Abstract
Lysine-specific demethylase 1 (LSD1) functions as a transcriptional coregulator by modulating histone methylation. Its role in neural stem cells has not been studied. We show here for the first time that LSD1 serves as a key regulator of neural stem cell proliferation. Inhibition of LSD1 activity or knockdown of LSD1 expression led to dramatically reduced neural stem cell proliferation. LSD1 is recruited by nuclear receptor TLX, an essential neural stem cell regulator, to the promoters of TLX target genes to repress the expression of these genes, which are known regulators of cell proliferation. The importance of LSD1 function in neural stem cells was further supported by the observation that intracranial viral transduction of the LSD1 small interfering RNA (siRNA) or intraperitoneal injection of the LSD1 inhibitors pargyline and tranylcypromine led to dramatically reduced neural progenitor proliferation in the hippocampal dentate gyri of wild-type adult mouse brains. However, knockout of TLX expression abolished the inhibitory effect of pargyline and tranylcypromine on neural progenitor proliferation, suggesting that TLX is critical for the LSD1 inhibitor effect. These findings revealed a novel role for LSD1 in neural stem cell proliferation and uncovered a mechanism for neural stem cell proliferation through recruitment of LSD1 to modulate TLX activity.
TLX is an orphan nuclear receptor that plays an important role in vertebrate brain functions (12, 14, 27, 28). We have shown that TLX is an essential regulator of neural stem cell maintenance and self-renewal in both embryonic and adult brains (8, 14, 18, 30). TLX acts by controlling the expression of a network of target genes to establish the undifferentiated and self-renewable state of neural stem cells. Elucidating molecular mechanisms underlying TLX regulation would be a significant advance in understanding neural stem cell self-renewal and neurogenesis. The transcription action of nuclear receptors is modulated by an extensive set of nuclear receptor cofactors (4, 10, 13). The identification and characterization of the coregulator complexes are essential for understanding the mechanistic basis of nuclear receptor-regulated events. Identifying TLX transcriptional coregulators in neural stem cells would represent a major step in uncovering TLX-mediated transcriptional regulation.
Histone modifications, such as acetylation, phosphorylation, and methylation, are switches that alter chromatin structure to form a binding platform for downstream “effector” proteins to allow transcriptional activation or repression (24). Each modification can affect chromatin architecture, yet the sum of these modifications may be the ultimate determinant of the chromatin state that regulates gene transcription (5, 17). Histone methylation has been linked to transcriptional activation and repression (29). Whether methylation leads to transcriptional activation or repression is influenced by a variety of factors, including the types of histone, the lysine acceptor, the histone location, and other contextual influences. In general, methylation of histone H3 lysine 9 (H3K9), H3K27, or H4K20 is linked to formation of tightly packed chromatin and gene silencing, whereas methylation on H3K4, H3K36, and H3K79 is associated with actively transcribed regions and gene activation (9). Lysine methylation exists in three different states, i.e., mono-, di-, or trimethylation, which brings about additional regulatory complexity.
The recent discovery of a large number of histone demethylases indicates that demethylases play a central role in the regulation of histone methylation dynamics (1-3, 6, 11, 16, 20, 22, 25). The first lysyl demethylase identified is lysine-specific demethylase 1 (LSD1), which demethylates H3K4 or H3K9 in a reaction that uses flavin as a cofactor. LSD1 is limited to mono- or dimethylated substrates (16). In 2005, it was predicted that there exists a second class of histone demethylases that contain a jumonji C (Jmjc) domain (19), a motif present in many proteins that are known to regulate transcription. The identification of the amino oxidase LSD1 and of the Jmjc domain-containing hydroxylases demonstrates that histone methylation is reversible and dynamically regulated (23).
We show here that the histone demethylase LSD1 is expressed in neural stem cells and plays an important role in neural stem cell proliferation. Both chemical inhibition of LSD1 activity and small interfering RNA (siRNA) knockdown of LSD1 expression led to marked inhibition of neural stem cell proliferation. Furthermore, LSD1 functions in neural stem cells through interaction with the stem cell regulator TLX. The inhibitory effect on neural stem cell proliferation by LSD1 siRNA was reduced dramatically in TLX siRNA-treated cells. LSD1 is recruited to the promoters of TLX downstream target genes along with histone deacetylase 5 (HDAC5) to repress TLX target gene expression. Moreover, treatment of adult mice with LSD1 siRNA or inhibitors resulted in dramatically reduced cell proliferation in the hippocampal dentate gyri of wild-type brains. However, the LSD1 inhibitors had almost no effect on cell proliferation in TLX-null brains. These results suggest that LSD1 is an important regulator of neural stem cell proliferation via modulation of TLX signaling.
MATERIALS AND METHODS
Plasmid DNAs and transient transfections.
The hemagglutinin (HA)-TLX construct was generated by cloning TLX cDNA into CMX-HA vector. p21-luc was generated by cloning a 3-kb natural p21 promoter containing the consensus TLX binding site and placing it upstream of a luciferase reporter gene. For reporter assays, HEK 293 and CV-1 cells were transfected as described previously (15). Neural stem cells were transfected using a Nucleofector kit (Amaxa) and program A33. Reporter luciferase activity was normalized by the level of β-galactosidase activity. Each transfection was carried out in triplicates.
Cell culture, IP, Western blotting, and ChIP.
