Abstract
During yeast sporulation, a forespore membrane (FSM) initiates at each spindle-pole body and extends to form the spore envelope. We used Schizosaccharomyces pombe to investigate the role of septins during this process. During the prior conjugation of haploid cells, the four vegetatively expressed septins (Spn1, Spn2, Spn3, and Spn4) coassemble at the fusion site and are necessary for its normal morphogenesis. Sporulation involves a different set of four septins (Spn2, Spn5, Spn6, and the atypical Spn7) that does not include the core subunits of the vegetative septin complex. The four sporulation septins form a complex in vitro and colocalize interdependently to a ring-shaped structure along each FSM, and septin mutations result in disoriented FSM extension. The septins and the leading-edge proteins appear to function in parallel to orient FSM extension. Spn2 and Spn7 bind to phosphatidylinositol 4-phosphate [PtdIns(4)P] in vitro, and PtdIns(4)P is enriched in the FSMs, suggesting that septins bind to the FSMs via this lipid. Cells expressing a mutant Spn2 protein unable to bind PtdIns(4)P still form extended septin structures, but these structures fail to associate with the FSMs, which are frequently disoriented. Thus, septins appear to form a scaffold that helps to guide the oriented extension of the FSM.
Yeast sporulation is a developmental process that involves multiple, sequential events that need to be tightly coordinated (59, 68). In the fission yeast Schizosaccharomyces pombe, when cells of opposite mating type (h+ and h−) are mixed and shifted to conditions of nitrogen starvation, cell fusion and karyogamy occur to form a diploid zygote, which then undergoes premeiotic DNA replication, the two meiotic divisions, formation of the spore envelopes (comprising the plasma membrane and a specialized cell wall), and maturation of the spores (74, 81). At the onset of meiosis II, precursors of the spore envelopes, the forespore membranes (FSMs), are formed by the fusion of vesicles at the cytoplasmic surface of each spindle-pole body (SPB) and then extend to engulf the four nuclear lobes (the nuclear envelope does not break down during meiosis), thus capturing the haploid nuclei, along with associated cytoplasm and organelles, to form the nascent spores (55, 68, 81). How the FSMs recognize and interact with the nuclear envelope, extend in a properly oriented manner, and close to form uniformly sized spherical spores is not understood, and study of this model system should also help to elucidate the more general question of how membranes obtain their shapes in vivo.
It has been shown that both the SPB and the vesicle trafficking system play important roles in the formation and development of the FSM and of its counterpart in the budding yeast Saccharomyces cerevisiae, the prospore membrane (PSM). In S. pombe, the SPB changes its shape from a compact dot to a crescent at metaphase of meiosis II (26, 29), and its outer plaque acquires meiosis-specific components such as Spo2, Spo13, and Spo15 (30, 57, 68). This modified outer plaque is required for the initiation of FSM assembly. In S. cerevisiae, it is well established that various secretory (SEC) gene products are required for PSM formation (58, 59). Similarly, proteins presumably involved in the docking and/or fusion of post-Golgi vesicles and organelles in S. pombe, such as the syntaxin-1A Psy1, the SNAP-25 homologue Sec9, and the Rab7 GTPase homologue Ypt7, are also required for proper FSM extension (34, 53, 54). Consistent with this hypothesis, Psy1 disappears from the plasma membrane upon exit from meiosis I and reappears in the nascent FSM.
Phosphoinositide-mediated membrane trafficking also contributes to the development of the FSM. Pik3/Vps34 is a phosphatidylinositol 3-kinase whose product is phosphatidylinositol 3-phosphate [PtdIns(3)P] (35, 72). S. pombe cells lacking this protein exhibit defects in various steps of FSM formation, such as aberrant starting positions for extension, disoriented extension and/or failure of closure, and the formation of spore-like bodies near, rather than surrounding, the nuclei, suggesting that Pik3 plays multiple roles during sporulation (61). The targets of PtdIns(3)P during sporulation appear to include two sorting nexins, Vps5 and Vps17, and the FYVE domain-containing protein Sst4/Vps27. vps5Δ and vps17Δ mutant cells share some of the phenotypes of pik3Δ cells (38). sst4Δ cells also share some of the phenotypes of pik3Δ cells but are distinct from vps5Δ and vps17Δ cells, consistent with the hypothesis that Pik3 has multiple roles during sporulation (62).
Membrane trafficking processes alone do not seem sufficient to explain how the FSMs and PSMs extend around and engulf the nuclei, suggesting that some other mechanism(s) must regulate and orient FSM/PSM extension. The observation that the FSM is attached to the SPB until formation of the immature spore is complete (68) suggests that the SPB may regulate FSM extension. In addition, the leading edge of the S. cerevisiae PSM is coated with a complex of proteins (the LEPs) that appear to be involved in PSM extension (51, 59). S. pombe Meu14 also localizes to the leading edge of the FSM, and deletion of meu14 causes aberrant FSM formation in addition to a failure in SPB modification (60). However, it has remained unclear whether the SPB- and LEP-based mechanisms are sufficient to account for the formation of closed FSMs and PSMs of proper size and position (relative to the nuclear envelope), and evidence from S. cerevisiae has suggested that the septin proteins may also be involved.
The septins are a conserved family of GTP-binding proteins that were first identified in S. cerevisiae by analysis of the cytokinesis-defective cdc3, cdc10, cdc11, and cdc12 mutants (41). Cdc3, Cdc10, Cdc11, and Cdc12 are related to each other in sequence and form an oligomeric complex that localizes to a ring in close apposition to the plasma membrane at the mother-bud neck in vegetative cells (12, 20, 25, 41, 47, 77). The septin ring appears to be filamentous in vivo (12), and indeed, the septins from both yeast (11, 20) and metazoans (31, 36, 69) can form filaments in vitro. The yeast septin ring appears to form a scaffold for the localization and organization of a wide variety of other proteins (8, 22), and it forms a diffusion barrier that constrains movement of membrane proteins through the neck region (7, 8, 73). In metazoan cells, the septins are involved in cytokinesis but are also implicated in a variety of other cellular processes, such as vesicular transport, organization of the actin and microtubule cytoskeletons, and oncogenesis (27, 70).
In S. cerevisiae, a fifth septin (Shs1) is also expressed in vegetative cells, but the remaining two septin genes, SPR3 and SPR28, are expressed at detectable levels only during sporulation (15, 17). In addition, at least some of the vegetatively expressed septins are also present in sporulating cells (17, 48), and one of them (Cdc10) is expressed at much higher levels there than in vegetative cells (32). The septins present during sporulation are associated with the PSM (15, 17, 48, 51), and their normal organization there depends on the Gip1-Glc7 protein phosphatase complex (71). However, it has been difficult to gain insight into the precise roles of the septins during sporulation in S. cerevisiae (59), because some septins are essential for viability during vegetative growth, and the viable mutants have only mild phenotypes during sporulation (15, 17), possibly because of functional redundancy among the multiple septins.
S. pombe seemed likely to provide a better opportunity for investigating the role of septins during spore formation. There are seven septin genes (spn1+ to spn7+) in this organism (23, 41, 63). Four of these genes (spn1+ to spn4+) are expressed in vegetative cells, and their products form a hetero-oligomeric complex that assembles during cytokinesis into a ring at the division site (2, 3, 10, 76, 79). The septin ring is important for proper targeting of endoglucanases to the division site (44), and septin mutants show a corresponding delay in cell separation (10, 41, 44, 76). However, even the spn1Δ spn2Δ spn3Δ spn4Δ quadruple mutant is viable and grows nearly as rapidly as the wild type (our unpublished results), a circumstance that greatly facilitates studies of the septins' role during sporulation.
spn5+, spn6+, and spn7+ are expressed at detectable levels only during sporulation (1, 45, 78; our unpublished results), and spn2+, like its orthologue CDC10 (see above), is strongly induced (45), but the roles of the S. pombe septins in sporulation have not previously been investigated. In this study, we show that the septins are important for the orientation of FSM extension, suggesting that the septins may have a more general role in dynamic membrane organization and shape determination.
MATERIALS AND METHODS
Strains, growth conditions, and plasmids.
S. pombe strains used in this study are listed in Table 1; strain constructions are described below or in the table. PCR primers are listed in Table 2. Yeast cells were grown in YE (rich) or EMM (synthetic) medium (50), and SSA (16) or MM-N (EMM without NH4Cl) medium was used to induce mating and/or sporulation. Except where noted, cells to be examined microscopically were grown for 12 to 18 h on an EMM plate at 30°C, transferred to an SSA plate at 30°C, and incubated for 8 to 14 h before examination. When appropriate, thiamine was added to media at 15 μM to repress expression of genes controlled by the nmt1 promoter.
TABLE 1.
S. pombe strains used in this studya
| Strain | Genotype | Source or reference |
|---|---|---|
| 968 | h90 (wild type) | 40 |
| 972 | h− (wild type) | 40 |
| 975 | h+ (wild type) | 40 |
| SG14 | h90leu1-32 | 24 |
| THP18 | h90ade6-M216 ura4-D18 leu1-32 | 38 |
| THP17 | h90ade6-M210 ura4-D18 leu1-32 | 38 |
| TW747 | h+/h−ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | 62 |
| ED665b | h−ade6-M210 ura4-D18 leu1-32 | P. Fantes |
| ED666 | h+ade6-M210 ura4-D18 leu1-32 | P. Fantes |
| JB60 | h−ade6-M210 ura4-D18 leu1-32 spn5Δ::ura4+ | O. Al-Awarc |
| JB61 | h+ade6-M210 ura4-D18 leu1-32 spn5Δ::ura4+ | O. Al-Awarc |
| JB68 | h90spn5+-GFP:kanMX6 | This studyd |
| JW193 | h+spn7+-GFP:kanMX6 | This studyd |
| TK144 | h90ade6-M216 ura4-D18 leu1-32 spn5Δ::ura4+ | See text |
| TK172 | h+/h-ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 spn5Δ::ura4+/spn5Δ::ura4+ | This studye |
| TK278 | h90ade6-M216 ura4-D18 leu1-32 spn5Δ::ura4+spn6Δ::LEU2 | See text |
| TK367 | h+/h−ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 spn6Δ::ura4+/spn6Δ::ura4+ | This studye |
| MO229 | h90ade6-M216 ura4-D18 leu1-32 spn7Δ::ura4+ | See text |
| MO246 | h90ade6-M216 ura4-D18 leu1-32 spn1Δ::ura4+ | See text |
| MO250 | h90ade6-M216 ura4-D18 leu1-32 spn2Δ::ura4+ | See text |
| MO277 | h90ade6-M216 ura4-D18 leu1-32 spn6Δ::ura4+ | See text |
| MO371 | h90ade6-M216 ura4-D18 leu1-32 spn3Δ::ura4+ | See text |
| MO372 | h90ade6-M216 ura4-D18 leu1-32 spn4Δ::ura4+ | See text |
| MO599 | h90ade6-M216 ura4-D18 leu1-32 pik1Δ117-198::ura4+ | See text |
| MO623 | h90ade6-M210 spn5+-GFP:kanMX6 | This studyf |
| MO629 | h90ade6-M210 spn7+-GFP:kanMX6 | This studyf |
| MO660 | h90ade6-M216 ura4-D18 leu1-32 spn2Δ::ura4+spn7Δ::LEU2 | See text |
| MO662 | h90ade6-M216 ura4-D18 leu1-32 spn5Δ::ura4+spn7Δ::LEU2 | See text |
| MO664 | h90ade6-M216 ura4-D18 leu1-32 spn6Δ::ura4+spn7Δ::LEU2 | See text |
| MO667 | h90ura4-D18 leu1-32 spn6Δ::ura4+ | This studyg |
| MO670 | h90ura4-D18 leu1-32 | This studyh |
| MO684 | h90ade6-M210 ura4-D18 leu1-32 spn2Δ::kanMX6 | See text |
| MO701 | h90ade6-M216 ura4-D18 leu1-32 spn2+-GFP:kanMX6 | See text |
| MO705 | h90/h90ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 spn2Δ::ura4+/spn2Δ::ura4+ | This studye |
| MO706 | h90/h90ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 spn7Δ::ura4+/spn7Δ::ura4+ | This studye |
| MO807 | h90spn4+-GFP:kanMX6 | This studyi |
| MO808 | h90spn1+-GFP:kanMX6 | This studyi |
| MO815 | h90leu1-32 spn2+-mRFP:kanMX6 | See text |
| MO817 | h90leu1+:GFP-psy1+ | See text |
| MO822 | h90ura4-D18 pik1Δ117-198::ura4+leu1+:GFP-psy1+ | This studyj |
| MO832 | h90ura4-D18 pik1Δ117-198::ura4+leu1+:GFP-psy1+spn2+-mRFP:kanMX6 | See text |
| MO900 | h90spn2+-mRFP:kanMX6 spn5+-GFP:kanMX6 | This studyk |
| MO902 | h90spn2+-mRFP:kanMX6 spn7+-GFP:kanMX6 | This studyk |
| MO905 | h90ade6-M216 ura4-D18 leu1-32 meu14Δ::ura4+ | This studyl |
| MO911 | h90ade6-M216 ura4-D18 leu1-32 meu14Δ::ura4+spn6Δ::ura4+ | This studym |
| SY13 | h90ade6-M216 ura4-D18 leu1-32 meu14+-GFP | This studyn |
| SY86 | h90ade6-M216 ura4-D18 leu1-32 meu14+-GFP spn2Δ::kanMX6 | This studyo |
| SY154 | h90ade6-M216 ura4-D18 leu1-32 meu14Δ::ura4+ | This studyn |
| SY175 | h90ade6-M216 ura4-D18 leu1-32 his2 spn7Δ::ura4+ | This studyp |
| SY202 | h90ade6-M210 ura4-D18 leu1-32 meu14Δ::ura4+spn2Δ::kanMX6 | This studyp |
| SY211 | h90ade6-M216 ura4-D18 leu1+:GFP-psy1+spn2+-mRFP:kanMX6 meu14Δ::ura4+ | This studyq |
| SY233 | h90ade6-M210 ura4-D18 leu1-32 spn5Δ::LEU2 | This studyr |
| SY284 | h90ade6-M216 ura4-D18 leu1-32 meu14+-GFP spn6Δ::ura4+ | This studyo |
| SY288 | h90ade6-M216 ura4-D18 leu1-32 meu14+-GFP spn5Δ::ura4+ | This studyo |
| SY292 | h90ade6-M216 ura4-D18 leu1-32 meu14+-GFP spn7Δ::ura4+ | This studyo |
| SY386 | h90ade6-M216 ura4-D18 leu1-32 meu14Δ::ura4+spn7Δ::ura4+ | This studyp |
| SY430 | h90ade6-M216 ura4-D18 leu1-32 meu14Δ::ura4+spn5Δ::LEU2 | This studyp |
All strains were derived from wild-type strains 968, 972, and 975.
