Abstract
An analysis of mRNA expression in T47D breast cancer cells treated with the synthetic progestin R5020 revealed a subset of progesterone receptor (PR) target genes that are enriched for E2F binding sites. Following up on this observation, we determined that PR-B acts in both direct and indirect manners to positively upregulate E2F1 expression in T47D cells. The direct effects of PR on E2F1 expression were confirmed by chromatin immunoprecipitation (ChIP) analysis, which indicated that the agonist-bound receptor was recruited to several enhancer elements proximal to the E2F1 transcript. However, we also noted that cycloheximide partially inhibits R5020 induction of E2F1 expression, indicating that the ligand-dependent actions of PR on this gene may involve additional indirect regulatory pathways. In support of this hypothesis, we demonstrated that treatment with R5020 significantly increases both hyperphosphorylation of Rb and recruitment of E2F1 to its own promoter, thus activating a positive feedback loop that further amplifies its transcription. Furthermore, we established that PR-mediated induction of Krüppel-like factor 15 (KLF15), which can bind to GC-rich DNA within the E2F1 promoter, is required for maximal induction of E2F1 expression by progestins. Taken together, these results suggest a new paradigm for multimodal regulation of target gene expression by PR.
The steroid hormone progesterone plays a central role in the development, growth, and differentiation of the female reproductive system. The biological functions of progesterone are mediated by the two progesterone receptor (PR) isoforms, PR-A and PR-B, which belong to the nuclear receptor (NR) superfamily of ligand-regulated transcription factors (for a review, see reference 15). In the absence of ligand, PR is sequestered by heat shock proteins and maintained in an inactive state in the cytoplasm of target cells. Upon ligand binding, PR undergoes a conformational change that leads to its dissociation from the heat shock protein complex, an event that facilitates receptor dimerization and translocation into the nucleus. The receptor dimer is then capable of interacting with specific progesterone-responsive elements (PREs) within target gene promoters. The DNA-bound receptor subsequently nucleates the assembly of large cofactor-containing protein complexes that can either positively or negatively affect gene transcription.
In addition to this classical pathway of transcriptional activation, extranuclear PR can indirectly regulate gene expression by rapidly activating other signaling pathways. For instance, the N-terminal domain of PR contains a polyproline motif that has been shown to directly interact with the SH3 domains of c-Src and mediate rapid, nongenomic activation of c-Src family tyrosine kinases and the downstream mitogen-activated protein kinase (MAPK) cascade (3). Additionally, progestins have been shown to rapidly activate the phosphoinositol 3-kinase (PI3K)/Akt/nuclear factor κB (NF-κB) cascade and the Janus family of tyrosine kinases (JAK)/signal transducer and activator of transcription (STAT) signaling pathway in breast cancer cells (25, 28). Thus, through activation of these extranuclear signaling pathways, PR can regulate gene expression in a manner that is completely independent of its classic nuclear activities.
While the nuclear and extranuclear actions of PR have been well studied in isolation, it is important to understand the mechanisms by which these pathways can interact and integrate to ultimately affect gene expression. Previous studies have established that the cross talk that occurs between PR and cytoplasmic signaling cascades is bidirectional and complex. On one hand, PR can activate rapid extranuclear signaling pathways such as that regulated by Src/MAPK and thereby modulate MAPK-dependent transcription; conversely, activated MAPKs can phosphorylate PR, or its attendant cofactors, and thereby modulate its ability to regulate target gene transcription (26). For example, MAPK kinase kinase 1 (MEKK1) has been shown to phosphorylate PR on Ser294, which results in increased progestin-mediated transcription as well as enhanced ligand-dependent receptor downregulation (30).
Phosphorylation of cofactors can also have a dramatic impact on the transcriptional program set in motion by nuclear receptors. For instance, it was recently reported that not only can phosphorylation of steroid receptor coactivator 3 (SRC-3) (also known as ACTR, AIB1, p/CIP, RAC3, or TRAM-1) affect its activity, but different patterns of phosphorylation on SRC-3 can dictate the specificity of SRC-3 for various transcription factors (38). Although not yet studied in detail, it is likely, by extrapolation from studies of other NRs, that cofactor phosphorylation will also have a dramatic effect on PR transcriptional activity in cells. Cumulatively, studies highlighting the importance of the cross talk that occurs between the nuclear and extranuclear functions of PR have provided the impetus to define the molecular mechanisms by which these pathways are integrated and how disruption in these events can have pathological consequences.
Given the recent interest in the cross talk that occurs between the PR and MAPK signaling pathways, we assessed the overall impact of MAPK inhibition on PR transcriptional activity. During the course of microarray and biochemical analyses that were undertaken to address this issue, we discovered that PR utilizes multiple pathways, both direct and indirect, to achieve regulation of E2F1 expression in T47D cells. Furthermore, our results support a paradigm for multimodal PR signaling in which PR and other regulatory proteins work in concert to achieve the desired regulation of downstream gene expression.