Neural stem cells were isolated from adult mouse brains as described previously (14) and cultured in Dulbecco modified Eagle medium-F12 medium with 1 mM l-glutamine, N2 supplement (Gibco-BRL), epidermal growth factor (EGF) (20 ng/ml), fibroblast growth factor 2 (FGF2) (20 ng/ml), and heparin (50 ng/ml). For differentiation, neural stem cells were exposed to N2-supplemented medium containing 1 μM retinoic acid and 0.5% fetal bovine serum for 7 days. Immunoprecipitation (IP) of neural stem cells was performed using TLX antibody followed by protein A-Sepharose incubation. IP of HEK293 cells was performed using anti-Flag antibody (Sigma) or anti-HA antibody (Santa Cruz). Western blotting was performed using antibodies specific for HA (1:1,000; Santa Cruz), Flag (1:6,000; Sigma), LSD1 (1:1,000; Cell Signaling Technology), HDAC5 (1:100; Santa Cruz), p21 (1:500; Oncogene), and Pten (1:1,000; Cell Signaling). Chromatin IP (ChIP) assays were performed using the EZ-ChIP kit (Upstate) and 5 μg antibody per reaction. Antibodies used in the ChIP assays included antibodies specific for TLX (Shi lab), LSD1 (Cell Signaling Technology), HDAC5 (Santa Cruz), monomethyl H3K4 (Abcam), dimethyl H3K4 (Abcam), and trimethyl H3K4 (Abcam). Primers for quantitative ChIP assays included p21 F (5′-AGT TCT GAT TTC TCA GGG ATA TG-3′) and p21 R (5′-CTG TTG CTG CTA CCC AGG AAG GA-3′), pten F (5′-GTA CGC TAG TTA TCA GCA TTT CT-3′) and pten R (5′-TGA GCG CGC CCC ACC TAC CGT AGA-3′), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) F (5′-TCT TCA CCA CCA TGG AGA AGG C-3′) and GAPDH R (5′-CTG ACA ATC TTG AGT GAG TTG T-3′).
RT-PCR and RNA interference assays.
Reverse transcription (RT) in RT-PCR analysis was performed using the Omniscript RT kit (Qiagen) as described previously (18). Primers for semiquantitative RT-PCR included LSD1 forward primer (5′-GTG TTA TGC TTT GAC CGT GTG T-3′) and LSD1 reverse primer (5′-CAA AGA AGA GTC TTG GGA TTG G-3′), TLX forward primer (5′-GGC TCT CTA CTT CCG TGG ACA-3′) and TLX reverse primer (5′-GCA TTC CGG AAA CTT CTC AGT TCA GTT CAG AAC CAC TG-3′), HDAC5 forward primer (5′-CTC AAG CAG CAG CAG CAG CTC CA-3′) and HDAC5 reverse primer (5′-CCT TCT GTT TAA GCC TCG AAC G-3′), p21 forward primer (5′-CAG GGT TTT CTC TTG CAG AAG A-3′) and p21 reverse primer (5′-ATG TCC AAT CCT GGT GAT GTC CG-3′), pten forward primer (5′-GTG GTC TGC CAG CTA AAG GTG A-3′) and pten reverse primer (5′-TCA GAC TTT TGT AAT TTG TGA ATG CT-3′), and actin forward primer (5′-ACC TGG CCG TCA GGC AGC TC-3′) and actin reverse primer (5′-CCG AGC GTG GCT ACA GCT TC-3′). Real-time RT-PCR was performed with the Applied Biosystems 7300 real-time PCR system using iTaq SYBR green Supermix with ROX (Bio-Rad). Data were analyzed with API software. The expression level of each analyzed gene was normalized against the actin expression level. Primers for real-time RT-PCR included p21 forward (5′-ATG TCC AAT CCT GGT GAT GTC CG-3′) and p21 reverse (5′-CAA AGT TCC ACC GTT CTC GG-3′), pten forward (5′-CAA TGT TCA GTG GCG GAA CTT G-3′) and pten reverse (5′-GAA CTT GTC CTC CCG CCG C-3′), and actin forward (5′-CCG AGC GTG GCT ACA GCT TC-3′) and actin reverse (5′-ACC TGG CCG TCA GGC AGC TC-3′). siRNA duplexes were transfected into neural stem cells using Transfectin (Bio-Rad) at a final concentration of 100 nM. Specifically, RNA duplexes were mixed with 1 μl Transfectin in 50 μl medium, incubated at room temperature for 20 min, and added dropwise to cells in 450 μl medium in 24-well plates to a total volume of 500 μl per well. The transfected cells were harvested at 48 h after transfection for subsequent analyses. The LSD1 siRNA duplex sense sequence is CAC AAG GAA AGC TAG AAG A (11).
BrdU treatment and immunofluorescence analysis.
Neural stem cells were treated with bromodeoxyuridine (BrdU) for 2 h. Cells were fixed and acid treated, followed by incubation with rat anti-BrdU antibody (1:10,000; Accurate). Quantitative studies were based on four or more replicates. Immunostaining of neural stem cells with antibodies for LSD1 (1:1,000; Cell Signaling Technology) was performed as described previously (14).
LSD1 inhibitor treatment in vivo.
Two LSD1 inhibitors, pargyline and tranylcypromine, were used to treat 4-week-old wild-type or TLX-null littermate mice. These mice were treated by daily intraperitoneal (i.p.) injection of either physiological saline (control, 100 μl/kg), pargyline at 330 μmol/kg, or tranylcypromine at 10 mg/kg for 10 days. Immediately after the drug treatment, BrdU was i.p. delivered at 50 mg/kg for 5 days. Treated mice were then perfused with 4% paraformaldehyde. Brains were sectioned for immunostaining with antibodies for NeuN (1:600; Chemicon) and BrdU (1:10,000; Accurate).
Viral production and infection.
The LSD1 siRNA or scrambled control siRNA-expressing lentiviruses were produced using 293T cells. For intracranial viral infection, 0.5 μl of concentrated lentivirus was injected into the hippocampal dentate gyri of 4-week-old ICR mice by stereotaxic injection. Two weeks after viral injection, the transduced mice were injected with BrdU intraperitoneally once daily at 50 mg/kg for 5 days. Brains were fixed 1 day after the last BrdU injection and processed for immunostaining as described previously (14). The coordinates for the dentate gyrus were as follows: AP, −1.8 mm; ML, ±1.5 mm; and DV, −2.5 mm.
RESULTS
LSD1 is expressed in neural stem cells and interacts with TLX.