American Type Culture Collection strain ATCC 96993.
Strain ED666 was transformed with a fragment of genomic DNA in which an EcoRV fragment spanning codons 91 to 299 of spn5 was replaced by ura4+, yielding strain JB61 (O. Al-Awar and J. R. Pringle, unpublished results). A cross of JB61 to ED665 yielded segregant JB60.
spn5 and spn7 were tagged at their C termini with GFP sequences in strains 968 and 975, respectively, using a PCR method (6).
Constructed either by mating a pair of segregants from crosses of the original mutants to h+ and h− wild-type strains (TK172 and TK367, respectively) or by mating the original haploid mutants to segregants from crosses of these mutants to strain THP17 (MO705 and MO706); interallelic complementation of the two ade6 alleles forces the maintenance of diploidy in medium lacking adenine (50).
Segregants from crosses of THP17 to JB68 and JW193.
Segregants from the cross of MO670 to MO277.
Segregant from a cross of SG14 to THP17.
Segregants from crosses of THP17 to JW183 and JW251 (J.-Q. Wu and J.R. Pringle, unpublished data). The chromosomal spn4+ and spn1+ loci had been tagged at their C termini with GFP sequences by a PCR method (6).
Segregant from a cross of MO599 to MO817.
Segregants from crosses of MO815 to MO623 and MO629. Ade+ Leu+ segregants were isolated and checked by PCR for the presence of both tagged septin genes.
Segregant from a cross of SY211 to THP17.
Segregant from a cross of MO905 to SY284.
Segregants from crosses of THP17 to h− ade6-M216 ura4-D18 leu1-32 meu14+-GFP and h− ade6-M216 ura4-D18 leu1-32 meu14Δ::ura4+ strains (gifts from H. Nojima, Osaka University).
Segregants from crosses of an h90 ade6-M210 ura4-D18 leu1-32 meu14+-GFP strain (obtained by the cross described in footnote n) to MO250, MO277, TK144, and MO229.
Derived by one or more crosses from MO229, SY154, MO684, and SY233.
Segregant from the cross of MO817 to an h90 ade6-M216 ura4-D18 leu1-32 spn2+-mRFP:kanMX6 meu14Δ::ura4+ strain constructed by crossing MO815 to the meu14Δ::ura4+ strain (see footnote n).
Segregant from a cross of THP18 to an h− ade6-M210 ura4-D18 leu1-32 spn5Δ::LEU2 strain (constructed as described in the legend to Fig. 1, but using a different parent strain for transformation).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)i |
|---|---|
| spn1Dfwa | CAGATCTCATGGCGTCAATGGTACTCG (BglII) |
| spn1Drva | CTCGAGTAAGGAGACTATTTAAAAAACTTCC (XhoI) |
| spn2Dfwa,g | CAGATCTAATGGAAGTTCCTTCTGCAGTTACC (BglII) |
| spn2Drva,g | CTCGAGGAAGAAGACTATTGAGCGGTG (XhoI) |
| spn3Dfwa | CAGATCTAATGTGGTATACATATAACGAAGAC (BglII) |
| spn3Drva | CTCGAGCACCATTAAATTGGTTATGACTGG (XhoI) |
| spn4Dfwa | AAACCCGGGATGAATGAAGAAGAGACAAACTTCG (SmaI) |
| spn4Drva | AAATCTAGACTAACGTTTGCGAGAGCTGGTAGC (XbaI) |
| spn5Dfwa,g | CAGATCTGATGGATAGCTCAAATTTGTCTTCATC (BglII) |
| spn5Drva,g | CTCGAGTTTATGCTAAGATGCCTGC (XhoI) |
| spn6Dfwa,g | TTAGATCTCACGATGTCTTTGACTGAAAACCTTCAATTAT (BglII) |
| spn6Drva,g | TTCCCGGGTCATTTTTTATGACCGCGGCCCTTGT (SmaI) |
| spn7Dfwa,g | CAGATCTGATGAATAAAGGCCCAAGACATCG (BglII) |
| spn7Drva,g | CTCGAGCACTCTCAAATGTTTAATTTAGC (XhoI) |
| pik1Dfwa | AAGGATCCGATGCCATCTTCGAATTCGGG (BamHI) |
| pik1Drva | TTGCCAAATCGCAGATCTTCTGCGC |
| spn2KDfwb | TACAATGTTGACTTGGCCAAAACCAACAAATTCCAAATTCATTCAATTCGCAACTTGCAACGGATCCCCGGGTTAATTAA |
| spn2KCfwc | ATGGGAAGTCCCGCACCTGTGTACCCTTCTGAACCACATCTCCATACAGCCACCGCTCAACGGATCCCCGGGTTAATTAA |
| spn2KDrvb,c | GACAGTCATACAAATGGTTTAGTCTTTGTTCTAAACATACTATATATTACCTTAGGAAGAGAATTCGAGCTCGTTTAAAC |
| spn7KCfwc | ACAATACGACAAAGGAATTGGAGATGAAGAAAATGGATGATTTGTCTCATGAGCGTTACGAAAACCTCCCTTTCTATCGCCGGATCCCCGGGTTAATTAA |
| spn7KDrvc | GATTAAGCAATTAAAAAGAAATCAAAAGACTTTTCATGAACTTATACTAAATTATTCTTTTGATTGTTTATAATTTGCACGAATTCGAGCTCGTTTAAAC |
| ura4checkrvd | TCTTTGGCTACTGGTTCCTACAC |
| kancheckrvd | CGGATAAAATGCTTGATGGTCGGAAGAGG |
| GFPfwe | CCCCTCGAGTATGAGTAAAGGAGAA (XhoI) |
| GFPrve | CCCGGATCCGTCGACTTGTATAGTTCATCCATGCCATGTGTAATCCC (BamHI) |
| psy1fwe | CCCGTCGACAATGAATAAAGCAAACGAT (SalI) |
| psy1rve | CCCGAGCTCATCTAACCGGCCATATCACT (SacI) |
| propsy1fwe | CCCCTCGAGTGGTGCATCCAATCTCTG (XhoI) |
| propsy1rve | CCCCTCGAGTTTGTTATATTTATCTGTTTAAA (XhoI) |
| spn2Gfwf | AAGGATCCAGCGCTAATTATTTGTGAATCTGG (BamHI) |
| spn2Grvf | TTGCGGCCGCTGAGCGGTGGCTGTATGGAG (NotI) |
| spn5Gfwf | AAGGATCCTCCCGCCACTTCACTAAG (BamHI) |
| spn5Grvf | TTGCGGCCGCGCTAAGATGCCTGCTTTTTTC (NotI) |
| spn6Gfwf | AAAGATCTATTCGATCCTTGACACTGTC (BglII) |
| spn6Grvf | TTGCGGCCGCTTTTTATGACCGCGGCCCTTG (NotI) |
| spn7Gfwf | AAAGATCTTAAACCTGCAGCTCAACTC (BglII) |
| spn7Grvf | TTGCGGCCGCCGATAGAAAGGGAGGTTTTCG (NotI) |
| spn2mQQh | AACCAACAGTTAATTCAACAAGGT |
Used to construct the plasmids for the gene disruptions described in Fig. 1.
Used to generate strain MO684 (spn2Δ::kanMX6).
Used to generate strains MO701 (spn2+-GFP::kanMX6), MO815 (spn2+-mRFP::kanMX6), MO832 (spn2+-mRFP::kanMX6), and JW193 (spn7+-GFP:kanMX6).
Used to confirm successful disruption/tagging by the ura4+ or kanMX6 marker cassette. ura4checkrv was used with the appropriate Dfw series primer; kancheckrv was used with Spn2Gfw to check the spn2 deletion and with Spn2Dfw to check the C-terminal tagging.
Used to construct plasmid pBR(GFP-Psy1).
Used to construct the pAL(spn-GFP) plasmids.
Used to amplify cDNAs for construction of pMAL(spnN) and pREP41(GFP-spn) plasmids.
Used for mutagenesis of spn2+ to generate spn24Q.
Underlining indicates restriction enzyme site (corresponding enzyme is indicated in parentheses).
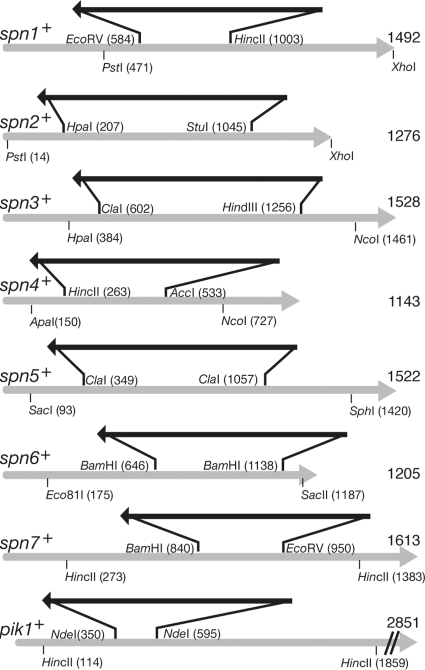
Strains containing mutant alleles of spn1 to spn7 and pik1 were constructed as follows. The genes were amplified from genomic DNA by PCR and cloned by TA cloning into vector pT7Blue (Novagen). Plasmids containing partially deleted genes were then constructed as shown in Fig. 1, using ura4+ and LEU2 marker cassettes obtained as HindIII fragments from plasmids KS-ura4 (6) and KS-LEU2. KS-LEU2 was constructed by cloning a HindIII fragment containing LEU2 from pART1 (46) into pBluescript II KS(+) (Stratagene). Fragments were released from the plasmids by digestion with the indicated restriction enzymes (Fig. 1) and were transformed into strain THP18, with selection for Ura+ or Leu+. The success of the constructions was checked by PCR and also (for spn6 and spn7) by showing that ura4+ and LEU2 were tightly linked to each other when the spnΔ::ura4+ and spnΔ::LEU2 strains were crossed. Double mutants were constructed by crosses among the single mutants.
FIG. 1.
Disruption of spn and pik1 genes. Gray arrows represent the genomic coding regions (including introns, where present) of the indicated genes; the total number of base pairs in each coding region (from the first base of the start codon to the last base of the stop codon) is indicated. Black arrows (not drawn to scale) represent the ura4+ and LEU2 marker cassettes (see Materials and Methods), which were inserted after cutting of the original plasmids at the indicated sites, ligation to HindIII linkers, and digestion with HindIII. Sites indicated below the gray arrows were used to release the fragments used for transformation; the XhoI sites used with spn1 and spn2 were included in the primers used to clone these genes (Table 2).