MATERIALS AND METHODS
Biochemicals.
Promegestone (R5020) was purchased from NEN Life Science Products (Boston, MA). Cycloheximide, U0126, and mithramycin A were obtained from Sigma-Aldrich (St. Louis, MO).
Plasmids.
The normalization vector pCMVβ-gal was obtained from Clontech (Palo Alto, CA). The E2F1 promoter luciferase constructs pGL2-hE2F1-Luc(−242), pGL2-hE2F1-Luc(−204), and pGL2-hE2F1-Luc(−122) and the KLF15 expression constructs pcDNA-hKLF15 and pcDNA-hKLF15-NΔ291 have been previously described (13, 23). pBKC-hPR-B was previously described (8), and pcDNA3 was purchased from Invitrogen (Carlsbad, CA). pcDNA3-hPR-B was constructed as follows. A BamHI fragment of hPR-B (amino acids [aa] 22 to 933) was cut out from pBKC-hPR-B and subcloned into pcDNA3 using the BamHI site to create pcDNA3-PR 22-933. Next, the 5′ region of PR was amplified using simian virus 40 (SV40)-hPR-B (7) as a template, using the sense primer 5′-GGGGTACCCCGGCGCGCCCATGACTGAGCTGAAG-3′ and the antisense primer 5′-AGGCCGGGAGCAGCAGCT-3′. This fragment was subsequently digested with KpnI and BstEII and then cloned into pcDNA3-PR 22-933 using KpnI/BstEII sites to create pcDNA3-hPR-B.
pENTR-hPR-B was constructed by cloning a KpnI-to-EcoRI fragment of pcDNA3-hPR-B into the pENTR-1A vector, purchased from Invitrogen. pENTR-hPR-B-C587A was created as follows. The fragment of hPR-B between the AscI and HindIII restriction sites was amplified by PCR using the PR DNA-binding mutant hPR-Bcys (a kind gift of K. Horwitz, University of Colorado, Denver, CO) as a template, using the sense primer 5′-TGCATCCTGTACAAAGCGGAGGG-3′ and the antisense primer 5′-ACTTGAAGCTTGACAAACTCCTGTGG-3′. This fragment was cloned into pENTR-hPR-B using the AscI and HindIII restriction sites. MSCV-GWb-Gal4DBD-IRES-EGFP, MSCV-GWb-hPR-B-IRES-EGFP, and MSCV-GWb-hPR-B-C587A-IRES-EGFP were created using the Invitrogen Gateway recombinase subcloning system according to the manufacturer's instructions. To do this, Gal4DBD, hPR-B, or hPR-B-C587A was shuttled from pENTR-Gal4DBD, pENTR-hPR-B, or pENTR-hPR-B-C587A to MSCV-IRES-EGFP that was converted to a Gateway destination vector.
Cell culture.
The T47D and BT483 human breast ductal carcinoma cell lines were purchased from the American Type Culture Collection (Manassas, VA). Human mammary epithelial cells (HMECs) (63NP1) were a kind gift from J. Marks (Duke University, Durham, NC). The T47D:A18 cell line was kindly provided by V. Jordan (Fox Chase Cancer Center, Philadelphia, PA) and has been previously described (21). The PR-negative T47D:C42 cell lines stably expressing LacZ, PR-A, PR-B, or PR-BmPro were kind gifts from D. Edwards (Baylor College of Medicine, Houston, TX).
To create T47D:C42-Gal4DBD, T47D:C42-hPR-B and T47D:C42-hPR-B-C587A stable cell lines, parental T47D:C42 cells provided by D. Edwards were infected with retrovirus expressing MSCV-GWb-Gal4DBD-IRES-EGFP (negative control), MSCV-GWb-hPR-B-IRES-EGFP, or MSCV-GWb-hPR-B-C587A-IRES-EGFP. Enhanced green fluorescent protein (EGFP)-positive cells were then selected through two rounds of cell sorting using flow cytometry, and hPR-B expression levels were confirmed by Western blot analysis (see Fig. S4 at http://mcdonnelllab.duhs.duke.edu).
Unless otherwise noted, all media and supplements were purchased from Invitrogen. The T47D, T47D:A18, and BT483 cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 8% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT), 0.1 mM nonessential amino acids (NEAA), and 1 mM sodium pyruvate (NaPyr). T47D:C42 cell lines provided by D. Edwards were cultured in CellBIND tissue culture flasks (Corning, Lowell, MA) using minimum essential medium (MEM) supplemented with 8% FBS, 10 mM HEPES, 25 μg/ml gentamicin, 50 U/ml penicillin-streptomycin (Pen/Strep), 0.1 mM NEAA, 60 μg/ml insulin, and 200 μg/ml zeocin. T47D:C42 stable cell lines created in our lab were maintained in MEM supplemented with 8% FBS, 10 mM HEPES, 25 μg/ml gentamicin, 50 U/ml Pen/Strep, and 0.1 mM NEAA. HMECs were maintained in mammary epithelial basal medium (MEBM) (Lonza, Basel, Switzerland) supplemented with Mammary epithelial cell growth medium (MEGM) SingleQuots (Lonza), 5 μg/ml transferrin (Sigma-Aldrich), and 10 μM isoproterenol (Sigma-Aldrich). All cell lines were grown in a 37°C incubator with 5% CO2.