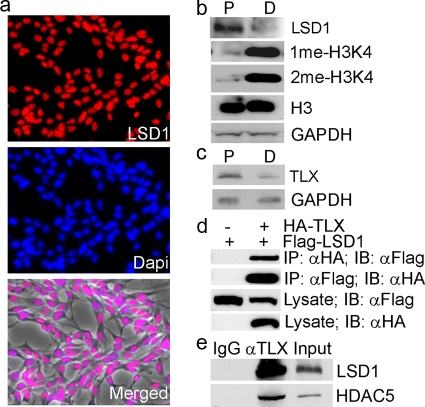
Stem cell proliferation and self-renewal are the result of transcription control in concert with chromatin remodeling and epigenetic modifications. Recently, LSD1 was identified as a histone demethylase that modulates histone methylation. LSD1 is highly expressed in the brain (21). To determine whether LSD1 plays a role in neural stem cell proliferation and differentiation, the expression of LSD1 in neural stem cells was first examined. Neural stem cells were isolated from adult mouse brains and cultured as described previously (14). They express the well-characterized neural progenitor marker nestin (data not shown). These cells are also self-renewable as revealed by clonal analysis (data not shown) and are mulitpotent, with the ability to differentiate into neurons (Tuj1+), astrocytes (GFAP+), and oligodendrocytes (O4+) (data not shown). Immunofluorescence analysis using an LSD1-specific antibody revealed that LSD1 is expressed in the nuclei of neural stem cells (Fig. 1a). To determine whether the expression of LSD1 is regulated during neural stem cell proliferation and differentiation, cell lysates were prepared from neural stem cells or differentiated cells and analyzed by Western blotting. LSD1 is highly expressed in proliferating neural stem cells. The LSD1 expression is reduced significantly in differentiated cells (Fig. 1b), similar to the expression profile of TLX (Fig. 1c). LSD1 has been shown to demethylate mono- or dimethylated H3K4 (16). Consistent with decreased expression of LSD1 in differentiated cells, increased levels of mono- and dimethylated H3K4 were observed in these cells, although no significant difference in total histone H3 levels was seen (Fig. 1b). The expression profile of LSD1 suggests that it may play a role in neural stem cell proliferation.
FIG. 1.
LSD1 is expressed in neural stem cells and interacts with TLX. (a) Expression of LSD1 in neural stem cells revealed by immunofluorescence analysis. LSD1 staining is shown in red, and DAPI (4′,6′-diamidino-2-phenylindole) staining is shown in blue. The merged image of LSD1 staining, DAPI staining, and phase-contrast image (gray) is shown at the bottom panel. (b) Expression of LSD1 in neural stem cells cultured under proliferation (P) or differentiation (D) conditions. The levels of monomethyl H3K4 (1me-H3K4) and dimethyl H3K4 (2me-H3K4) are shown in parallel. Total histone H3 levels were included as a control. GAPDH was included as a loading control. (c) Expression of TLX in neural stem cells cultured under proliferation (P) or differentiation (D) conditions. GAPDH was included as a loading control. (d) TLX interacts with LSD1 in HEK 293 cells as revealed by reciprocal coimmunoprecipitation analysis. Lysates of HA-TLX- and Flag-LSD1-transfected cells were immunoprecipitated with anti-HA or anti-Flag antibody, followed by immunoblotting with anti-Flag or anti-HA antibody. Protein expression in cell lysates was shown by immunoblotting with anti-HA or anti-Flag antibody. IP, immunoprecipitation; IB, immunoblotting. (e) TLX interacts with LSD1 and HDAC5 in neural stem cells, analyzed by immunoprecipitation analysis. Lysates of neural stem cells were immunoprecipitated with TLX-specific antibody (αTLX), followed by immunoblotting with anti-LSD1 or anti-HDAC5 antibody. IgG was included as a negative control. Ten percent input was loaded on the gel and included as a control.
We have shown that TLX plays an important role in neural stem cell proliferation and self-renewal (8, 14, 18, 30). To determine whether LSD1 interacts with TLX, HA-tagged TLX was transfected into HEK 293 cells with Flag-tagged LSD1. Cell lysates were subjected to reciprocal immunoprecipitation analysis. TLX was found to be associated with LSD1 in their immunocomplexes (Fig. 1c), suggesting that TLX interacts with LSD1. To test whether TLX interacts with LSD1 in neural stem cells, lysates of neural stem cells were immunoprecipitated with a TLX-specific antibody. The presence of LSD1 in the TLX immunocomplex was examined by Western blot analysis using an LSD1-specific antibody. LSD1 was detected in the TLX immunocomplex in neural stem cells, along with HDAC5, suggesting that TLX interacts with LSD1 and HDAC5 in neural stem cells (Fig. 1d).
Inhibition of LSD1 activity leads to reduced neural stem cell proliferation.
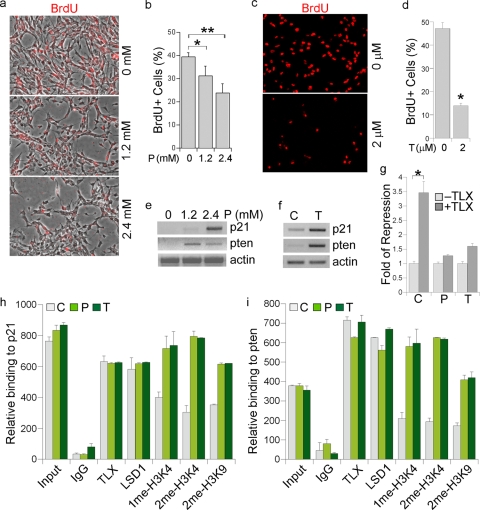
To probe the function of LSD1 in neural stem cells, we treated neural stem cells with solvent or an LSD1 inhibitor, pargyline. Pargyline is a monoamine oxidase inhibitor that has been shown to block LSD1-mediated demethylation (11). BrdU treatment was performed to label actively dividing cells. The percentage of BrdU-positive cells in total living cells was compared between solvent- and pargyline-treated cells. A significant, dose-dependent decrease in the percentage of BrdU-positive cells was seen in pargyline-treated neural stem cells (Fig. 2a and b). In addition to pargyline, tranylcypromine has also been shown to act as a potent inhibitor of LSD1-mediated demethylation (7). Neural stem cells were treated with solvent or tranylcypromine, followed by BrdU labeling to determine cell proliferation. A remarkable decrease in the rate of BrdU labeling was seen in tranylcypromine-treated cells (Fig. 2c and d). Similarly, when neural stem cells were stained with Ki67, another proliferative marker, a significant decrease in the percentage of Ki67-positive cells was observed in both pargyline- and tranylcypromine-treated cells (data not shown). The LSD1 inhibitors exhibited minimal effects on cell death (data not shown). These results together further support the idea that the LSD1 inhibitors exert a strong inhibitory effect on neural stem cell proliferation.