In addition, the complete spn2 coding region was deleted or tagged at its C terminus with the GFP or mRFP1 (13) sequence by a PCR method (6) using pFA6a-kanMX6, pFA6a-GFP(S65T)-kanMX6, or pFA6a-mRFP-kanMX6 (62) as template and THP17, THP18, SG14, or MO822 as the recipient strain, yielding strains MO684, MO701, MO815, and MO832, respectively. Proper integration at the spn2 locus was confirmed by PCR (6).
A strain expressing green fluorescent protein (GFP)-tagged Psy1 was constructed as follows. First, plasmid pTN381 was constructed by inserting a SacI linker into the BamHI site of pBR(leu1) (56). Second, GFPS65T sequences were amplified by PCR from plasmid pEG5 (a gift from Y. Hiraoka, National Institute of Information and Communications Technology, Kobe, Japan), digested with XhoI and BamHI (sites in the primers), and cloned into XhoI/BamHI-digested pBluescript II, yielding pBS(GFP). The psy1+ open reading frame (ORF) was then amplified by PCR from genomic DNA, digested with SalI and SacI (sites in the primers), and ligated into SalI/SacI-digested pBS(GFP), yielding pBS(GFP-psy1). The psy1+ promoter region was then amplified from genomic DNA by PCR, digested with XhoI (sites in the primers), and ligated into XhoI-digested pBS(GFP-psy1), yielding pBS(psy1pro-GFP-psy1). The orientation of the promoter fragment was checked by restriction enzyme digestion. The ApaI-SacI fragment of pBS(psy1pro-GFP-psy1) was then ligated into ApaI/SacI-digested pTN381, yielding pBR(GFP-Psy1). Finally, pBR(GFP-Psy1) was linearized by digestion at the SnaBI site in the leu1+ 3′-noncoding region and transformed into strain SG14, yielding strain MO817, which was checked by confirming that Leu+ and Leu− progeny segregated 2:2 in crosses and that the expected GFP labeling of the FSM was seen.
Plasmids for the expression of septins tagged by GFP or maltose-binding protein (MBP) were constructed as follows. For GFP tagging, the genomic spn loci, including promoters, were amplified by PCR and inserted as BamHI-NotI or BglII-NotI fragments (sites in the primers) into BamHI/NotI-digested pAL(GFP) (62), yielding pAL(spn2-GFP), pAL(spn5-GFP), pAL(spn6-GFP), and pAL(spn7-GFP). The tagged proteins appeared to be functional based on their ability to rescue the sporulation defects of the corresponding mutants (data not shown). For MBP tagging, plasmid pMALBB was constructed by inserting a BglII linker into the BamHI site of expression vector pMAL-cRI (New England Biolabs). cDNAs of the spn2+, spn5+, spn6+, and spn7+ coding regions were amplified by reverse transcription-PCR (RT-PCR), using RNA extracted (see below) from strain THP18, and were cloned by TA cloning into vector pT7Blue. The N-terminal portions of the cDNAs were then isolated using a BglII site (included in the primer) and an appropriate site within each gene, as follows (numbers indicate base pairs of the cDNAs): spn2+, AccI (1-408); spn5+, AccII (1-601); spn6+, BamHI (1-585); and spn7+, BamHI (1-594). These fragments were cloned into BglII/HincII- or BglII-cut pMALBB, yielding pMAL(spn2N), pMAL(spn5N), pMAL(spn6N), and pMAL(spn7N), respectively. Site-directed mutagenesis of spn2+ was done in plasmid pAL(spn2-GFP) and in the pT7Blue spn2+ cDNA plasmid as described previously (67). The mutagenized pAL(spn2-GFP) plasmid was then cut with BamHI and NotI, and the spn2 fragment was recloned into BamHI/NotI-cut pAL(GFP), whereas the mutagenized spn2 cDNA from pT7Blue(spn2+) was cloned into pMALBB as described above.
Other plasmids were as follows. pAL(spo3-GFP), pAL(spo3-HA), and pREP81(GFP-psy1) were described previously (53), and pAUSK+ is like pALSK+ (75) except that it contains S. cerevisiae URA3 instead of LEU2. pAU(Spo3-HA) was created by inserting a BamHI-SacI fragment from pAL(spo3-HA) into pAUSK+. pREP41(GFP-spn2) and pREP41(GFP-spn5) were constructed by ligating the BglII-XhoI fragments from the pT7Blue(spn2+) and pT7Blue(spn5+) cDNA clones (see above), respectively, into BglII/XhoI-digested pGFT41 (a gift from Y. Watanabe) (constructed as described for pGFT81 [82], but using pREP41 [9] as the parent plasmid). These plasmids express full-length GFP-Spn2 and GFP-Spn5 under the control of the medium-strength nmt41 promoter; the tagged proteins appeared to be functional based on their ability to rescue the sporulation defects of the corresponding mutants in media lacking thiamine (data not shown). To construct pREP41(GFP-PHOsh2-dimer), a SmaI-SacI fragment containing GFP-PHOsh2-PHOsh2-GFP was excised from pTL511 (66) and ligated into SmaI/SacI-digested pGADT7 (Clontech). An NdeI-XhoI (both sites from pGADT7) fragment from the resulting plasmid was then subcloned into NdeI/XhoI-digested pREP41 (9).
RNA isolation.
Total RNA was extracted using a simplified hot-phenol method based on that described previously (42). Briefly, the frozen cell pellet (∼100 μl) was thawed on ice, suspended in 500 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5), mixed with 500 μl of phenol-chloroform by vortexing, and incubated at 65°C for 1 h with periodic vortexing. After centrifugation, the aqueous phase was removed and extracted as before, using phenol-chloroform. Finally, RNA was precipitated with ethanol and dissolved in diethyl pyrocarbonate-treated water.
Light microscopy.
Cells to be examined by phase-contrast microscopy were suspended in phosphate-buffered saline (PBS) without fixation. Except where noted, cells to be examined by fluorescence microscopy were growing exponentially in EMM liquid culture or were collected from a sporulation plate and suspended in MM-N medium. In either case, cells were usually fixed with formaldehyde, washed, stained for DNA with 4′,6-diamidino-2-phenylindole (DAPI), and (where appropriate) immunostained for Spo3-hemagglutinin (Spo3-HA) essentially as described previously (62). In one case, DAPI staining was accompanied by staining of F-actin using tetramethyl rhodamine isocyanate (TRITC)-phalloidin (Sigma) at 2 μM. In another case, cells were stained with DAPI after fixation with 30% ethanol for ≥15 min at room temperature. In another case, cells were fixed with methanol (8 min at −20°C) and acetone (1 min at 4°C) and washed twice in PBS; DNA was then stained with bisbenzimide H33258 (Sigma) at 50 μg/ml in PBS. Fluorescence of the GFP-PHOsh2 dimer was observed using unfixed cells because fixation destroyed the GFP signal, and some observations of GFP-red fluorescent protein (RFP) double labeling were made using cells that were suspended in PBS without fixation or DAPI staining. Cells were observed using a Zeiss Axiovert 100 fluorescence microscope with a Plan Apochromat 100×/1.40-numerical-aperture (NA) objective (most images), a Nikon Microphot SA microscope with a Plan Apo 60×/1.40-NA objective (see Fig. 4A), an Olympus IX71 inverted microscope with a UPlan Apo 100×/1.35-NA objective (see Fig. 6C and D and 10C), or a Nikon Eclipse 600-FN microscope with a Plan Apo 100×/1.40-NA objective (see Fig. 10B and 11D). To obtain three-dimensional (3D) reconstructed images, IPLab 3.6 software (Scanalytics) was used. The maximum projections of deconvoluted images (see Fig. 11D) were created from z series of eight 0.5-μm steps, using MetaMorph software 7.0 (MDS Analytical Technologies). Time-lapse imaging was performed using MM-N medium and the Axiovert 100 microscope essentially as described previously (62); the glass-bottomed dish was made by gluing a coverslip to the bottom of a Falcon 35-mm plastic dish through which a hole had been drilled.
FIG. 4.
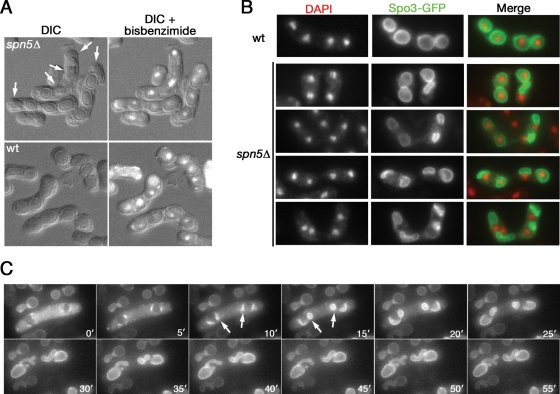
Disoriented FSM extension and defective spore formation in septin mutant cells. (A) Strains JB60 (h− spn5Δ) and JB61 (h+ spn5Δ) were grown overnight in YE medium at 30°C, mixed in equal numbers on an MM-N plate, incubated for 3 days at 25°C, collected, fixed with methanol and acetone, stained with bisbenzimide (see Materials and Methods), and visualized by DIC (left) and by DIC plus fluorescence (right). h− and h+ wild-type strains were handled identically and are shown for comparison. Arrows, immature or defective spores with poorly defined coats and apparently without enclosed nuclei. (B) Strains THP18 (wild type) and TK144 (spn5Δ) harboring pAL(spo3-GFP) were sporulated, fixed, and examined by fluorescence microscopy. (C) Strain TK144 harboring plasmid pREP81(GFP-psy1) was grown on an EMM plate, transferred to an SSA plate, and incubated for 13 h before the beginning of time-lapse observations (see Materials and Methods). Times are indicated in minutes since the beginning of observation. Arrows, FSMs showing disoriented extension.
FIG. 6.
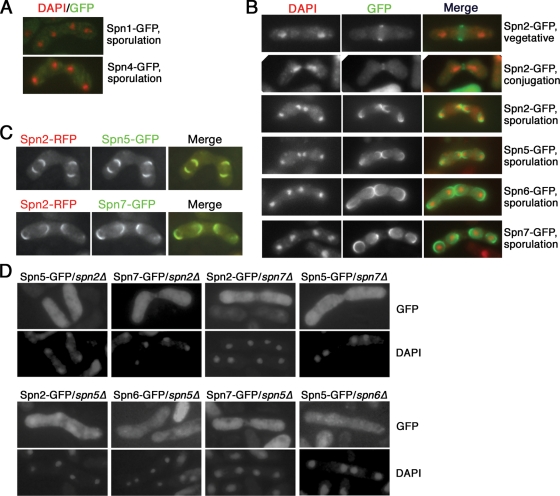
Septin localization during conjugation and sporulation. (A) Absence of Spn1 and Spn4 localization during sporulation. Strains MO808 (spn1+-GFP) and MO807 (spn4+-GFP) were sporulated, stained with DAPI, and observed by fluorescence microscopy. (B) Formation of septin rings at the site of cell fusion during conjugation and around developing spores. Strains MO250 (spn2Δ), TK144 (spn5Δ), MO277 (spn6Δ), and MO229 (spn7Δ) were transformed with plasmids pAL(spn2-GFP), pAL(spn5-GFP), pAL(spn6-GFP), and pAL(spn7-GFP), respectively, and then sporulated, stained with DAPI, and observed by fluorescence microscopy. (Top two rows) Formation of Spn2 rings at the site of septation in vegetative cells (from a population growing in EMM liquid medium) and at the site of cell fusion during conjugation. (Bottom four rows) Localization of Spn2, Spn5, Spn6, and Spn7 to a ring that forms around each developing spore. (C) Colocalization of septins during sporulation. Strains MO900 (spn2+-mRFP spn5+-GFP) and MO902 (spn2+-mRFP spn7+-GFP) were sporulated and observed by fluorescence microscopy without fixation or DAPI staining. (D) Interdependence of septin localization during sporulation. Strains MO250 (spn2Δ), TK144 (spn5Δ), MO277 (spn6Δ), and MO229 (spn7Δ) were transformed with plasmids expressing the indicated GFP-tagged septins and then observed as described for the lower rows in panel B.
FIG. 10.
Seemingly independent localization of septins and LEPs. (A) Localization of Spn2 and Psy1 in meu14Δ cells. Strain SY211 (spn2+-mRFP GFP-psy1+ meu14Δ) was sporulated and observed without fixation. (B) Localization of Meu14 in spnΔ cells. Strains SY13 (meu14+-GFP), SY86 (spn2Δ meu14+-GFP), SY288 (spn5Δ meu14+-GFP), SY284 (spn6Δ meu14+-GFP), and SY292 (spn7Δ meu14+-GFP) were sporulated, fixed, and stained with DAPI. Representative cells in anaphase II (wild type and spn2Δ mutant) or metaphase (spn5Δ, spn6Δ, and spn7Δ mutants) are shown.