Microarray.
Oligonucleotide microarray analysis was conducted on two biological replicate cultures of T47D cells. For each biological replicate, T47D cells were seeded into one well of a six-well plate per treatment in phenol red-free DMEM supplemented with 10% charcoal-stripped fetal bovine serum (CS-FBS) (HyClone, Logan, UT) for 72 h. Cells were pretreated for 10 min with vehicle or 10 μM U0126 and then treated for 24 h with vehicle or 10 nM R5020. After treatment, the culture medium was removed from each of the wells and the entire plate was frozen at −80°C until further processing. RNA was isolated from the frozen dishes by adding RLT lysis buffer (Qiagen, Valencia, CA) to each well and then processed using RNeasy mini columns (Qiagen) following the manufacturer's recommended procedure. The quantity and purity of the extracted RNA were evaluated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and its integrity measured using an Agilent Bioanalyzer. For microarray hybridizations, 1 μg of total RNA was amplified and labeled with a fluorescent dye (either Cy3 or Cy5) using the Low RNA Input Linear Amplification Labeling kit (Agilent Technologies, Palo Alto, CA) following the manufacturer's protocol. The amount and quality of the fluorescently labeled cRNA were assessed using a NanoDrop ND-1000 spectrophotometer and an Agilent Bioanalyzer. Equal amounts of Cy3- or Cy5-labeled cRNA were hybridized to the Agilent Human Whole Genome 44K Oligo microarray for 17 h prior to washing and scanning. Data were extracted from scanned images using Agilent's Feature Extraction software. Gene expression data were loaded into the Rosetta Resolver gene expression analysis system. Fluorophore reversal hybridization data were combined using an error-weighted average for each treated sample. PR-regulated probe sets were identified as those with a P value of <0.001 and an absolute fold change of >1.3.
Transient-transfection assays.
For reporter gene assays, T47D:A18 cells were seeded in 24-well plates in phenol red-free DMEM containing 8% CS-FBS, 0.1 mM NEAA, and 1 mM NaPyr at 24 h before transfection. DNA was introduced into the cells using Lipofectin (Invitrogen)-mediated transfection as described by the manufacturer. Briefly, triplicate transfections were performed using 3 μg of total DNA; within each experiment, the total amount (in μg) of DNA used to transfect each plate was kept constant by addition of the corresponding empty expression vector DNA lacking a cDNA insert. Cells were incubated with the DNA-Lipofectin mixture for 24 h. Next, the transfection mix was replaced with fresh medium containing the appropriate ligands. Following overnight treatment, luciferase and β-galactosidase (β-gal) activities were assayed on a Fusion Alpha-FP HT universal microplate reader (Perkin-Elmer, Danvers Grove, IL). Each experiment was repeated at least three times, and results are expressed as relative luciferase activity (normalized to β-gal for transfection efficiency) for one representative experiment performed in triplicate. Error bars indicate the standard error of the mean (SEM) for the triplicate wells.
For studies involving transient transfection of small interfering RNA (siRNA), validated Stealth siRNA directed against a control luciferase sequence (siLuc) or two different regions of KLF15 were obtained from Invitrogen (see Table S1 at http://mcdonnelllab.duhs.duke.edu for siRNA sequences). T47D:A18 cells were plated in phenol red-free DMEM containing 8% CS-FBS, 0.1 mM NEAA, and 1 mM NaPyr in the presence of 40 nM siLuc or siKLF15 using DharmaFECT-1 (Dharmacon, Lafayette, CO) as the transfection agent according to the manufacturer's recommendations. After 48 h of knockdown, cells were serum starved in phenol red-free DMEM containing 0.1% CS-FBS, 0.1 mM NEAA, and 1 mM NaPyr for 24 h and then treated with the appropriate ligand and harvested for quantitative PCR (qPCR) analysis as described below.
RNA isolation and quantitative PCR.
BT483, T47D:A18, or T47D:C42 cells were seeded in six-well plates in phenol red-free medium containing 8% CS-FBS and the appropriate supplements for 48 h. Next, cells were serum starved for 24 h as described above and treated with the appropriate ligand. After the indicated time period, cells were harvested and total RNA was isolated using the Aurum total RNA minikit (Bio-Rad, Hercules, CA). One microgram of RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). The Bio-Rad iCycler real-time PCR system was used to amplify and quantitate levels of target gene cDNA. qPCRs were performed with 1 μl cDNA, 10 μM specific primers (see Table S1 at http://mcdonnelllab.duhs.duke.edu for qPCR primer sequences), and iQ SYBR green Supermix (Bio-Rad). Data are normalized to the 36B4 housekeeping gene and presented as fold induction over vehicle. Data are the means ± SEMs for triplicate amplification reactions from one representative experiment. Each experiment was repeated at least three independent times with very similar results.