FIG. 2.
LSD1 inhibitor treatment inhibits neural stem cell proliferation. (a) Cell proliferation was revealed by BrdU labeling (red) in neural stem cells treated with different concentrations (0, 1.2, and 2.4 mM) of pargyline. The images shown are BrdU staining merged with phase contrast images. (b) Percentage of BrdU-positive (BrdU+) cells in 0, 1.2 mM, and 2.4 mM pargyline-treated neural stem cells. Error bars are standard deviations of the means; assays were repeated three times. *, P < 0.01; **, P < 0.001 (by Student's t test). (c) Cell proliferation was revealed by BrdU labeling (red) in solvent (0 μM)- and tranylcypromine (2 μM)-treated neural stem cells. (d) Percentage of BrdU-positive cells in solvent- and tranylcyptomine-treated neural stem cells. Error bars are standard deviations of the means; assays were repeated three times. *, P < 0.001 by Student's t test. (e) Gene expression regulated by the LSD1 inhibitor pargyline, revealed by RT-PCR analysis. Actin was included as a loading control. (f) Gene expression regulated by the LSD1 inhibitor tranylcypromine, revealed by RT-PCR analysis. (g) Pargyline and tranylcypromine treatment relieve TLX-mediated repression of p21 promoter-driven luciferase (p21-luc) activity. CV-1 cells were transfected with p21-luc along with the LSD1 expression vector and a control vector (−TLX) or with the LSD1 expression vector and the TLX-expressing vector (+TLX). The transfected cells were treated with solvent (C), pargyline (P), or tranylcypromine (T). Fold repression was determined by dividing luciferase activity in TLX-transfected cells (+TLX) with luciferase activity in control vector-transfected cells (−TLX) for each treatment. *, P < 0.01 by Student's t test. (h and i) Pargyline and tranylcypromine treatment lead to increased mono- and dimethyl H3K4 (1me-H3K4 and 2me-H3K4) levels on p21 (h) and pten (i) promoters in neural stem cells, analyzed by quantitative ChIP assays. Input, DNA input; IgG was included as a negative control. Antibodies specific for TLX, LSD1, 1me-H3K4, 2me-H3K4, and 2me-H3K9 were included in the assay. C, solvent control; P, pargyline; T, tranylcypromine.
We have identified p21 and pten as TLX target genes, the expression of which is repressed by TLX (18). To test whether LSD1 contributes to the repression of p21 and pten gene expression by TLX in neural stem cells, we treated neural stem cells with solvent or the LSD1 inhibitors pargyline and tranylcypromine. Gene expression analysis revealed significantly induced p21 and pten gene expression in pargyline- or tranylcypromine-treated neural stem cells (Fig. 2e and f), suggesting that LSD1 plays an important role in repressing p21 and pten gene expression, presumably through its interaction with TLX. The induced p21 and pten expression is consistent with reduced cell proliferation in pargyline- or tranylcypromine-treated neural stem cells (Fig. 2a to d).
To further determine whether LSD1 regulates TLX-mediated repression of p21 gene expression, we transfected a p21 promoter-driven luciferase reporter gene (p21-luc) along with LSD1 in the absence or presence of a TLX expression vector. The transfected cells were treated with solvent, pargyline, or tranylcypromine. Cotransfection of TLX and LSD1 with p21-luc led to a marked repression of the luciferase reporter. Treatment of pargyline or tranylcypromine relieved TLX-mediated repression significantly (Fig. 2g), suggesting that LSD1 is important for TLX-mediated repression of p21 gene expression.
LSD1 has been shown to play a role in demethylation of mono- and dimethylated H3K4 (16). Next we determined the methylated histone H3K4 levels at the TLX binding sites of p21 and pten promoters. Treatment of neural stem cells with either pargyline or tranylcypromine led to a marked increase in levels of mono- and dimethylated H3K4 histones on both p21 and pten promoters (Fig. 2h and i), consistent with induced p21 and pten expression in these cells (Fig. 2e and f), whereas no effect was detected on a non-TLX target, GAPDH (data not shown). Increased histone H3K9me2 levels were also observed in the LSD1 inhibitor-treated neural stem cells (Fig. 2h and i), consistent with the observation that LSD1 can also demethylate H3K9 in certain cellular contexts (11).
siRNA knockdown of LSD1 inhibits neural stem cell proliferation.