FIG. 11.
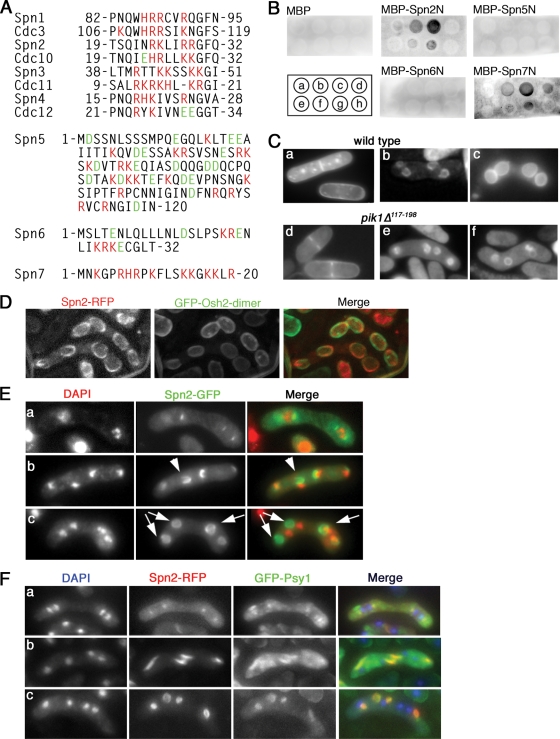
Interaction of Spn2 and Spn7 with phosphoinositides. (A) Clusters of basic amino acids near the N termini of some septins. Spn1 to Spn4 were aligned with their S. cerevisiae orthologues to show the conservation of this aspect of septin structure. For Spn5 to Spn7 (which do not have clear orthologues), the N-terminal sequences are shown without comparisons. Red, basic amino acids; green, acidic amino acids; numbers, amino acid positions relative to the N termini. (B) Binding of Spn2 and Spn7 to PtdIns(4)P and PtdIns(5)P in a protein-lipid overlay assay. Phosphoinositide spots on a nitrocellulose membrane were incubated with purified MBP or the indicated MBP fusion proteins (see Materials and Methods). The fusion proteins contained N-terminal fragments (136 to 200 amino acids in length) of the respective septin. Bound proteins were detected using an anti-MBP antibody. The positions of lipid spots were as follows: a, PtdIns; b, PtdIns(4)P; c, PtdIns(5)P; d, PtdIns(4,5)P2; e, PtdIns(3)P; f, PtdIns(3,4)P2; g, PtdIns(3,5)P2; and h, PtdIns(3,4,5)P3. Small, uneven signals (such as seen for MBP-Spn2N [g]) appear to be nonspecific background because they were independent of the dose of lipid on the membrane (not shown). (C) Presence of PtdIns(4)P in normal FSMs and in the abnormal FSMs of a pik1 mutant. Wild-type strain THP18 (a to c) and pik1Δ117-198 strain MO599 (d to f) were transformed with pREP41(GFP-PHOsh2-dimer), grown in EMM-plus-thiamine liquid medium overnight, shifted to EMM liquid medium (a and d) or to an SSA plate (b, c, e, and f) for 12 h, and examined by fluorescence microscopy. (D) Distinct localizations of septin structures and PtdIns(4)P. Strain MO815 (spn2+-mRFP) was transformed with pREP41(GFP-PHOsh2-dimer) and treated as in panel C. Maximum projections of deconvoluted images are shown. (E) Formation of septin rings that do not surround the nuclei in sporulating pik1 mutant cells. Strain MO599 (pik1Δ117-198) was transformed with plasmid pAL(spn2-GFP), sporulated, and examined by fluorescence microscopy. Cells in metaphase (a) and anaphase (b) and after completion of meiosis II (c) are shown. Arrowhead, a horseshoe-shaped structure that does not surround the adjacent nucleus (seen at 17 to 36% of nuclei at this stage in three independent experiments); arrows, seemingly completed septin rings that do not surround the adjacent nuclei (seen at 76 to 89% of nuclei at this stage in three experiments). (F) Association between abnormal septin rings and abnormal FSMs in pik1 mutant cells. Strain MO832 (pik1Δ117-198 spn2+-mRFP GFP-psy1+) was sporulated and examined by fluorescence microscopy.
Electron microscopy.
Cells were grown on an EMM plate for 12 h at 30°C, transferred to an MM-N plate, incubated for 6 h at 30°C, scraped from the plate, mounted directly on a copper grid to form a thin layer, and plunged into liquid propane cooled with liquid nitrogen (−196°C) in a Leica EM CPC cryoworkstation (Leica Microsystems). The frozen cells were transferred to anhydrous acetone containing 2% OsO4 at −80°C in a Leica EM AFS automatic freeze-substitution apparatus, held at −80°C for 78 h, warmed gradually to 0°C over a period of 11.4 h, held at 0°C for 1.5 h, warmed gradually to 23°C (room temperature) over a period of 3.9 h, and held at 23°C for 2 h. After being washed three times with anhydrous acetone, samples were infiltrated with increasing concentrations of Spurr's resin in anhydrous acetone and, finally, with 100% Spurr's resin. After polymerization (5 h at 50°C plus 60 h at 60°C) in capsules, ultrathin sections were cut on a Leica Ultracut UCT microtome and stained with uranyl acetate and lead citrate. The sections were then viewed on a Hitachi H-7600 electron microscope at 100 kV.
Protein-lipid overlay assay.
Escherichia coli strain BL21 (Novagen) harboring plasmids encoding MBP fusion proteins was cultured at 15°C for 2 days, and the fusion proteins were purified by standard methods (43). A total of 2.5 μl of lipid solution containing 100 picomoles of a phosphoinositide (Wako Chemicals) dissolved in 1:1 chloroform-methanol was spotted onto a Hybond-C-Extra membrane (GE Healthcare) and dried at room temperature. The membrane was incubated with 3% fatty-acid-free bovine serum albumin (BSA) in TBS-T (10 mM Tris-HCl, pH 8.0, containing 150 mM NaCl and 0.1% Tween 20) at ∼23°C for 1 h and then with TBS-T containing 0.5 μg/ml of the indicated protein overnight at 4°C. The membrane was then washed three times (10 min per wash) with TBS-T at ∼23°C. Proteins interacting with the phosphoinositides were detected with an anti-MBP monoclonal antiserum (New England Biolabs) and a horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody (BioSource International).
Expression, purification, and analysis of septin complexes.
cDNAs of the full-length coding regions of spn2+ and spn5+ were cloned into the multicloning sites of the tandem-expression vector pET-Duet1 (Novagen) to achieve expression of untagged Spn2 and His6-tagged Spn5. Similarly, spn6+ and spn7+ sequences were cloned into pCOLA-Duet1 (Novagen) to create two plasmids; one of these expressed untagged Spn6 and untagged Spn7, whereas the other expressed His6-tagged Spn6 and untagged Spn7. To analyze complexes lacking one septin, similar plasmids were constructed that expressed a single septin. Pairs of plasmids were cotransformed into E. coli strain BL21(DE3) (Novagen). The transformants were grown in LB plus ampicillin and kanamycin at 37°C to an optical density at 600 nm (OD600) of 1.0, and expression was induced by adding isopropyl-β-d-thiogalactopyranoside for 16 h at 16°C. Preparation of high-speed supernatants and purification of the septin complexes were done exactly as described previously (36), in the presence of 0.5 M KCl. Polymerization of septin complexes was induced by diluting the solution in the same buffer without KCl.
RESULTS
Defective mating and sporulation in septin mutants.
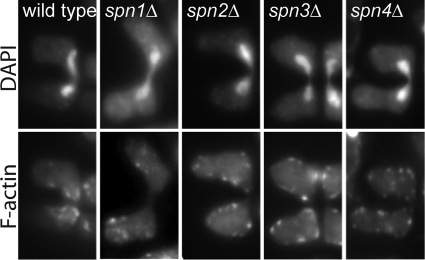
To investigate septin function during mating and sporulation in S. pombe, we constructed septin mutants by deleting part of the septin ORF in a homothallic strain background (Fig. 1). As expected (see the introduction), all mutants were viable. When mating and sporulation were induced, a wild-type control strain formed mating tubes that fused to form zygotes; the initially narrow conjugation bridges then expanded to the full cell diameter, and four spores formed in most asci (Fig. 2A and B). Some apparent asci contained no visible spores, but very few asci (<5%) contained 1 to 3 spores (Fig. 2B). The spn1Δ, spn3Δ, and spn4Δ mutants also formed mostly four-spored asci (Fig. 2A and B); taken together with the protein localization data (see below), these results suggest that these septins are not involved in spore formation. However, these mutants and the spn2Δ mutant showed a defect in the reshaping of the conjugation bridge, which remained narrow even after the spores had formed (Fig. 2A). Consistent with the relative severities of the vegetative-cell defects in mutants lacking these septins (3; our unpublished data), the conjugation bridge defect was most severe in the spn1Δ and spn4Δ mutants and less so in the spn2Δ and spn3Δ mutants (Fig. 2A). The failure to reshape the conjugation bridge was associated with a defect in actin organization at that site. In wild-type cells, F-actin localizes to cortical patches (presumably representing sites of endocytosis [33]) in the projection tip and conjugation bridge during cell fusion (64) (Fig. 3); these patches then disperse after karyogamy. In contrast, the F-actin patches in the spn1Δ, spn2Δ, spn3Δ, and spn4Δ mutants were dispersed before karyogamy (Fig. 3). Taken together with the protein localization data (see below), these mutant phenotypes suggest that the vegetative complex of Spn1, Spn2, Spn3, and Spn4 (3) continues to function during conjugation, perhaps to direct and/or anchor the exocytic and endocytic machinery to the conjugation site.
FIG. 2.
Dependence of normal spore formation on Spn2, Spn5, Spn6, and Spn7 but not on Spn1, Spn3, or Spn4. (A) Phase-contrast images of sporulating strains. Strains THP18 (wild type; wt), MO246 (spn1Δ), MO250 (spn2Δ), MO371 (spn3Δ), MO372 (spn4Δ), TK144 (spn5Δ), MO277 (spn6Δ), and MO229 (spn7Δ) were incubated on SSA plates for 2 days. Arrowheads, asci with <4 spores; arrows, abnormally narrow mating bridges. (B) Percentages of asci with various numbers of spores. Strains and methods were as described for panel A, with the addition of the double mutant strains MO660, MO662, MO664, and TK278. More than 200 cells that appeared to be asci (whether or not they contained detectable spores) were counted for each strain. The frequencies of asci with no, four, and aberrant numbers of spores are shown in the white, black, and hatched areas, respectively; lines in the hatched areas indicate the numbers of asci with one, two, and three spores. (C) Normal meiotic divisions in septin mutants. Diploid strains TW747 (wild type), MO705 (spn2Δ/spn2Δ), TK172 (spn5Δ/spn5Δ), TK367 (spn6Δ/spn6Δ), and MO706 (spn7Δ/spn7Δ) were grown to exponential phase in EMM medium and then washed and resuspended in MM-N medium. Cells were collected at intervals, fixed with ethanol, and stained with DAPI (see Materials and Methods). More than 200 total cells (whether or not they were clearly asci) were counted in each sample to determine the percentages of cells with four distinct nuclear DNA masses.
FIG. 3.
Failure of cortical F-actin patches to accumulate at cell junctions during mating and karyogamy in spn1, spn2, spn3, and spn4 mutant cells. Strains THP18 (wild type), MO246 (spn1Δ), MO250 (spn2Δ), MO371 (spn3Δ), and MO372 (spn4Δ) were incubated on an SSA plate for 6 h, fixed, stained with DAPI and TRITC-phalloidin, and observed by fluorescence microscopy as described in Materials and Methods. Representative images are shown.
In contrast, the spn5Δ, spn6Δ, and spn7Δ mutants displayed no obvious defects in conjugation bridge enlargement (Fig. 2A) or in actin patch localization to that site (data not shown), consistent with the evidence that these genes are not expressed until after conjugation bridge enlargement has occurred. However, these mutants, and to a lesser extent the spn2Δ mutant, showed a strong defect in spore formation in which the majority of asci contained fewer than four spores (Fig. 2A and B). Double mutants and even an spn5Δ spn6Δ spn7Δ triple mutant did not show a more severe defect (Fig. 2B; data not shown), suggesting that Spn2, Spn5, Spn6, and Spn7 all function in the same process during sporulation. Homozygous spn2Δ, spn5Δ, spn6Δ, and spn7Δ diploids also formed many asci with fewer than four visible spores (data not shown) but had no obvious abnormality in meiotic nuclear division (Fig. 2C), suggesting that the defect is indeed in the process of spore formation.