Virus production and infections.
Adenoviruses expressing β-gal and hPR-B were generated using the ViraPower adenoviral expression system (Invitrogen) and were amplified and purified by CsCl2 centrifugation. For adenovirus infection, HMECs were seeded in six-well plates in normal medium for 48 h and then serum starved with 0.001% serum medium without epidermal growth factor (EGF) for 36 h. Cells were infected at a multiplicity of infection (MOI) of 150 in the absence or presence of hormone added 90 min postinfection, and RNA was isolated 16 h after treatment.
Western blotting.
T47D:A18 cells were seeded in 10-cm plates in phenol red-free DMEM containing 8% CS-FBS, 0.1 mM NEAA, and 1 mM NaPyr for 48 h, after which the cells were serum starved for 24 h as described above. Following treatment with the appropriate compound for the indicated time periods, cells were harvested in ice-cold phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, 1 mM EDTA, 10% glycerol, 50 mM NaF, 2 mM Na3VO4, and 1× protease inhibitor mixture [EMD Chemicals, Inc., San Diego, CA]) while rotating at 4°C for 30 min. Twenty micrograms of whole-cell extract was resolved by SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad), and probed with the appropriate antibodies. The mouse monoclonal E2F1 KH95 antibody and the goat polyclonal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) V-18 antibody were from Santa Cruz Biotechnology (Santa Cruz, CA). The mouse monoclonal PR 1294 antibody was a kind gift from D. Edwards (Baylor College of Medicine, Houston, TX), and the mouse monoclonal HisG antibody (used to probe for His-tagged KLF15) was from Invitrogen. The mouse monoclonal Rb 4H1 antibody and rabbit polyclonal antibodies against p44/42 MAPK (Erk1/2), phospho-Rb Ser780, and phospho-Rb Ser807/811 were all from Cell Signaling Technology (Danvers, MA). The anti-mouse, anti-rabbit, and anti-goat secondary antibody-horseradish peroxidase conjugates were from Amersham Biosciences (Buckinghamshire, United Kingdom). Results shown are representative blots.
ChIP.
T47D:A18 cells were seeded in 15-cm dishes using DMEM supplemented with 8% FBS, 0.1 mM NEAA, and 1 mM NaPyr for 24 h. Cells were grown to 90% confluence in phenol red-free DMEM supplemented with 8% CS-FBS, 0.1 mM NEAA, and 1 mM NaPyr for 48 h, after which the cells were serum starved for 24 h as described above. Following treatment with the appropriate ligand for the indicated time periods, cells were subjected to chromatin immunoprecipitation (ChIP) analysis as previously described (6), with the following modifications. Immunoprecipitation was performed overnight at 4°C with 10 μg PR-specific antibody (1294; D. Edwards, Baylor College of Medicine), 10 μg E2F1-specific antibody (KH95; Santa Cruz Biotechnology), or 10 μg mouse IgG control (Santa Cruz Biotechnology). After immunoprecipitation, 70 μl protein A/G-plus-agarose beads (Santa Cruz) (50% slurry in 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) was added and allowed to incubate for 3 h at 4°C. qPCR analysis was performed as described above (see Table S1 at http://mcdonnelllab.duhs.duke.edu for ChIP primer sequences). Data are normalized to the input for the immunoprecipitation.
Microarray data accession number.
All microarray experimental results have been deposited in the Gene Expression Omnibus database under accession number GSE18276.
RESULTS
Gene regulation by progestins is significantly altered by inhibition of MAPK.
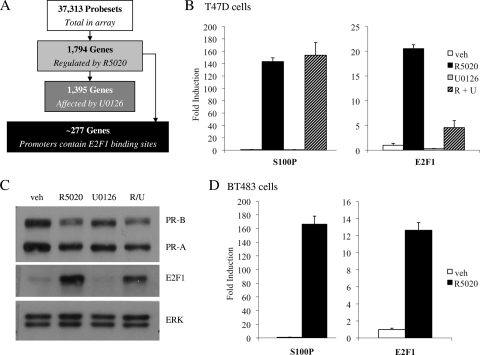
In order to evaluate the degree to which the PR and MAPK signaling pathways converge at the level of gene transcription, we performed a microarray analysis to assess genome-wide changes in PR-dependent gene transcription in the presence of the MEK 1/2 inhibitor U0126 in T47D breast cancer cells. Gene expression profiling resulted in the identification of 2,510 probe sets that were differentially expressed in response to treatment with R5020 for 24 h (Fig. 1A). These probe sets mapped to 1,794 unique transcripts, of which 1,104 were upregulated and 690 were downregulated. Surprisingly, we observed that pretreatment with U0126 altered progestin-mediated regulation of 1,395 genes.