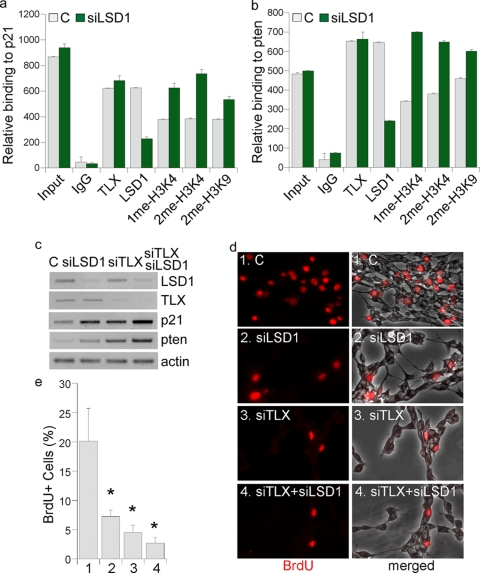
In addition to blocking LSD1 activity using chemical inhibitors, we also knocked down LSD1 expression in neural stem cells using its sequence-specific siRNA. Chromatin immunoprecipitation (ChIP) analysis revealed reduced LSD1 levels on p21 and pten promoters in LSD1 siRNA-treated cells, along with increased mono- and dimethylated H3K4 levels at the TLX binding sites on p21 and pten promoters (Fig. 3a and b). A mild increase in H3K9me2 levels was also observed in the LSD1 siRNA-treated cells (Fig. 3a and b).
FIG. 3.
Knockdown of LSD1 expression leads to induced p21 and pten gene expression and reduced neural stem cell proliferation. (a and b) LSD1 siRNA treatment leads to increased mono- and dimethyl H3K4 (1me-H3K4 and 2me-H3K4) levels on p21 (a) and pten (b) promoters in neural stem cells as analyzed by quantitative ChIP assays. Input, DNA input; IgG, IgG control. Antibodies specific for TLX, LSD1, 1me-H3K4, 2me-H3K4, and 2me-H3K9 were included in the assay. (c) Gene expression regulated by LSD1-specific siRNA treatment as revealed by RT-PCR analysis. Neural stem cells were treated with control siRNA (C), LSD1 siRNA (siLSD1), TLX siRNA (siTLX), or the combination of both (siTLX/siLSD1). Actin was included as a loading control. (d) LSD1 siRNA reduces neural stem cell proliferation as revealed by decreased BrdU labeling (red). Neural stem cells were transfected with control siRNA (C, panels 1), LSD1 siRNA (siLSD1, panels 2), TLX siRNA (siTLX, panels 3), or the combination of LSD1 siRNA and TLX siRNA (siTLX+siLSD1, panels 4). (e) Percentages of BrdU-positive (BrdU+) cells in control (bar 1)-, LSD1 siRNA (bar 2)-, TLX siRNA (bar 3)-, and TLX siRNA plus LSD1 siRNA (bar 4)-treated neural stem cells. Error bars are standard deviations of the means; assays were repeated three times. *, P < 0.001 by one-way ANOVA.
Treatment of LSD1 siRNA induced both p21 and pten gene expression (Fig. 3c, compare lanes 1 and 2), similar to the effect in TLX siRNA-treated cells (Fig. 3c, compare lanes 2 and 3). However, the induction of p21 and pten gene expression by LSD1 siRNA was reduced dramatically in TLX siRNA-treated cells (Fig. 3c, compare lanes 3 and 4), suggesting that the effect of LSD1 on p21 and pten gene expression is mediated primarily through TLX, presumably via TLX-LSD1 interaction.
Cell proliferation in control and LSD1 siRNA-treated cells was examined by BrdU labeling analysis. Treatment with LSD1 siRNA led to a 64% decrease of BrdU labeling (Fig. 3d and e, compare panels 1 and 2), suggesting that LSD1 plays an important role in neural stem cell proliferation. Treatment of neural stem cells with TLX siRNA also led to a marked reduction of neural stem cell proliferation (Fig. 3d and e, compare panels 1 and 3). However, in TLX siRNA-treated neural stem cells, no significant further inhibition of cell proliferation was detected upon LSD1 siRNA treatment (Fig. 3d and e, compare panels 3 and 4), suggesting that the proliferation-inhibitory effect of LSD1 siRNA is mediated largely through TLX. Treatment with the LSD1 siRNA also led to significantly reduced numbers of Ki67-positive cells (data not shown) but had a minimal effect on cell death. These results together suggest that LSD1 plays an important role in neural stem cell proliferation and that TLX is an essential effector of LSD1 function in neural stem cells.
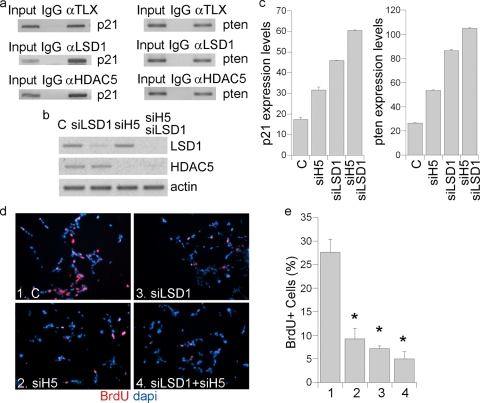
ChIP analyses revealed that both LSD1 and HDAC5 were recruited to the promoters of TLX downstream target genes p21 and pten (Fig. 4a). To determine the contribution of LSD1 and HDAC5 to TLX target gene repression and neural stem cell proliferation, neural stem cells were treated with LSD1 siRNA, HDAC5 siRNA, or a combination of both (Fig. 4b). Knockdown of either LSD1 or HDAC5 led to induced p21 and pten gene expression (Fig. 4c). Accordingly, knockdown of either LSD1 or HDAC5 led to reduced neural stem cell proliferation (Fig. 4d and e). Double knockdown led to more dramatic inhibition of neural stem cell proliferation than with individual LSD1 and HDAC5 knockdown (Fig. 4d and e). Taken together, these data suggest that both LSD1 and HDAC5 are important for TLX-mediated transcriptional repression and neural stem cell proliferation.
FIG. 4.