Disoriented FSM extension in septin mutants.
In wild-type cells, differential interference contrast (DIC) microscopy clearly showed the uniform, thick walls of the mature spores (Fig. 4A). In contrast, in septin mutants, some presumptive spores did not appear to have mature walls (Fig. 4A, arrows); these structures always lacked nuclei (Fig. 4A, upper right panel), suggesting that engulfment of a nucleus is a prerequisite for spore maturation. The use of Spo3-GFP as a marker for the FSM revealed uniform cup-shaped or spherical FSMs, each of which engulfed a nucleus, in wild-type cells (53) (Fig. 4B, top panels). In contrast, in septin mutants, the FSMs were frequently misshapen and failed to engulf a nucleus (Fig. 4B). Time-lapse analysis using GFP-Psy1, another FSM marker (53, 55), revealed the defect in more detail (Fig. 4C; see Movie S1 in the supplemental material). At the onset of FSM formation, although some FSMs appeared to behave normally, others extended in inappropriate directions and formed small spheres at an early stage (Fig. 4C, arrows). These FSMs then continued to grow to seemingly normal size, but with aberrant shapes (Fig. 4C, 20′ to 50′). In the spn5Δ strain, 15 of 36 FSMs observed (in nine independent asci) extended in such a disoriented manner. Similar time-lapse observations were made with spn6Δ (three asci) and spn5Δ spn6Δ (two asci) strains (data not shown).
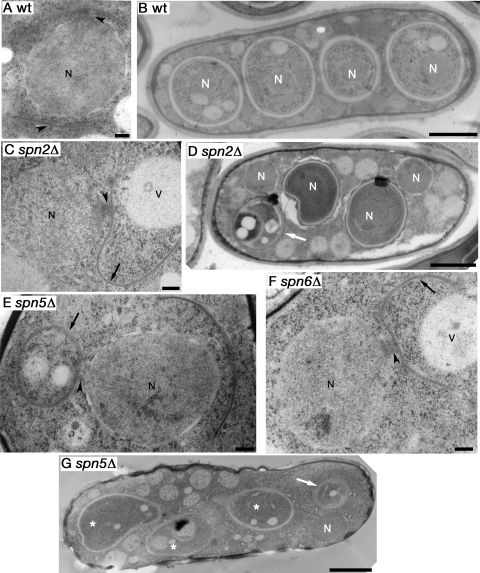
Similar results were obtained by electron microscopy. In wild-type cells, as expected, the nuclei were always engulfed by the FSMs (Fig. 5B). In contrast, the spn2Δ, spn5Δ, and spn6Δ mutants frequently left some nuclei bare (Fig. 5D, F, and G). Although the FSMs appeared to begin extension close to the SPBs in both wild-type and mutant cells (Fig. 5A, C, E, and F, arrowheads), in the mutant cells they frequently curved away from the nuclei, extended asymmetrically from the SPBs, or both (Fig. 5C, E, and F, arrows), and they eventually formed spore-like structures without nuclei (Fig. 5D and G, arrows).
FIG. 5.
Electron microscopic analysis of aberrant FSM and spore wall formation in septin mutants. Diploid strains TW747 (wild type) (A and B), MO705 (spn2Δ/spn2Δ) (C and D), TK172 (spn5Δ/spn5Δ) (E and G), and TK367 (spn6Δ/spn6Δ) (F) were grown, sporulated, and prepared for electron microscopy as described in Materials and Methods. Representative images are shown for each strain. N, nuclei; V, vacuoles; arrowheads, SPBs with FSMs extending from their cytoplasmic faces; black arrows, FSMs extending away from the nuclear envelopes and, in some cases, presumably closing without enclosing a nucleus; white arrows, aberrant spore-like structures presumably formed by aberrant FSM extension and closure; asterisks, FSMs that might possibly enclose nuclei that are not visible in this plane of section. Bars in panels A, C, E, and F, 0.25 μm; bars in panels B, D, and G, 2 μm.
Taken together, the results suggest that the septins are important for the proper orientation of FSM extension and that the formation of aberrant numbers of spores in septin mutants results from the reduced fidelity of this process.
Formation of a novel septin complex that localizes to a ring associated with the FSM.
To investigate how the septins regulate the direction of FSM extension, we examined septin localization during sporulation, using septins tagged at their C termini and expressed from their normal promoters, septins tagged at their N termini and expressed from the nmt41 promoter, or both. As expected (see the introduction), Spn1, Spn2, Spn3, and Spn4 all localized to a ring at the division site in vegetative cells (Fig. 6B, top row; data not shown). In addition, these four proteins also localized to an apparent ring at the site of cell fusion during conjugation (Fig. 6B, second row; data not shown); these rings disappeared by the time that expansion of the conjugation bridges was complete (data not shown). These localization data are consistent with the phenotypic data suggesting that the vegetative septin complex is involved in conjugation bridge expansion (see above).
In sporulating cells, we observed no localization of Spn1, Spn3, and Spn4 (Fig. 6A; data not shown), consistent with the genetic evidence that these proteins are not involved in spore formation (see above). In contrast, Spn2, Spn5, Spn6, and Spn7 colocalized to partial or complete ring-like structures that curled around each haploid nucleus (Fig. 6B, last four rows, and C). Localization of the septins to these structures was interdependent (Fig. 6D), suggesting that in sporulating cells, as in vegetative cells, a septin complex (albeit one composed of a different set of septins) is the functional unit.
To test this hypothesis biochemically, we first tried to purify the putative septin complex from extracts of sporulating cells by using the tandem affinity purification (TAP) system. Surprisingly, however, we were unable to purify any of the TAP-tagged septin proteins, and indeed, these proteins did not appear to be soluble even in the presence of 0.5 M KCl (data not shown). These results differ from those obtained previously with many other septin complexes (3, 19, 20, 36). Indeed, the same Spn2-TAP protein could be purified successfully from vegetative cells (our unpublished results; also see reference 3), suggesting that the septin complexes in vegetative and sporulating cells have different properties.
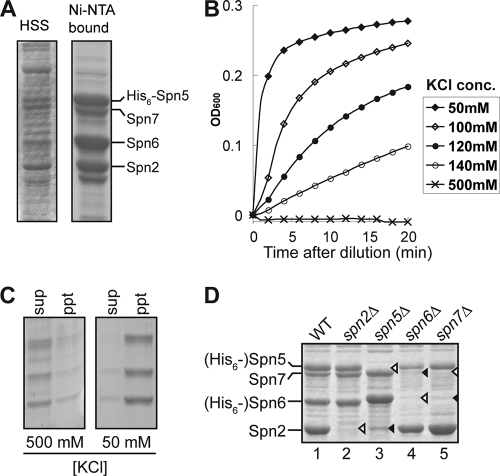
To overcome this problem, we coexpressed the four septins in bacteria as described previously (11). When His6-Spn5 was coexpressed with Spn2, Spn6, and Spn7, complexes were purified that contained the four septins in nearly 1:1:1:1 stoichiometry (Fig. 7A and D, lane 1), indicating that these septins are indeed capable of forming a complex. These complexes appeared to polymerize when the KCl concentration was decreased (Fig. 7B and C), suggesting that septin function during sporulation involves the assembly of higher-order structures and that Spn2, Spn5, Spn6, and Spn7 are sufficient to form a functional complex that is capable of such assembly.
FIG. 7.
Formation of complexes by sporulation septins. (A) His6-Spn5, Spn2, Spn6, and Spn7 were coexpressed in bacteria and extracted as described in Materials and Methods. The high-speed supernatant (HSS) and the complex purified by Ni-nitrilotriacetic acid (Ni-NTA) binding were analyzed by SDS-PAGE and Coomassie staining. (B and C) Polymerization of the purified septin complexes from panel A at lowered KCl concentrations was monitored by measuring turbidity (A600) as a function of time (B) and by centrifuging samples from the 10-min time point at 100,000 × g for 1 h and analyzing the supernatants (sup) and precipitates (ppt) as in panel A (C). (D) Analysis of septin complexes lacking a subunit. His6-Spn5 was coexpressed with Spn2, Spn6, and Spn7 (lane 1), with Spn6 and Spn7 (lane 2), with Spn2 and Spn7 (lane 4), or with Spn2 and Spn6 (lane 5), and His6-Spn6 was coexpressed with Spn2 and Spn7 (lane 3). The complexes were analyzed as in panel A. Open arrowheads, septins not expressed in particular samples; closed arrowheads, septins that were present but did not copurify with the His6-tagged subunit.
When Spn2 was omitted from the expression system, Spn5, Spn6, and Spn7 still formed a stoichiometric complex (Fig. 7D, lane 2), and when Spn5 was omitted, Spn6 and Spn7 still formed a stoichiometric complex (Fig. 7D, lane 3). These data suggest that the sporulation septins may form a linear core tetramer of Spn2-Spn5-(Spn6, Spn7), similar to the linear complexes proposed to form from the septins expressed in vegetative cells (3, 11, 47). The similarity extends even to the presence of Spn2 at an end of the tetramer, where its homotypic association could form the octamers that would in turn be the subunits for assembly of longer filaments (11, 47). However, a problem for this model is that when either Spn6 or Spn7 was omitted from the expression system, the other was also lost from the complexes, which contained only Spn2 associated with His6-Spn5 (Fig. 7D, lanes 4 and 5). A possible explanation could be that an interaction between Spn6 and Spn7 is necessary for one (or conceivably both) of them to bind to Spn5.
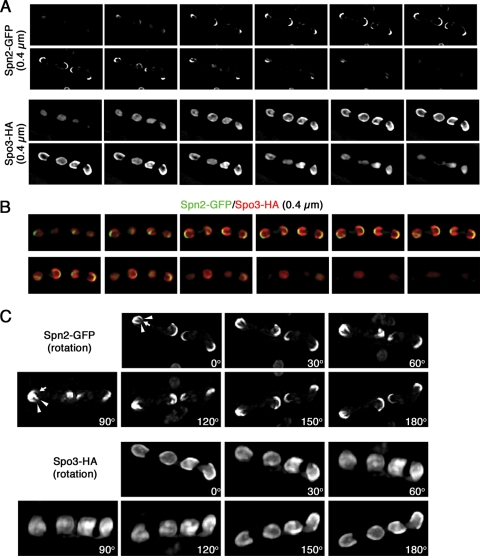
Time-lapse observations revealed that the septin structures, like the FSMs, extended from the vicinity of the SPBs to surround the nuclei (Fig. 8A; see Movie S2 in the supplemental material), and double labeling of a septin and a bona fide FSM marker confirmed the close association of the septin structure with the FSM throughout FSM formation (Fig. 8B). Surprisingly, however, the double labeling also revealed that the septin structure was not coterminous with the FSM at any stage (Fig. 8B, panels a to d). Examination of serial z sections (Fig. 9A and B; see Movies S3 to S5 in the supplemental material) and of rotated images (Fig. 9C; see Movies S6 and S7 in the supplemental material) after three-dimensional deconvolution indicated that whereas the FSM itself is a cup-shaped structure that eventually forms a sphere enclosing the haploid nucleus, the septins appear to form a horseshoe-shaped structure that eventually becomes a ring (compare the images in Fig. 9 to those in Fig. 8A and B, panels c and d; see Fig. 13A). In some cases, there appeared to be an additional projection of labeled septin material from the vicinity of the SPB (Fig. 9C, arrows), suggesting that the nucleation of septin assemblies in this region might sometimes be more complex. However, these structures were short and observed around <5% of the SPBs, and we were unable to characterize them further.
FIG. 8.
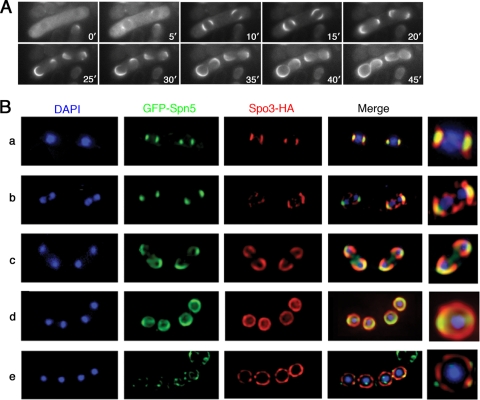
Formation of septin ring during sporulation via a horseshoe-shaped structure that extends along but is not coterminous with the FSM. (A) Wild-type strain THP18 harboring plasmid pREP41(GFP-spn2) was sporulated and observed by time-lapse microscopy. Times are indicated in minutes after the beginning of observation. (B) Wild-type strain THP18 harboring plasmids pREP41(GFP-spn5) and pAU(spo3-HA) was sporulated and observed by immunofluorescence with an anti-HA antibody (see Materials and Methods); Spo3 provides a marker for the entire FSM (53). Two-dimensional projections of three-dimensional deconvoluted images captured as 0.2-μm z sections were ordered according to the stages of meiosis II and spore formation, as follows: a, metaphase; b and c, anaphase; d, telophase; e, a mature ascus in which the Spo3-HA signal has begun to disappear. Magnified images of individual nuclear regions are shown on the right.