FIG. 1.
Induction of endogenous E2F1 RNA/protein by R5020. (A) Flow chart schematic depicting breakdown of genes analyzed in T47D microarray. (B and C) Synchronized T47D:A18 cells were pretreated with vehicle (veh) or 10 μM U0126 (U) for 30 min prior to addition of vehicle or 100 pM R5020 (R) for 18 h. (C) After treatment, cells were harvested and 20 μg whole-cell extract was resolved by SDS-PAGE; transferred to PVDF; and subjected to immunoblotting for PR, E2F1, or ERK 1/2 as a loading control. A representative blot is shown. (D) Synchronized BT483 cells were treated with vehicle or 10 nM R5020 for 18 h. (B and D) After treatment, cells were lysed and RNA was isolated and reverse transcribed. S100P or E2F1 mRNA levels were quantified using qPCR and normalized to the housekeeping gene 36B4. Results are expressed as mean fold induction over vehicle-treated cells ± SEM (n = 3).
To determine how many of these genes are potential direct PR target genes, we utilized Patser (10) to scan the 2-kb upstream promoter regions with the PR position weight matrix (32) and found that 634 of the progestin-regulated genes have promoters that contain putative progesterone response elements (PREs) (see Fig. S8 at http://mcdonnelllab.duhs.duke.edu). Interestingly, an additional unbiased transcription factor enrichment analysis carried out using oPOSSUM (11) also detected a significant overrepresentation of E2F1 binding sites in the promoters of PR target genes; in fact, further analyses using Patser identified potential E2F1 binding sites in the promoters of 277 progestin-regulated genes (Fig. 1A; see Fig. S8 at http://mcdonnelllab.duhs.duke.edu). Furthermore, the microarray analysis showed that progestin treatment stimulated the transcription of classic E2F1 target genes such as those for CDC2, CDC6, cyclin E, and CDK2. These findings suggested that PR may indirectly affect transcription in cells by positively upregulating the expression and/or activity of E2F1, a key transcription factor involved in the regulation of the cell cycle.
Progestins induce expression of endogenous E2F1 mRNA and protein.
Our hypothesis that PR could regulate the expression of E2F1 was supported by the microarray data, which indicated a 2.2-fold induction of E2F1 expression after treatment with R5020. To validate our microarray studies, we utilized qPCR to examine progestin-mediated regulation of endogenous E2F1 gene transcription in T47D:A18 cells. In order to reduce overall background levels of E2F, T47D cells were arrested in G0 by serum starvation for 24 h. This cell cycle arrest was verified by propidium iodide cell cycle analysis (data not shown). In Fig. 1B, we demonstrate that synchronized T47D:A18 cells treated with R5020 for 18 h show an approximately 20-fold increase in E2F1 mRNA levels. While pretreatment with U0126 did not affect regulation of the PR target gene S100P by R5020, inhibition of MAPK did reduce both progestin-mediated induction and basal expression of E2F1 mRNA levels. Western immunoblot analysis confirmed these results at the protein level; treatment with R5020 for 18 h dramatically increased E2F1 protein levels, and pretreatment with U0126 partially blocked this effect (Fig. 1C).
In addition, we confirmed that progestin treatment stimulates the transcription of classic E2F1 target genes such as those for CDC2, CDC6, cyclin E1, and CDK2 (see Fig. S1 at http://mcdonnelllab.duhs.duke.edu), suggesting that the E2F1 protein induced by PR is functional and active. However, we have not eliminated the possibility that PR may also exert direct effects on the expression of these genes. Importantly, we also observed a 12-fold increase in E2F1 mRNA levels after treatment with R5020 in PR-positive BT483 breast cancer cells (Fig. 1D), indicating that the regulatory activities of PR on this target gene are not restricted to T47D cells.
Finally, all of the experiments in this study were performed using concentrations of R5020 in the range of 100 pM to 10 nM, depending on the cell line and assay. In the course of these experiments, it was noted that in general, treatment of cells with 100 pM R5020 led to a greater induction of E2F1 mRNA and protein levels than higher doses such as 10 nM R5020 (see Fig. S2 at http://mcdonnelllab.duhs.duke.edu). Because the focus of this study was to define the mechanisms underlying PR regulation of E2F1, the elucidation of the biphasic nature of E2F1 induction by R5020 will be addressed in a separate study.
PR-B is necessary for progestin-dependent regulation of E2F1 expression.