Simultaneous knockdown of LSD1 and HDAC5 led to dramatic inhibition of neural stem cell proliferation. (a) TLX recruitment of LSD1 and HDAC5 to the promoters of p21 and pten genes, analyzed by ChIP assays. Input, DNA input; IgG was included as a negative control. (b) The knockdown effect of the LSD1 siRNA (siLSD1) and the HDAC5 siRNA (siH5) in neural stem cells was analyzed by semiquantitative RT-PCR. Actin was included as a loading control. (c) Induction of p21 and pten gene expression in neural stem cells treated with LSD1 siRNA (siLSD1) or HDAC5 siRNA (siH5) or the combination of both (siLSD1/siH5) as analyzed by quantitative real-time RT-PCR. The expression levels of p21 and pten were normalized by actin expression levels and plotted. (d) Treatment of LSD1 siRNA and/or HDAC5 siRNA reduces neural stem cell proliferation. Cell proliferation was analyzed by BrdU labeling (red). Nuclear DAPI staining is shown in blue. Neural stem cells were transfected with control siRNA (C, panel 1), HDAC5 siRNA (siHDAC5, panel 2), LSD1 siRNA (siLSD1, panel 3), or the combination of LSD1 and HDAC5 siRNA (siLSD1+siHDAC5, panel 4). (e) Percentages of BrdU-positive cells in control (bar 1)-, HDAC5 siRNA (bar 2)-, LSD1 siRNA (bar 3)-, and LSD1 siRNA+ HDAC5 siRNA (bar 4)-treated neural stem cells. Error bars are standard deviations of the mean; assays were repeated three times. *, P < 0.01 by one-way ANOVA.
LSD1 siRNA or inhibitor treatment reduces cell proliferation in the dentate gyri of adult mouse brains.
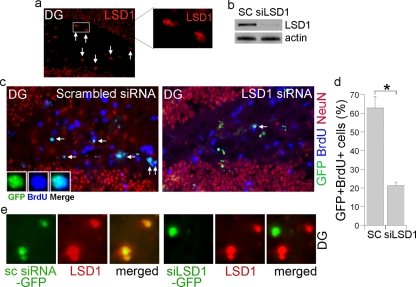
The subgranular zone (SGZ) of the hippocampal dentate gyrus is a well-characterized adult neurogenic area where neural progenitor cells reside. Immunostaining analysis revealed that LSD1 is expressed in the SGZ of the dentate gyrus (Fig. 5a). To determine whether LSD1 plays a role in neural progenitor cell proliferation in vivo, we prepared LSD1 siRNA-expressing lentivirus. Marked knockdown of LSD1 expression was detected in the LSD1 siRNA-transduced neural stem cells in culture (Fig. 5b). Next, the LSD1 siRNA-expressing lentivirus was transduced into the SGZ of the dentate gyrus by intracranial lentiviral injection. Viral expression of the LSD1 siRNA was monitored by a coexpressed green fluorescent protein (GFP). Transduction of the LSD1 siRNA led to significantly reduced cell proliferation, as revealed by the decreased number of BrdU-positive and GFP-positive cells, compared to that in the scrambled control siRNA-transduced brains (Fig. 5c and d) (P < 0.01 by Student's t test). Knockdown of LSD1 expression by the LSD1 siRNA in the SGZ of the dentate gyrus was shown by dramatically reduced LSD1 immunostaining (Fig. 5e, right panels), whereas LSD1 expression was not affected by the scrambled control siRNA transduction (Fig. 5e, left panels). These results further strengthen the concept that LSD1 is important for neural progenitor cell proliferation in adult mouse brains.
FIG. 5.
LSD1 siRNA treatment leads to reduced cell proliferation in the dentate gyri of adult mouse brains. (a) Expression of LSD1 in the subgranular zone (SGZ) of the dentate gyri of wild-type adult mouse brains as revealed by immunostaining. The LSD1 staining is shown in red. Cells indicated by arrows are examples of the LSD1-positive cells in the SGZ. An enlarged image of the LSD1-positive cells in the boxed region is shown on the right. (b) Knockdown of LSD1 expression using the LSD1 siRNA-expressing lentivirus in cultured neural stem cells. SC, scrambled siRNA control. Actin was included as a loading control. (c) Reduced BrdU staining of LSD1 siRNA-transduced cells in the dentate gyri (DG) of adult brains. DG sections from scrambled siRNA control- or LSD1 siRNA-transduced and BrdU-treated mice were immunostained for BrdU (blue). NeuN staining (red) was included to show the structure of the DG. The virus-transduced cells are shown in green due to the expression of a GFP marker. (d) Percentages of doubly GFP-positive and BrdU-positive cells out of GFP+ cells in the dentate gyrus SGZ of the LSD1 siRNA lentivirus-infected mice. SC, scrambled control siRNA; siLSD1, LSD1 siRNA. Data are represented as means ± standard deviations. *, P < 0.01 by Student's t test. (e) Examples of scrambled siRNA control (sc siRNA-GFP)- or LSD1 siRNA (siLSD1-GFP)-transduced cells in the dentate gyrus (DG) SGZ. LSD1 staining is shown in red. The virus-transduced cells are shown in green due to the expression of a GFP marker. The merged images are shown on the right.
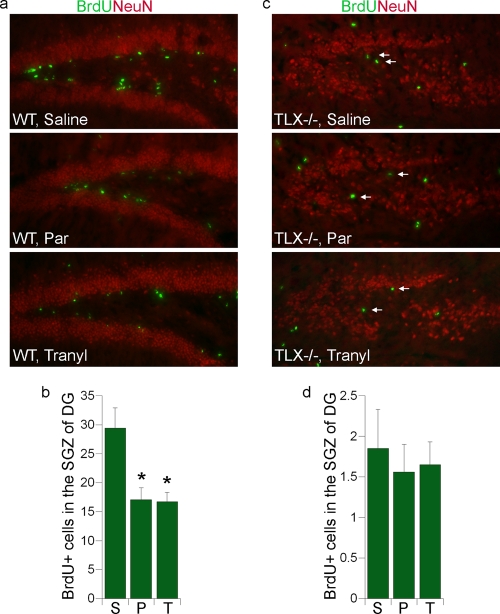
To examine the effect of the LSD1 inhibitors pargyline and tranylcypromine on cell proliferation in the dentate gyri of adult mouse brains, we injected wild-type adult mice with pargyline or tranylcypromine for 10 days, followed by 5 days of BrdU injection to label dividing neural progenitor cells. Saline treatment was included as a control. Pargyline- or tranylcypromine-injected mice exhibited a marked reduction in BrdU labeling in the SGZ of the dentate gyrus compared to that in saline-injected mice (Fig. 6a and b). Quantitative analysis of BrdU labeling revealed a significant decrease (P < 0.001 by one-way analysis of variance [ANOVA]) in the number of dividing cells in the dentate gyrus SGZ of pargyline- or tranylcypromine-treated mice compared to saline-treated mice. The LSD1 inhibitor-mediated decrease in cell proliferation in the dentate gyri of wild-type brains is consistent with the effect of pargyline and tranylcypromine on proliferation of neural stem cells in vitro.