FIG. 9.
Comparison of septin localization to that of FSM marker Spo3, using three-dimensional deconvoluted images viewed as serial z sections at 0.4-μm intervals (A and B) or as images rotated by increments of 30° (C). Strain MO701 (spn2+-GFP) was transformed with plasmid pAL(spo3-HA), sporulated, and observed as described in the legend to Fig. 8B. Panels A and C are from the same deconvoluted image. Arrowheads, the arms of the septin horseshoe; arrows, an apparent additional extended septin structure.
FIG. 13.
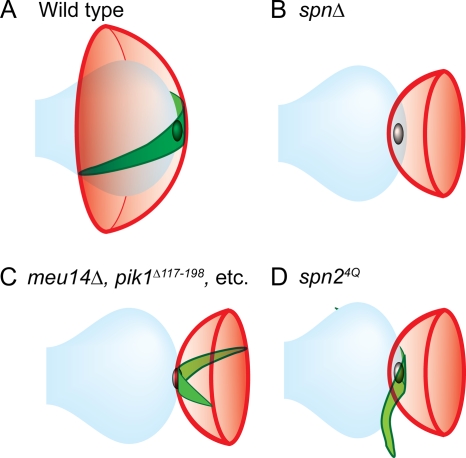
Summary of spatial relationships between the nuclear envelope (blue), the septin structure (green), and the FSM (red) in various strains. See the text for details.
Apparent independence of septin and LEP function.
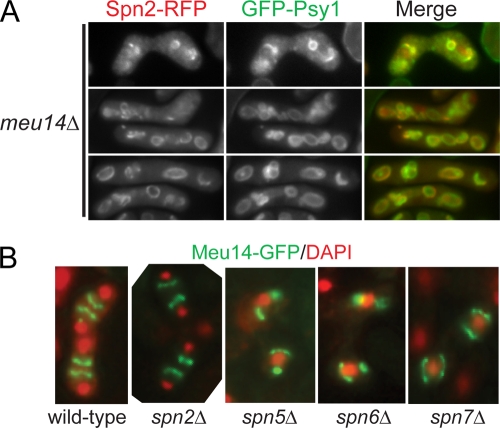
The abnormalities in FSM formation seen in septin mutants (see above) are similar to those seen in mutants lacking the LEP Meu14 (60) (Fig. 10A). This suggested that the septins might function through the LEPs or vice versa. However, the localization of the septins and LEPs appeared to be independent of each other: a labeled septin was consistently associated with the FSMs in a meu14Δ strain, even when the FSMs were abnormal (Fig. 10A; see Fig. 13C), and the localization of Meu14 in septin mutants seemed as normal as could be expected given the abnormalities of FSM structure in these mutants (Fig. 10B). Moreover, the spore formation defect of each spnΔ meu14Δ double mutant was significantly more severe than that of either single-mutant parent (Table 3). Taken together, these data suggest that the septins and the LEPs contribute to FSM morphogenesis in parallel pathways rather than a linear pathway.
TABLE 3.
Genetic interactions between septins and Meu14a
| Expt | Strain | Genotype | % Asci with four spores | % Asci with no spores |
|---|---|---|---|---|
| 1 | THP18 | Wild type | 85 | 2 |
| SY154 | meu14Δ | 65 | 14 | |
| MO684 | spn2Δ | 57 | 5 | |
| SY202 | spn2Δ meu14Δ | 32 | 21 | |
| SY233 | spn5Δ | 17 | 11 | |
| SY430 | spn5Δ meu14Δ | 5 | 62 | |
| SY175 | spn7Δ | 21 | 20 | |
| SY386 | spn7Δ meu14Δ | 3 | 71 | |
| 2 | MO670 | Wild type | 89 | 9 |
| MO905 | meu14Δ | 25 | 20 | |
| MO667 | spn6Δ | 4 | 34 | |
| MO911 | spn6Δ meu14Δ | 1 | 69 |
Data are from two separate experiments. In experiment 1, cells of the indicated strains were incubated on an SSA plate at 30°C for 2 days and examined by phase-contrast microscopy using an Olympus microscope. In experiment 2, cells of the indicated strains were incubated on an MM-N plate at 30°C for 1 day and examined by DIC microscopy using a Nikon Eclipse microscope.
Binding of septins to PtdIns(4)P, a phospholipid enriched in the FSM.
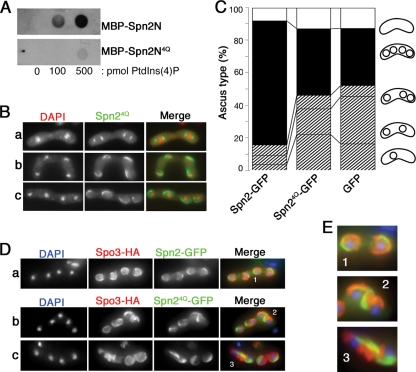
Mammalian SEPT4 and S. cerevisiae Cdc3, Cdc10, Cdc11, and Cdc12 have all been reported to bind directly to phosphoinositides in vitro via the polybasic regions near their N termini (14, 83). To ask if the S. pombe septins might behave similarly, we first examined their sequences. Unambiguous polybasic regions were found near the N termini of five of the seven proteins; in Spn1, Spn2, Spn4, and perhaps Spn3, the sequences were similar to those in the S. cerevisiae orthologues (Fig. 11A). We then used purified MBP-septin fusion proteins and a protein-lipid overlay assay to investigate phosphoinositide binding by the septins that are involved in sporulation. Spn2 and Spn7 bound to PtdIns(4)P and PtdIns(5)P but did not bind convincingly to the other phosphoinositides examined, whereas the less-basic septins Spn5 and Spn6 showed no detectable binding to any of the compounds tested (Fig. 11B). If the four sporulation septins indeed form a complex in vivo, then binding of two septins to phosphoinositides may be sufficient for the entire complex to associate with a membrane.
Because PtdIns(5)P has not been detected in S. pombe (49), we postulated that Spn2 and/or Spn7 might bind to PtdIns(4)P in the FSM. To visualize PtdIns(4)P, we used the GFP-PHOsh2 dimer (see Materials and Methods) as a fluorescent probe; in S. cerevisiae, this probe reveals both the Golgi and plasma membrane pools of PtdIns(4)P (66). In vegetative S. pombe cells, this probe gave a weak signal at the plasma membrane and stronger signals at newly formed septa and at intracellular foci that presumably represent the Golgi apparatus (Fig. 11C, panel a). As expected, these signals were not affected by deletion of the sole S. pombe PtdIns 3-kinase gene, pik3+/vps34+ (data not shown), whereas the presumed Golgi signal disappeared in a mutant defective in the putative Golgi-targeted PtdIns 4-kinase Pik1 (Fig. 11C, panel d; also see below), suggesting that the localization of the GFP-PHOsh2 dimer indeed reflects the distribution of PtdIns(4)P. During sporulation in wild-type cells, the GFP-PHOsh2 dimer was enriched at FSMs, while the signals at the plasma membrane and Golgi apparatus declined or disappeared (Fig. 11C, panels b and c), suggesting that PtdIns(4)P is indeed enriched in the FSMs during sporulation. Because the GFP-PHOsh2 dimer and Spn2-RFP signals were not coterminous (Fig. 11D), PtdIns(4)P does not appear to be enriched specifically in the zone of contact between the FSM and the septin structure but is instead distributed throughout the FSM; thus, the distribution of PtdIns(4)P cannot be solely responsible for determining the architecture of the septin structure (or vice versa).
To ask if PtdIns(4)P in the FSM is important for septin binding, we wished to mutate the relevant PtdIns kinase(s). S. cerevisiae has two type III PtdIns 4-kinases, Pik1 and Stt4, which synthesize PtdIns(4)P in the Golgi apparatus and plasma membrane, respectively (4, 5, 28), and the S. pombe genome encodes apparent orthologues of both enzymes. Although pik1 null mutations are lethal in S. cerevisiae (4), the S. pombe pik1 mutation that we constructed (Fig. 1) was nonlethal, although vegetative cells of the mutant contained only ∼60 to 70% as much total PtdIns(4)P as wild-type cells, with apparently a much more severe reduction in the level of PtdIns(4)P in the Golgi apparatus (Fig. 11C, panel d; our unpublished data). Given the apparent lack of PtdIns(4)P in the Golgi apparatus and the construction of the FSM from Golgi complex-derived vesicles (see the introduction), we were surprised that staining of sporulating pik1 mutant cells with the GFP-PHOsh2 dimer revealed FSM-like structures containing PtdIns(4)P (Fig. 11C, panels e and f). However, these structures were clearly smaller than normal FSMs and were usually abnormal in shape (Fig. 11C, panels e and f), and essentially no recognizable spores were formed (none seen in >200 asci examined [our unpublished data]). Although Spo3 failed to localize to these abnormal FSM-like structures (and was apparently retained in the Golgi apparatus [data not shown]), Psy1 did appear to localize to them (Fig. 11F), and examination of this marker revealed that the FSM-like structures frequently failed to enclose the adjacent nuclei. This phenotype resembles that of the septin mutants, except for the smaller size of the FSM-like structures in the pik1 mutant (which presumably reflects the absence of Spo3 and/or inefficient vesicle transport from the Golgi apparatus to the FSM). Remarkably, the septins did associate with the abnormal FSM-like structures, beginning in what appeared to be a normal location near the poles of the meiosis II spindles (Fig. 11E, panel a, and F, panel a), and thus also formed structures that failed to enclose the adjacent nuclei (Fig. 11E, panels b and c, and F, panels b and c; also see Fig. 13C). These results suggest that the PtdIns(4)P content of the FSM-like membranes is sufficient for septin association but that septin association alone is not sufficient for the normal oriented growth of the FSMs. Attempts to generate strains in which the PtdIns(4)P content of the FSM is more severely reduced have thus far been blocked by the lethality of stt4 knockout strains (data not shown) and the unavailability as yet of a conditional stt4 mutant.
Importance of PtdIns(4)P binding by Spn2 for septin association with the FSM.
As another approach to assessing the importance of septin-phosphoinositide binding for FSM organization, we substituted glutamines for the four conserved basic amino acids in Spn2. The resulting protein, Spn24Q, showed only very weak PtdIns(4)P binding in the protein-lipid overlay assay (Fig. 12A). When expressed in spn2Δ cells, Spn24Q-GFP was found in structures that began to assemble near the spindle poles (Fig. 12B, panel a) and continued to form at least nascent rings (Fig. 12B, panels b and c); thus, it appears that the mutant protein can function as a subunit of the septin complex and that binding to phosphoinositides is dispensable for septin assembly. However, spn24Q did not complement the sporulation defect of spn2Δ cells (Fig. 12C), suggesting that a weakened interaction between the septin assemblage and the FSM resulted in a failure to encapsulate nuclei. Indeed, when we examined the spatial relationship between Spn24Q and the FSM (marked by Spo3) in spn2Δ cells, we found that the FSMs and septin assemblages often extended independently of each other and that neither structure regularly encapsulated the nuclei (Fig. 12D and E and 13D). These results suggest (i) that the septin assemblage can form an extended higher-order structure even when not associated with the FSM, (ii) that phosphoinositide binding by Spn2 is important for septin-FSM association, and (iii) that this association is important for both the septins and the FSMs to extend in a properly oriented manner.
FIG. 12.
Importance of Spn2-phosphoinositide interaction for septin-FSM association and for properly oriented septin and FSM extension and nuclear enclosure. (A) Weak PtdIns(4)P binding in vitro by Spn2 lacking the N-terminal basic amino acids (Spn24Q). Wild-type and mutant Spn2 N-terminal regions were tested for PtdIns(4)P binding by a protein-lipid overlay assay as described in the legend to Fig. 11B, using membranes spotted with the indicated amounts of PtdIns(4)P. (B) Formation of horseshoe-shaped septin structures in spn2Δ cells expressing Spn24Q. Strain MO250 (spn2Δ) was transformed with plasmid pAL(spn24Q-GFP), sporulated, and examined by fluorescence microscopy. Cells in metaphase (a) and anaphase (b) and after completion of meiosis II (c) are shown. (C) Inability of Spn24Q to support normal spore formation. Strain MO250 (spn2Δ) harboring plasmid pAL(spn2-GFP), pAL(spn24Q-GFP), or pAL(GFP) was incubated on an SSA plate for 2 days, and asci with different numbers of spores were counted as described in the legend to Fig. 2B. (D) Inability of Spn24Q to support septin-FSM association or normal septin or FSM extension and enclosure of nuclei. Strain MO684 (spn2Δ) was transformed with plasmid pAU(spo3-HA) and either pAL(spn2-GFP) (a) or pAL(spn24Q-GFP) (b and c), sporulated, and examined by immunofluorescence microscopy using an anti-HA antibody. (E) Magnified images of regions 1, 2, and 3 in panel D.