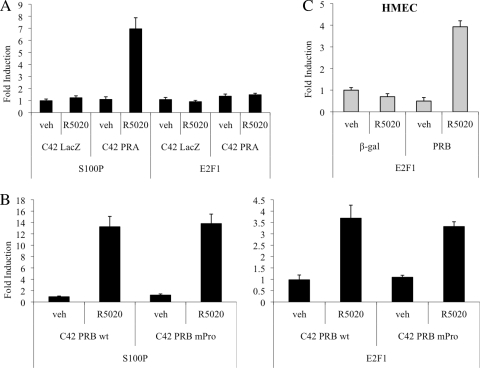
To determine whether PR is necessary for R5020-mediated induction of E2F1 transcription, we examined the effects of progestin treatment on E2F1 expression in T47D:C42 cells (a PR-negative T47D subclone) that stably express a LacZ reporter gene (control cells), wild-type human PR-A, or PR-B (2). qPCR analysis demonstrated that R5020 does not induce E2F1 transcription in control cells or those expressing PR-A alone (Fig. 2A). However, induction of E2F1 expression was observed in cells in which wild-type PR-B was expressed (Fig. 2B).
FIG. 2.
PR-B mediates induction of E2F1 expression by R5020. (A and B) The indicated T47D:C42 cells were synchronized and treated with vehicle (veh) or 10 nM R5020 for 18 (A) or 16 (B) h. wt, wild type. (C) Human mammary epithelial cells (HMECs) were infected with a β-gal (negative control)- or PR-B-expressing adenovirus and subsequently treated with vehicle or 10 nM R5020 for 16 h. (A to C) After treatment, cells were lysed and RNA was isolated and reverse transcribed. S100P or E2F1 mRNA levels were quantified using qPCR and normalized to the housekeeping gene 36B4. Results are expressed as mean fold induction over vehicle-treated cells ± SEM (n = 3).
Given that R5020-mediated induction of E2F1 can be partially inhibited by U0126, we initially thought that the rapid, nongenomic actions of PR signaling through Src family kinases and the downstream MAPK pathway might be partly responsible for its regulation of E2F1. To further investigate this issue, we compared R5020 induction of E2F1 transcription in T47D:C42 cells that stably express wild-type PR-B or PR-BmPro, a mutant form of PR-B in which three key proline residues in the polyproline motif were replaced with alanines. This mutant PR receptor is unable to mediate rapid, nongenomic activation of Src family kinases or downstream MAPK, but its classical genomic functions remain intact (3). Interestingly, we determined that R5020 induces equal expression of E2F1 mRNA in cells expressing wild-type PR-B or the mutant PR-BmPro version (Fig. 2B). From these data, we conclude that although MAPK activity affects regulation of E2F1 expression, its activation is not dependent on direct PR signaling through Src family kinases.
Finally, treatment with R5020 has no effect on E2F1 mRNA levels in ER−/PR− human mammary epithelial cells (HMECs) infected with a control β-gal adenovirus, but infection with PR-B restores the ability of progestins to induce transcription of E2F1 in these cells (Fig. 2C). Collectively, these studies confirm that the PR-B isoform is both necessary and sufficient for progestin-mediated induction of E2F1 gene expression.
Direct regulation of E2F1 transcription by PR.
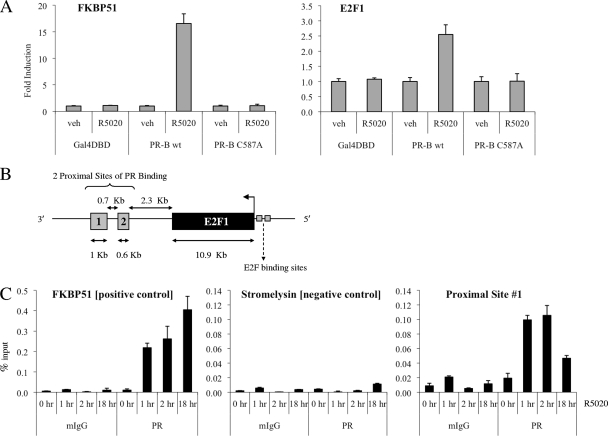
Next, we set out to define the mechanism(s) by which PR regulates E2F1 expression. Given that R5020 is able to stimulate an increase in E2F1 mRNA levels as early as 4 h posttreatment (see Fig S3 at http://mcdonnelllab.duhs.duke.edu), we suspected that the E2F1 gene might be a direct transcriptional target of PR. To investigate whether PR regulates E2F1 expression through the classic direct pathway of transcriptional regulation, we generated T47D:C42 cell lines that stably express wild-type PR-B or PR-B C587A, a zinc-finger mutant of PR-B that is unable to bind DNA (see Fig. S4 at http://mcdonnelllab.duhs.duke.edu). While R5020 treatment induced E2F1 expression in cells expressing wild-type PR-B, no significant change in E2F1 mRNA levels was evident in cells expressing the DNA-binding mutant of PR-B (Fig. 3A). Therefore, we conclude that the DNA-binding capacity of PR is required for progestin regulation of E2F1.
FIG. 3.