FIG. 6.
LSD1 inhibitor treatment leads to reduced cell proliferation in the dentate gyri of adult mouse brains. (a) Representative images of hippocampal dentate gyrus brain sections of wild-type adult mice that were injected with saline, pargyline (Par) or tranylcypromine (Tranyl) and treated with BrdU. Brain sections were stained with BrdU (green) to measure cell proliferation. NeuN staining (red) was included to show the structure of dentate gyrus. (b) Average numbers of BrdU-positive (BrdU+) cells in the subgranular zone (SGZ) of the dentate gyrus (DG) in one field of 20-μm wild-type brain sections. (n = 6 mice for each treatment group). S, saline; P, pargyline; T, tranylcypromine. Error bars are standard deviations of the means. *, P < 0.001 by one-way ANOVA. (c) Representative images of hippocampal dentate gyrus brain sections of TLX−/− adult mice that were injected with saline, pargyline (Par), or tranylcypromine (Tranyl) and treated with BrdU. Brain sections were stained with BrdU (green) and NeuN (red). The BrdU-positive cells that are located in the SGZ of DG are indicated by arrows. (d) Average numbers of BrdU-positive (BrdU+) cells in the SGZ of the DG in one field of 20-μm TLX−/− brain section. (n = 6 mice for each treatment group). S, saline; P, pargyline; T, tranylcypromine. Error bars are standard deviations of the means.
In parallel to the treatment of wild-type adult mice, TLX-null adult mice were treated similarly with the LSD1 inhibitors. BrdU labeling was decreased substantially in the dentate gyrus SGZ of TLX-null mice (Fig. 6c and d, top panel) compared to wild-type mice (Fig. 6a and b, top panel). The dentate gyrus is reduced considerably in TLX−/− adult brains, consistent with our previous observation (14). Interestingly, no significant further decrease in cell proliferation was induced by either pargyline or tranylcypromine in the dentate gyrus SGZ of TLX-null brains (Fig. 6c and d), suggesting that the effect of the LSD1 inhibitors on neural progenitor proliferation in the dentate gyrus SGZ is mediated largely through TLX signaling.
DISCUSSION
In this study we showed that the histone demethylase LSD1 plays an important role in neural stem cell proliferation. Either inhibition of LSD1 activity or knockdown of LSD1 expression led to dramatically reduced neural stem cell proliferation. We also identified TLX as a critical effector of LSD1 function in neural stem cells. This idea was supported by several pieces of evidence. First, LSD1 is recruited to the promoters of TLX target genes p21 and pten by TLX to repress their expression. Second, knockdown of TLX expression diminished LSD1 siRNA-mediated inhibition of neural stem cell proliferation significantly. Furthermore, treatment of adult mice with the LSD1 siRNA or the LSD1 inhibitors pargyline and tranylcypromine led to dramatically reduced neural progenitor proliferation in the hippocampal dentate gyri of wild-type mouse brains; however, the inhibitor treatment had almost no effect on cell proliferation in TLX−/− mouse brains.
The data presented here demonstrated that the function of TLX is modulated by LSD1 in neural stem cells. The association of LSD1 with TLX in neural stem cells led to transcriptional repression of TLX target gene expression. Both LSD1 inhibitor treatment and siRNA knockdown resulted in an increase in H3K4 methylation levels on TLX target gene promoters and a concomitant induction of TLX target gene expression, suggesting that LSD1 mediates transcriptional repression of TLX target genes via histone demethylation. These results mirrored the observation in retinoblastoma cells, in which LSD1 was found to be present in the TLX immunocomplex to modulate its activity (26).
LSD1 has been shown to catalyze demethylation of mono- and dimethyl H3K4 through a flavin adenine dinucleotide (FAD)-dependent oxidative reaction (16). Treatment with both LSD1 inhibitors and siRNA led to increased monomethyl and dimethyl H3K4 levels on p21 and pten promoters, providing strong evidence that LSD1 participates in the demethylation of TLX target gene promoters. Because LSD1 is unable to remove trimethyl H3K4, no induction of trimethyl H3K4 levels was detected on the p21 promoter in LSD1 siRNA-treated cells, as expected. No change in TLX binding was detected in LSD1 siRNA-treated cells, although a slight decrease of TLX binding was seen in LSD1 inhibitor-treated cells, which may be due to an indirect effect.
To determine the role of LSD1 in neural stem cells, we treated neural stem cells with the LSD1 inhibitors pargyline and tranylcypromine, which resulted in significant inhibition of cell proliferation. However, chemical inhibition may have pleiotropic effects. We therefore knocked down LSD1 expression using its sequence-specific siRNA. Both induction of TLX target gene expression and reduced neural stem cell proliferation were observed upon LSD1 knockdown, similar to the effect of LSD1 inhibitor treatment.
We demonstrated that TLX recruits both LSD1 and HDAC5 to its target genes in neural stem cells. Knockdown of either LSD1 or HDAC5 led to induction of p21 and pten gene expression and inhibition of neural stem cell proliferation. When LSD1 and HDAC5 were knocked down simultaneously, a more dramatic effect on both induction of TLX target gene expression and inhibition of neural stem cell proliferation was detected, compared to individual siRNA treatment. Similarly, when TLX was knocked down, substantial induction of p21 and pten gene expression and inhibition of neural stem cell proliferation were observed. These results suggest that TLX recruits both LSD1 and HDAC5, presumably in a corepressor complex, to mediate its cellular function.