DISCUSSION
During formation of the yeast spore envelope, the initially small FSM/PSM sac extends in an oriented fashion so as to enclose a nucleus, other organelles, and cytoplasm in a double sphere of membrane; the spore wall then forms between the inner and outer membranes. Earlier work has shown that FSM/PSM extension involves the modified cytoplasmic face of the SPB; the proteins involved in the formation, docking, and fusion of post-Golgi vesicles; phosphoinositide-mediated membrane trafficking; and the LEP complex at the leading edge of the extending membrane sac (see the introduction). However, it has remained unclear whether these mechanisms are sufficient to explain the precise orientation of FSM/PSM extension and its eventual closure to form nascent spores of uniform size. Indeed, studies of S. cerevisiae have indicated that the septins are also involved, although their precise role has been difficult to elucidate because of the inviability of some septin mutants in this species and the relatively mild sporulation phenotypes of the viable mutants. In this study, we have gained deeper insight into the role of septins in spore formation through studies of S. pombe, in which all septin mutants are viable and some have relatively strong sporulation phenotypes. The results also extend our understanding of how septins assemble and associate with membranes and provide insight into the mechanisms by which intracellular membrane systems can acquire specific shapes and three-dimensional organizations.
Role of vegetative septin complex during conjugation.
In vegetative S. pombe cells, Spn1, Spn2, Spn3, and Spn4 form a complex at the division site that seems orthologous to the S. cerevisiae complex of Cdc3, Cdc10, Cdc11, and Cdc12. spn1Δ and spn4Δ mutants have the strongest cell division phenotypes, suggesting that Spn1 and Spn4 are central to complex formation (3, 47). This complex apparently continues to function during conjugation and zygote formation. We found that Spn1, Spn2, Spn3, and Spn4 all localized to an apparent ring at the site of cell fusion and that in the absence of any of these septins, the narrow conjugation bridge failed to expand as it does in wild-type cells. As in vegetative cells, the defects in spn1Δ and spn4Δ mutants were significantly more severe than those in spn2Δ and spn3Δ mutants. Since all four septins are presumably involved in the formation of octamers and higher-order structures (3, 47), it is unclear why some mutants would have weaker phenotypes than others, although it may reflect an ability of septin complexes to form partially functional filaments even when lacking certain subunits (11, 47). Based on the premature dispersal of actin patches from the conjugation bridge in the septin mutants, it seems likely that the septin complex serves to anchor components that are involved in the reshaping of the cell membrane and/or cell wall at that site. Since the actin-nucleating formin Fus1 also localizes to the conjugation bridge (39, 65), it would be interesting to examine its possible interactions with the septins.
Role of a novel septin complex in FSM extension.
Remarkably, in sporulating cells, Spn1, Spn3, and Spn4 are expressed at low levels (45), fail to localize, or both, and spn1Δ, spn3Δ, and spn4Δ mutations have no detectable effect on sporulation efficiency. Instead, efficient spore formation depends on an alternative set of septins, Spn2, Spn5, Spn6, and Spn7, which can form a complex in vitro and assemble interdependently in vivo into a structure associated with the FSM. In the absence of the septins, the FSM appears to begin assembly and extension normally, but it fails, in ∼50% of cases, to extend in a properly oriented fashion and to envelop the associated nucleus (Fig. 13B). Interestingly, although we could not detect localization of the other septins in the spn2Δ mutant, its spore formation defect was less severe than those of the spn5Δ, spn6Δ, and spn7Δ mutants, suggesting that like the case in vegetative and conjugating cells, Spn2 might be less critical than other septins to the formation of a functional septin complex. Indeed, Spn5, Spn6, and Spn7 still formed a complex in vitro in the absence of Spn2. Another interesting feature of the sporulation septin complex is that Spn7 appears to be an integral component despite its lack of features that characterize most septins, namely, a well-defined GTP-binding site (which in other septins appears to be critical for septin-septin interaction [47, 52, 69]) and a C-terminal coiled-coil domain.
Also remarkable is the overall form of the septin structure. Rather than a cup-shaped structure (like the FSM/PSM) or a ring associated with the lip of the FSM/PSM cup (like the LEP complex), the septins appear to form a curved, two-dimensional bar (a “horseshoe”) whose midpoint is at or near the SPB (Fig. 13A). As the FSM extends and closes to form the spherical prespores, the septin structure appears to extend and close to form an equatorial ring. It seems possible that an intrinsic propensity of septin assemblies to form curved bars that can become rings or spirals (18, 21, 36, 47, 80) has been exploited in this system to help ensure appropriate curvature of the extending FSM. Although it is not yet clear whether the septin structure lies on the inner or outer face of the FSM, the former possibility seems more likely because, in S. cerevisiae, tagged molecules of Cdc10 that were found in the PSM-associated structures could later be found at the bud necks in germinating spores (48), implying that they were present in the spore cytoplasm after PSM closure. This model predicts that the septin filaments are nucleated at or near the SPB and therefore are likely to constitute two (or occasionally more) (Fig. 9C) distinct bars rather than a continuous horseshoe; in this case, septin organization along the FSM in S. pombe might be more similar to that thought to exist along the PSM in S. cerevisiae (59) than is apparent from the images available to date. However, we were unable to detect a gap in the septin staining near the SPB by fluorescence microscopy, and analysis at a higher resolution (e.g., by electron microscopy) will presumably be needed to resolve this issue.
Further analysis of the sporulation septins should contribute to a general understanding of the organization and assembly of septin complexes, particularly if the sporulation complexes are distinct from those in vegetative cells, as suggested both by Neiman (59) and by our biochemical data (insolubility of the complexes in sporulating cells and interdependent binding of Spn6 and Spn7 in vitro). It should also be instructive to explore further the apparent differences between the septin structures observed in S. pombe and those seen in S. cerevisiae (17, 37, 51, 59, 71).
Another interesting question is why properly oriented FSM extension and nuclear envelopment fail only about half of the time in the septin mutants. Similarly, it is remarkable that at least some of the FSMs that do not have contact with a septin structure and do not surround a nuclear envelope nonetheless appear to close to form quasispherical structures. These observations suggest that another system, such as the LEP complex, is also capable of guiding FSM extension and closure but is not robust in function in the absence of the septin structure. Since the available data suggest that the septins and the LEPs function in parallel (59, 71; see Results), it is possible that a differential ability of the LEPs to function in the absence of septins may account for the differential severities of septin mutant sporulation phenotypes in S. cerevisiae (15, 17, 59) and S. pombe (this study).
Role of phosphoinositide binding in septin-FSM association and normal FSM extension.
Although many of the biological roles of the septins involve a close association with membranes (12, 27, 41), the mechanisms of septin-membrane association have remained poorly understood. It has been reported that both mammalian and S. cerevisiae septins can bind specific phosphoinositides via clusters of basic amino acids near their N termini (14, 83), suggesting that septins might associate with membranes by direct interaction with the membrane phospholipids (47). Our results support and significantly extend this model. First, we found that Spn2 and Spn7, which have well-defined basic clusters, bound phosphoinositides in vitro, whereas Spn5 and Spn6, which lack such clusters, did not (at least with the assay used here). Second, phosphoinositide binding by Spn2 was essentially eliminated by replacing its cluster of basic amino acids by uncharged residues (Spn24Q). Third, we detected binding specifically to PtdIns(4)P and PtdIns(5)P but not to other phosphoinositides and showed that PtdIns(4)P was highly enriched in the FSM and thus could potentially account for the association of the septins with that structure. Fourth, although the pik1Δ117-198 mutation reduced the total cellular PtdIns(4)P level and produced gross alterations of FSM development, it did not greatly reduce the PtdIns(4) content of the FSM, and the septins still associated with the aberrant FSMs (Fig. 13C). Finally, although spn2Δ cells expressing Spn24Q could at least assemble nascent septin horseshoe/ring structures, these structures did not consistently associate with the FSMs, and neither the septin structures nor the FSMs efficiently enclosed the adjacent nuclei (Fig. 13D). Thus, the data suggest (i) that the basic amino acid cluster is critical for septin-phosphoinositide binding in vivo as well as in vitro, (ii) that such binding is critical for septin-membrane association, and (iii) that such association is critical for both the septin structure and the FSM to extend with a normal orientation (although each structure is capable of extension in the absence of association with the other).
Three additional points should be noted. First, we cannot entirely rule out the possibility that the Spn24Q protein has a subtly altered structure that in turn alters the structure of the septin complex and in this way blocks its association with the membrane. However, this seems unlikely given that Spn24Q appears fully competent to assemble into extended septin structures and that the septin-FSM association otherwise seems quite robust (as evidenced by its persistence even when the FSM is abnormal in meu14Δ and pik1 mutants). Second, although all of our data, like those of Casamayor and Snyder (14), are consistent with PtdIns(4)P being the phosphoinositide to which the septins bind in yeast membranes, it is not clear that the in vitro phospholipid-binding assays faithfully reflect binding in vivo. Thus, it remains possible, as McMurray and Thorner (47) have suggested, that binding in vivo is actually to a different phospholipid, such as PtdIns(4,5)P2. Indeed, localization data obtained using a different fluorescent marker (GFP-PHPLCδ) suggest that PtdIns(4,5)P2 is also enriched in the FSM (our unpublished observations). Finding ways to selectively reduce PtdIns(4)P or PtdIns(4,5)P2 in the FSM would allow us to address this question. Third, although the pik1Δ117-198 cells displayed substantial amounts of PtdIns(4)P in the FSM and septin association with that structure, their defect in FSM development and spore formation was actually more severe than that of the septin deletion and spn24Q mutants. Thus, Pik1-produced PtdIns(4)P must be important in some other way for FSM extension. Because the presumed Golgi PtdIns(4)P signal was lost in the pik1Δ117-198 mutant (see Results) and the aberrant FSM structures were consistently smaller in this mutant than in the septin mutants (our unpublished results), it seems likely that at least part of the role of Pik1-produced PtdIns(4)P is in the delivery of membrane from the Golgi apparatus to the developing FSM. However, further studies will clearly be necessary to clarify the role of Pik1.
Supplementary Material
Acknowledgments
We thank Y. Watanabe (University of Tokyo), H. Nojima (Osaka University), and T. Levine (University College London) for plasmids and strains and Sadayuki Yamada of the Y. Fukui laboratory for help with strain construction. Some reagents were provided via Japan's National BioResource Project—Yeast.
This work was supported by a postdoctoral fellowship from the Uehara Memorial Foundation (to M.O.), by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan (to Y.F.), and by National Institutes of Health grant GM-31006 (to J.R.P.).
Footnotes
Published ahead of print on 1 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abe, H., and C. Shimoda. 2000. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 154:1497-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Awar, O. S. 1996. The role of septins in the morphogenesis of Schizosaccharomyces pombe and Drosophila melanogaster. Ph.D. thesis. University of North Carolina, Chapel Hill, NC.
- 3.An, H., J. L. Morrell, J. L. Jennings, A. J. Link, and K. L. Gould. 2004. Requirements of fission yeast septins for complex formation, localization, and function. Mol. Biol. Cell 15:5551-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Audhya, A., M. Foti, and S. D. Emr. 2000. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11:2673-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Audhya, A., and S. D. Emr. 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2:593-605. [DOI] [PubMed] [Google Scholar]
- 6.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 7.Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841-851. [DOI] [PubMed] [Google Scholar]
- 8.Barral, Y. 2008. Yeast septins: a cortical organizer, p. 101-123. In P. A. Hall, S. E. H. Russell, and J. R. Pringle (ed.), The septins. Wiley Blackwell, London, United Kingdom.