Evidence for direct regulation of E2F1 by PR. (A) The indicated T47D:C42 cells were synchronized and treated with vehicle (veh) or 10 nM R5020 for 24 h. Cells were lysed, and RNA was isolated and reverse transcribed. FKBP51 or E2F1 mRNA levels were quantified using qPCR and normalized to the housekeeping gene 36B4. Results are expressed as mean fold induction over vehicle-treated cells ± SEM (n = 3). (B) Schematic depicting the locations of two proximal enhancer sites located around E2F1 that were identified in ChIP-chip experiments as potential PR-binding sites. (C) Synchronized T47D:A18 cells were treated with vehicle or 10 nM R5020 for the indicated times. Cells were harvested after cross-linking and subjected to immunoprecipitation with either a mouse IgG control (mIgG) or PR antibody. Following reversal of cross-linking, DNA was isolated and subjected to qPCR analysis using primers spanning a region in FKBP51 (positive control), stromelysin (negative control), or the potential PR-binding region proximal to E2F1 (proximal site 1). The results are presented as mean percent input ± SEM for triplicate amplification reactions from one representative experiment (n = 3).
We were unable to identify any putative progesterone response elements (PREs) within the promoter sequence surrounding E2F1 using Transcription Element Search software (TESS) (29). Furthermore, ChIP-chip analysis of T47D cells treated with progesterone did not identify any PR-binding sites within the 2-kb upstream promoter region of the E2F1 gene (chromosome 20:31737871-31739871) (our unpublished data). However, a genome-wide ChIP-chip analysis did reveal that progesterone-activated PR is recruited to two proximal enhancer sites, located ∼2.3 kb downstream of E2F1 (Fig. 3B). We noted that sites 1 and 2 are located within the XB51 locus; however, although R5020 treatment led to a 20- to 30-fold induction of E2F1 mRNA, XB51 was consistently induced less than 2-fold (data not shown).
Next, we performed ChIP studies to test whether R5020-activated PR is recruited to these proximal enhancer elements. Recruitment of PR to a previously characterized intronic PRE within FKBP51 was used as a positive control for PR binding (18). Our ChIP analysis confirmed that ligand-bound PR associates with site 1, with a 5-fold increase in recruitment at 1 to 2 h after treatment with R5020 (Fig. 3C). Moreover, PR remains associated with site 1 as late as 18 h posttreatment. Unfortunately, we were unable to ascertain whether PR binds to site 2 due to poor PCR efficiency despite attempts with multiple sets of PCR primers.
In addition to the proximal enhancer elements, the ChIP-chip data also identified four distal enhancer sites located ∼29.5 kb upstream of E2F1 (see Fig. S5A at http://mcdonnelllab.duhs.duke.edu). Our subsequent ChIP studies confirmed significant recruitment of PR to all four distal sites in a ligand-dependent manner (see Fig. S5B at http://mcdonnelllab.duhs.duke.edu). Sites 5 and 6 are located within intronic regions of ZNF341, a gene that is weakly regulated by PR; sites 3 and 4 are, respectively, located within intronic and promoter regions of PXMP4, a gene that is positively regulated by R5020 treatment (data not shown). Studies are currently ongoing to determine whether recruitment of PR to these distal sites is involved in progestin regulation of E2F1; however, TESS analysis indicates that all six sites contain putative PREs. Thus, we have identified both proximal and distal enhancer elements to which PR binds and possibly directly regulates expression of E2F1.
To further verify that E2F1 is a direct target of PR action, we pretreated T47D:A18 cells with or without the translational inhibitor cycloheximide, followed by addition of vehicle or R5020 for 18 h. Using qPCR, we determined that cycloheximide did not inhibit induction of SGK1 (serum- and glucocorticoid-regulated kinase), an established primary target of PR (2). In contrast, we observed that pretreatment with cycloheximide partially inhibits R5020-mediated induction of E2F1 transcription (see Fig. S6 at http://mcdonnelllab.duhs.duke.edu), signifying that nascent protein synthesis is required to achieve maximal PR induction of E2F1 expression. Furthermore, while R5020 can upregulate E2F1 mRNA levels by early time points such as 4 to 6 h posttreatment, maximal induction of E2F1 transcription by R5020 is not achieved until 18 h posttreatment (data not shown). These data prompted us to consider that the ligand-dependent actions of PR on the E2F1 gene may involve additional indirect regulatory pathways.
R5020 treatment increases phosphorylation of Rb and recruitment of E2F1 to its own promoter.
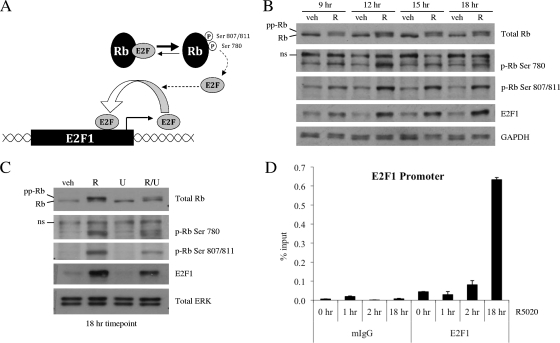
It is well known that E2F1 can upregulate its own expression by binding to previously defined E2F binding sites within its own promoter (13). Therefore, we hypothesized that E2F1 protein produced as a result of direct PR regulation could act to further amplify progestin-induced E2F1 transcription by activating a positive feedback loop. Since the ability of E2F family members to influence transcription of target genes is regulated by the phosphorylation status of the retinoblastoma protein Rb, we first examined the effects of progestin treatment on the phosphorylation of Rb. While a cascade of phosphorylation events regulates Rb activity, we chose to focus on the phosphorylation of three sites in particular. Prior studies indicate that sequential phosphorylation of Rb on Ser780, followed by Ser807/811, is important for release of E2F from Rb and optimal activation of downstream E2F target gene transcription, respectively (Fig. 4A) (17).