In summary, this study has uncovered a novel role for LSD1 in neural stem cell proliferation. The findings presented here revealed a mechanism by which recruitment of the histone demethylase LSD1 and the histone deacetylase HDAC5 enables TLX to mediate transcriptional repression in neural stem cells. The partnership of TLX with LSD1 and HDAC5 links transcriptional repression and epigenetic modulation at TLX target genes to neural stem cell proliferation. Stem cells provide great hope for the treatment of a variety of human diseases that lack efficacious therapies to date. We show here that LSD1 inhibitors, such as pargyline and tranylcypromine, control the demethylase activity of LSD1, thereby regulating TLX signaling. Considering the essential role of TLX in neural stem cell proliferation and self-renewal, specific modulation of LSD1 activity may become a promising therapeutic tool for the treatment of neurodegenerative diseases, such as Alzheimer's and Parkinson's diseases.
Acknowledgments
We thank J. Zaia for critical reading of the manuscript.
G.S. is a Herbert Horvitz postdoctoral fellow. R.S. is a Ford Foundation predoctoral fellow. W.L. is supported by a training grant from the California Institute for Regenerative Medicine. This work was supported by NIH NINDS grant R01 NS059546 (to Y.S.).
Footnotes
Published ahead of print on 1 February 2010.
REFERENCES
- 1.Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. [DOI] [PubMed] [Google Scholar]
- 2.Cloos, P. A., J. Christensen, K. Agger, A. Maiolica, J. Rappsilber, T. Antal, K. H. Hansen, and K. Helin. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442:307-311. [DOI] [PubMed] [Google Scholar]
- 3.Fodor, B. D., S. Kubicek, M. Yonezawa, R. J. O'Sullivan, R. Sengupta, L. Perez-Burgos, S. Opravil, K. Mechtler, G. Schotta, and T. Jenuwein. 2006. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20:1557-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 5.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 6.Klose, R. J., K. Yamane, Y. Bae, D. Zhang, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442:312-316. [DOI] [PubMed] [Google Scholar]
- 7.Lee, M. G., C. Wynder, D. M. Schmidt, D. G. McCafferty, and R. Shiekhattar. 2006. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 13:563-567. [DOI] [PubMed] [Google Scholar]
- 8.Li, W., G. Sun, S. Yang, Q. Qu, K. Nakashima, and Y. Shi. 2008. Nuclear receptor TLX regulates cell cycle progression in neural stem cells of the developing brain. Mol. Endocrinol. 22:56-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 10.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocrinol. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 11.Metzger, E., M. Wissmann, N. Yin, J. M. Muller, R. Schneider, A. H. Peters, T. Gunther, R. Buettner, and R. Schule. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436-439. [DOI] [PubMed] [Google Scholar]
- 12.Monaghan, A. P., D. Bock, P. Gass, A. Schwager, D. P. Wolfer, H. P. Lipp, and G. Schutz. 1997. Defective limbic system in mice lacking the tailless gene. Nature 390:515-517. [DOI] [PubMed] [Google Scholar]
- 13.Ordentlich, P., M. Downes, and R. M. Evans. 2001. Corepressors and nuclear hormone receptor function. Curr. Top. Microbiol. Immunol. 254:101-116. [DOI] [PubMed] [Google Scholar]
- 14.Shi, Y., D. Chichung Lie, P. Taupin, K. Nakashima, J. Ray, R. T. Yu, F. H. Gage, and R. M. Evans. 2004. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427:78-83. [DOI] [PubMed] [Google Scholar]
- 15.Shi, Y., M. Downes, W. Xie, H. Y. Kao, P. Ordentlich, C. C. Tsai, M. Hon, and R. M. Evans. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 15:1140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, and R. A. Casero. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 17.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 18.Sun, G., R. T. Yu, R. M. Evans, and Y. Shi. 2007. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 104:15282-15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trewick, S. C., P. J. McLaughlin, and R. C. Allshire. 2005. Methylation: lost in hydroxylation? EMBO Rep. 6:315-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811-816. [DOI] [PubMed] [Google Scholar]
- 21.Wang, J., K. Scully, X. Zhu, L. Cai, J. Zhang, G. G. Prefontaine, A. Krones, K. A. Ohgi, P. Zhu, I. Garcia-Bassets, F. Liu, H. Taylor, J. Lozach, F. L. Jayes, K. S. Korach, C. K. Glass, X. D. Fu, and M. G. Rosenfeld. 2007. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446:882-887. [DOI] [PubMed] [Google Scholar]
- 22.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467-481. [DOI] [PubMed] [Google Scholar]
- 23.Wissmann, M., N. Yin, J. M. Muller, H. Greschik, B. D. Fodor, T. Jenuwein, C. Vogler, R. Schneider, T. Gunther, R. Buettner, E. Metzger, and R. Schule. 2007. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 9:347-353. [DOI] [PubMed] [Google Scholar]
- 24.Wysocka, J., T. A. Milne, and C. D. Allis. 2005. Taking LSD 1 to a new high. Cell 122:654-658. [DOI] [PubMed] [Google Scholar]
- 25.Yamane, K., C. Toumazou, Y. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125:483-495. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama, A., S. Takezawa, R. Schule, H. Kitagawa, and S. Kato. 2008. Transrepressive function of TLX requires the histone demethylase LSD1. Mol. Cell. Biol. 28:3995-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, R. T., M. McKeown, R. M. Evans, and K. Umesono. 1994. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature 370:375-379. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, C. L., Y. Zou, W. He, F. H. Gage, and R. M. Evans. 2008. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature 451:1004-1007. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]
- 30.Zhao, C., G. Sun, S. Li, and Y. Shi. 2009. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16:365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]