pestova, T. V., J. R. Lorsch, and C. U. T. Hellen. 2007. The mechanism of translation initiation in eukaryotes, p. 87-128. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Press, Woodbury, NY.
- 38.Pettitt, J., B. Muller, I. Stansfield, and B. Connolly. 2008. Spliced leader trans-splicing in the nematode Trichinella spiralis uses highly polymorphic, noncanonical spliced leaders. RNA 14:760-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pouchkina-Stantcheva, N. N., and A. Tunnacliffe. 2005. Spliced-leader RNA mediated trans-splicing in phylum Rotifera. Mol. Biol. Evol. 22:1482-1489. [DOI] [PubMed] [Google Scholar]
- 40.Prévôt, D., D. Decimo, C. H. Herbreteau, F. Roux, J. Garin, J. L. Darlix, and T. Ohlmann. 2003. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 22:1909-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ptushkina, M., T. von der Haar, M. M. Karim, J. M. Hughes, and J. E. McCarthy. 1999. Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J. 18:4068-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross, L. H., J. H. Freedman, and C. S. Rubin. 1995. Structure and expression of novel spliced leader RNA genes in Caenorhabditis elegans. J. Biol. Chem. 270:22066-22075. [DOI] [PubMed] [Google Scholar]
- 43.Rutkowska-Wlodarczyk, I., J. Stepinski, M. Dadlez, E. Darzynkiewicz, R. Stolarski, and A. Niedzwiecka. 2008. Structural changes of eIF4E upon binding to the mRNA 5′ monomethylguanosine and trimethylguanosine Cap. Biochemistry 47:2710-2720. [DOI] [PubMed] [Google Scholar]
- 44.Sonenberg, N., and A. G. Hinnebusch. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tessier, L. H., M. Keller, R. L. Chan, R. Fournier, J. H. Weil, and P. Imbault. 1991. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 10:2621-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, J. D., R. C. Conrad, and T. Blumenthal. 1988. The C. elegans trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell 54:533-539. [DOI] [PubMed] [Google Scholar]
- 47.Ullu, E., and T. W. Nilsen. 1995. Molecular biology of protozoan and helminth parasites, p. 1-17. In J. Marr and M. Muller (ed.), Biochemistry of parasitic organisms and its molecular foundations. Academic Press, London, United Kingdom.
- 48.Vandenberghe, A. E., T. H. Meedel, and K. E. Hastings. 2001. mRNA 5′-leader trans-splicing in the chordates. Genes Dev. 15:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Doren, K., and D. Hirsh. 1990. mRNAs that mature through trans-splicing in Caenorhabditis elegans have a trimethylguanosine cap at their 5′ termini. Mol. Cell. Biol. 10:1769-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von der Haar, T., J. D. Gross, G. Wagner, and J. E. McCarthy. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11:503-511. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson, K. A., E. J. Merino, and K. M. Weeks. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610-1616. [DOI] [PubMed] [Google Scholar]
- 52.Wu, L., J. Fan, and J. G. Belasco. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U. S. A. 103:4034-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanagiya, A., Y. V. Svitkin, S. Shibata, S. Mikami, H. Imataka, and N. Sonenberg. 2009. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol. Cell. Biol. 29:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoffe, Y., J. Zuberek, M. Lewdorowicz, Z. Zeira, C. Keasar, I. Orr-Dahan, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and M. Shapira. 2004. Cap-binding activity of an eIF4E homolog from Leishmania. RNA 10:1764-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zayas, R. M., T. D. Bold, and P. A. Newmark. 2005. Spliced-leader trans-splicing in freshwater planarians. Mol. Biol. Evol. 22:2048-2054. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, H., Y. Hou, L. Miranda, D. A. Campbell, N. R. Sturm, T. Gaasterland, and S. Lin. 2007. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. U. S. A. 104:4618-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.