- 9.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 10.Berlin, A., A. Paoletti, and F. Chang. 2003. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J. Cell Biol. 160:1083-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertin, A., M. A. McMurray, P. Grob, S. Park, G. Garcia III, I. Patanwala, H. Ng, T. Alber, J. Thorner, and E. Nogales. 2008. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA 105:8274-8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byers, B., and L. Goetsch. 1976. A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69:717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casamayor, A., and M. Snyder. 2003. Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol. Cell. Biol. 23:2762-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Virgilio, C., D. J. DeMarini, and J. R. Pringle. 1996. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology 142:2897-2905. [DOI] [PubMed] [Google Scholar]
- 16.Egel, R., and M. Egel-Mitani. 1974. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88:127-134. [DOI] [PubMed] [Google Scholar]
- 17.Fares, H., L. Goetsch, and J. R. Pringle. 1996. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 132:399-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farkasovsky, M., P. Herter, B. Voss, and A. Wittinghofer. 2005. Nucleotide binding and filament assembly of recombinant yeast septin complexes. Biol. Chem. 386:643-656. [DOI] [PubMed] [Google Scholar]
- 19.Field, C. M., O. Al-Awar, J. Rosenblatt, M. L. Wong, B. Alberts, and T. J. Mitchison. 1996. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133:605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frazier, J. A., M. L. Wong, M. S. Longtine, J. R. Pringle, M. Mann, T. J. Mitchison, and C. Field. 1998. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gale, C., M. Gerami-Nejad, M. McClellan, S. Vandoninck, M. S. Longtine, and J. Berman. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell 12:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gladfelter, A. S., J. R. Pringle, and D. J. Lew. 2001. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4:681-689. [DOI] [PubMed] [Google Scholar]
- 23.Gladfelter, A. S., and P. Sudbery. 2008. Septins in four model fungal systems: diversity in form and function, p. 125-145. In P. A. Hall, S. E. H. Russell, and J. R. Pringle (ed.), The septins. Wiley Blackwell, London, United Kingdom.
- 24.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe. Handb. Genet. 1:395-446. [Google Scholar]
- 25.Haarer, B. K., and J. R. Pringle. 1987. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol. Cell. Biol. 7:3678-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagan, I., and M. Yanagida. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall, P. A., S. E. H. Russell, and J. R. Pringle. 2008. The septins. Wiley Blackwell, London, United Kingdom.
- 28.Hama, H., E. A. Schnieders, J. Thorner, J. Y. Takemoto, and D. B. DeWald. 1999. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:34294-34300. [DOI] [PubMed] [Google Scholar]
- 29.Hirata, A., and C. Shimoda. 1994. Structural modification of spindle pole bodies during meiosis II is essential for the normal formation of ascospores in Schizosaccharomyces pombe: ultrastructural analysis of spo mutants. Yeast 10:173-183. [DOI] [PubMed] [Google Scholar]
- 30.Ikemoto, S., T. Nakamura, M. Kubo, and C. Shimoda. 2000. S. pombe sporulation-specific coiled-coil protein Spo15p is localized to the spindle pole body and essential for its modification. J. Cell Sci. 113:545-554. [DOI] [PubMed] [Google Scholar]
- 31.John, C. M., R. K. Hite, C. S. Weirich, D. J. Fitzgerald, H. Jawhari, M. Faty, D. Schläpfer, R. Kroschewski, F. K. Winkler, T. Walz, Y. Barral, and M. O. Steinmetz. 2007. The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 26:3296-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaback, D. B., and L. R. Feldberg. 1985. Saccharomyces cerevisiae exhibits a sporulation-specific temporal pattern of transcript accumulation. Mol. Cell. Biol. 5:751-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaksonen, M., C. P. Toret, and D. G. Drubin. 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell. Biol. 7:404-414. [DOI] [PubMed] [Google Scholar]
- 34.Kashiwazaki, J., T. Nakamura, T. Iwaki, K. Takegawa, and C. Shimoda. 2005. A role for fission yeast Rab GTPase Ypt7p in sporulation. Cell Struct. Funct. 30:43-49. [DOI] [PubMed] [Google Scholar]
- 35.Kimura, K., S. Miyake, M. Makuuchi, R. Morita, T. Usui, M. Yoshida, S. Horinouchi, and Y. Fukui. 1995. Phosphatidylinositol-3 kinase in fission yeast: a possible role in stress responses. Biosci. Biotechnol. Biochem. 59:678-682. [DOI] [PubMed] [Google Scholar]
- 36.Kinoshita, M., C. M. Field, M. L. Coughlin, A. F. Straight, and T. J. Mitchison. 2002. Self- and actin-templated assembly of mammalian septins. Dev. Cell 3:791-802. [DOI] [PubMed] [Google Scholar]
- 37.Knop, M., and K. Strasser. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19:3657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koga, T., M. Onishi, Y. Nakamura, A. Hirata, T. Nakamura, C. Shimoda, T. Iwaki, K. Takegawa, and Y. Fukui. 2004. Sorting nexin homologues are targets of phosphatidylinositol 3-phosphate in sporulation of Schizosaccharomyces pombe. Genes Cells 9:561-574. [DOI] [PubMed] [Google Scholar]
- 39.Kurahashi, H., Y. Imai, and M. Yamamoto. 2002. Tropomyosin is required for the cell fusion process during conjugation in fission yeast. Genes Cells 7:375-384. [DOI] [PubMed] [Google Scholar]
- 40.Leupold, U. 1970. Genetical methods for Schizosaccharomyces pombe. Methods Cell Physiol. 4:169-177. [Google Scholar]
- 41.Longtine, M. S., D. J. DeMarini, M. L. Valencik, O. S. Al-Awar, H. Fares, C. De Virgilio, and J. R. Pringle. 1996. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8:106-119. [DOI] [PubMed] [Google Scholar]
- 42.Lyne, R., G. Burns, J. Mata, C. J. Penkett, G. Rustici, D. Chen, C. Langford, D. Vetrie, and J. Bähler. 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maina, C. V., P. D. Riggs, A. G. Grandea III, B. E. Slatko, L. S. Moran, J. A. Tagliamonte, L. A. McReynolds, and C. D. Guan. 1988. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene 74:365-373. [DOI] [PubMed] [Google Scholar]
- 44.Martín-Cuadrado, A. B., J. L. Morrell, M. Konomi, H. An, C. Petit, M. Osumi, M. Balasubramanian, K. L. Gould, F. Del Rey, and C. R. Vázquez de Aldana. 2005. Role of septins and the exocyst complex in the function of hydrolytic enzymes responsible for fission yeast cell separation. Mol. Biol. Cell 16:4867-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mata, J., R. Lyne, G. Burns, and J. Bähler. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32:143-147. [DOI] [PubMed] [Google Scholar]
- 46.McLeod, M., M. Stein, and D. Beach. 1987. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 6:729-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMurray, M. A., and J. Thorner. 2008. Biochemical properties and supramolecular architecture of septin hetero-oligomers and septin filaments, p. 49-100. In P. A. Hall, S. E. H. Russell, and J. R. Pringle (ed.), The septins. Wiley Blackwell, London, United Kingdom.
- 48.McMurray, M. A., and J. Thorner. 2008. Septin stability and recycling during dynamic structural transitions in cell division and development. Curr. Biol. 18:1203-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitra, P., Y. Zhang, L. E. Rameh, M. P. Ivshina, D. McCollum, J. J. Nunnari, G. M. Hendricks, M. L. Kerr, S. J. Field, L. C. Cantley, and A. H. Ross. 2004. A novel phosphatidylinositol(3,4,5)P3 pathway in fission yeast. J. Cell Biol. 166:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 51.Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko, A. Shevchenko, and M. Knop. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20:6946-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagaraj, S., A. Rajendran, C. E. Jackson, and M. S. Longtine. 2008. Role of nucleotide binding in septin-septin interactions and septin localization in Saccharomyces cerevisiae. Mol. Cell. Biol. 28:5120-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamura, T., M. Nakamura-Kubo, A. Hirata, and C. Shimoda. 2001. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding syntaxin-like protein. Mol. Biol. Cell 12:3955-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura, T., J. Kashiwazaki, and C. Shimoda. 2005. A fission yeast SNAP-25 homologue, SpSec9, is essential for cytokinesis and sporulation. Cell Struct. Funct. 30:15-24. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura, T., H. Asakawa, Y. Nakase, J. Kashiwazaki, Y. Hiraoka, and C. Shimoda. 2008. Live observation of forespore membrane formation in fission yeast. Mol. Biol. Cell 19:3544-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakamura-Kubo, M., T. Nakamura, A. Hirata, and C. Shimoda. 2003. The fission yeast spo14+ gene encoding a functional homologue of budding yeast Sec12 is required for the development of forespore membranes. Mol. Biol. Cell 14:1109-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakase, Y., M. Nakamura-Kubo, Y. Ye, A. Hirata, C. Shimoda, and T. Nakamura. 2008. Meiotic spindle pole bodies acquire the ability to assemble the spore plasma membrane by sequential recruitment of sporulation-specific components in fission yeast. Mol. Biol. Cell 19:2476-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neiman, A. M. 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:565-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okuzaki, D., W. Satake, A. Hirata, and H. Nojima. 2003. Fission yeast meu14+ is required for proper nuclear division and accurate forespore membrane formation during meiosis II. J. Cell Sci. 116:2721-2735. [DOI] [PubMed] [Google Scholar]
- 61.Onishi, M., T. Koga, R. Morita, Y. Nakamura, T. Nakamura, C. Shimoda, K. Takegawa, A. Hirata, and Y. Fukui. 2003. Role of phosphatidylinositol 3-phosphate in formation of forespore membrane in Schizosaccharomyces pombe. Yeast 20:193-206. [DOI] [PubMed] [Google Scholar]
- 62.Onishi, M., M. Iida, T. Koga, S. Yamada, A. Hirata, T. Iwaki, K. Takegawa, Y. Fukui, and H. Tachikawa. 2007. Schizosaccharomyces pombe Sst4p, a conserved Vps27/Hrs homolog, functions downstream of phosphatidylinositol 3-kinase Pik3p to mediate proper spore formation. Eukaryot. Cell 6:2343-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pan, F., R. L. Malmberg, and M. Momany. 2007. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 7:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petersen, J., O. Nielsen, R. Egel, and I. M. Hagan. 1998. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J. Cell Sci. 111:867-876. [DOI] [PubMed] [Google Scholar]
- 65.Petersen, J., O. Nielsen, R. Egel, and I. M. Hagan. 1998. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141:1217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roy, A., and T. P. Levine. 2004. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 279:44683-44689. [DOI] [PubMed] [Google Scholar]
- 67.Sawano, A., and A. Miyawaki. 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28:E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shimoda, C. 2004. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 117:389-396. [DOI] [PubMed] [Google Scholar]
- 69.Sirajuddin, M., M. Farkasovsky, F. Hauer, D. Kühlmann, I. G. Macara, M. Weyand, H. Stark, and A. Wittinghofer. 2007. Structural insight into filament formation by mammalian septins. Nature 449:311-315. [DOI] [PubMed] [Google Scholar]
- 70.Spiliotis, E. T., and W. J. Nelson. 2006. Here come the septins: novel polymers that coordinate intracellular functions and organization. J. Cell Sci. 119:4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takegawa, K., D. B. DeWald, and S. D. Emr. 1995. Schizosaccharomyces pombe Vps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J. Cell Sci. 108:3745-3756. [DOI] [PubMed] [Google Scholar]
- 73.Takizawa, P. A., J. L. DeRisi, J. E. Wilhelm, and R. D. Vale. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290:341-344. [DOI] [PubMed] [Google Scholar]
- 74.Tanaka, K., and A. Hirata. 1982. Ascospore development in the fission yeasts Schizosaccharomyces pombe and S. japonicus. J. Cell Sci. 56:263-279. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka, K., T. Yonekawa, Y. Kawasaki, M. Kai, K. Furuya, M. Iwasaki, H. Murakami, M. Yanagida, and H. Okayama. 2000. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20:3459-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tasto, J. J., J. L. Morrell, and K. L. Gould. 2003. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 160:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vrabioiu, A. M., and T. J. Mitchison. 2007. Symmetry of septin hourglass and ring structures. J. Mol. Biol. 372:37-49. [DOI] [PubMed] [Google Scholar]
- 78.Watanabe, T., K. Miyashita, T. T. Saito, T. Yoneki, Y. Kakihara, K. Nabeshima, Y. A. Kishi, C. Shimoda, and H. Nojima. 2001. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 29:2327-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu, J.-Q., J. R. Kuhn, D. R. Kovar, and T. D. Pollard. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell 5:723-734. [DOI] [PubMed] [Google Scholar]
- 80.Xie, H., M. Surka, J. Howard, and W. S. Trimble. 1999. Characterization of the mammalian septin H5: distinct patterns of cytoskeletal and membrane association from other septin proteins. Cell Motil. Cytoskel. 43:52-62. [DOI] [PubMed] [Google Scholar]
- 81.Yamamoto, M., and Y. Watanabe. 1997. Mating and sporulation in Schizosaccharomyces pombe, p. 1037-1106. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 82.Yamashita, A., Y. Watanabe, N. Nukina, and M. Yamamoto. 1998. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95:115-123. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, J., C. Kong, H. Xie, P. S. McPherson, S. Grinstein, and W. S. Trimble. 1999. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr. Biol. 9:1458-1467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.