FIG. 4.
R5020 further amplifies E2F1 transcription by activating a positive feedback loop. (A) Schematic depicting hyperphosphorylation of Rb and subsequent release of E2F, which allows E2F to bind its own promoter and increase transcription in a positive feedback loop. (B and C) Synchronized T47D:A18 cells were treated with vehicle (veh) or 100 pM R5020 (R) for the indicated times (B) or pretreated with vehicle or 10 μM U0126 (U) for 30 min prior to addition of vehicle or 100 pM R5020 for 18 h (C). After treatment, cells were harvested and 20 μg whole-cell extract was resolved by SDS-PAGE, transferred to PVDF, and subjected to immunoblotting for total Rb, Rb phosphorylated on Ser780 (p-Rb Ser 780), Rb phosphorylated on Ser807/811 (p-Rb Ser 807/811), E2F1, or GAPDH or ERK 1/2 as a loading control. ns, nonspecific band. A representative blot is shown (n = 3). (D) Synchronized T47D:A18 cells were treated with vehicle or 100 pM R5020 for the indicated times. Cells were harvested after cross-linking and subjected to immunoprecipitation with either mouse IgG control (mIgG) or E2F1 antibody. Following reversal of cross-linking, DNA was isolated and subjected to qPCR analysis using primers spanning a region in the E2F1 promoter containing E2F binding sites (depicted in Fig. 3B). The results are presented as percent input ± SEM for triplicate amplification reactions from one representative experiment (n = 3).
Western blot analysis with total and phospho-specific Rb antibodies shows that treatment with progestins for 9 to 18 h led to an increase in phosphorylation of Rb at Ser780 and Ser807/811, as well as an overall increase in total levels of hyperphosphorylated Rb (Fig. 4B). However, we saw no increase in phosphorylation of Rb at Ser780 and Ser807/811 or change in total levels of hyperphosphorylated Rb at any of the earlier time points that we examined (data not shown). Furthermore, we discovered that this progestin-mediated increase in Rb phosphorylation can be partially inhibited by pretreatment with U0126, and this corresponds with a reduction in the amount of E2F1 protein induced by an 18 h treatment with R5020 (Fig. 4C).
Since we observed an increase in Rb phosphorylation at 9 to 18 h after treatment with R5020, we hypothesized that any progestin-mediated increase in recruitment of E2F1 to its own promoter might correspondingly occur within this time frame. To address this question, we performed ChIP experiments with T47D:A18 cells to measure E2F1 occupancy at its own promoter. As expected, treatment with R5020 for 1 to 2 h did not result in a significant increase in E2F1 recruitment to the region of the E2F1 promoter containing E2F-binding sites (Fig. 4D). In contrast, while ligand-bound PR is already recruited to enhancer elements near the E2F1 gene at these early time points (Fig. 3C; see Fig. S5B at http://mcdonnelllab.duhs.duke.edu), Rb remains hypophosphorylated and bound to E2F1, thereby preventing it from binding to the promoters of target genes. However, by 18 h posttreatment, Rb has become hyperphosphorylated, which frees E2F1 and enables it to interact with its cognate response element in the E2F1 promoter. Correspondingly, ChIP studies showed a significant progestin-mediated increase in recruitment of E2F1 to its own promoter at this later time point (Fig. 4D). Collectively, these data indicate that PR acts indirectly to further amplify expression of E2F1 by stimulating phosphorylation of Rb and recruitment of E2F1 to its own promoter. Inhibition of MAPK decreases the ability of PR to stimulate hyperphosphorylation of Rb; this is one possible mechanism by which U0126 can act to impair progestin-mediated induction of E2F1 expression.
GC-rich DNA within the E2F1 promoter is important for progestin-mediated induction of E2F1 expression.
During our search for an indirect pathway through which PR could modulate E2F1 expression, we searched for additional regulatory elements located within the E2F1 promoter that might be involved in this response. In addition to the previously mentioned E2F binding sites, the E2F1 promoter also contains many GC-rich regions of DNA, which commonly serve as binding sites for members of the specificity protein/Krüppel-like factor (Sp/KLF) transcription factor superfamily (16). Previous studies have suggested that a member of the Sp/KLF superfamily may play a role in the regulation of the E2F1 promoter; more specifically, the loss of a small, 82-bp region (positions −204 to −122 in Fig. 5A) that contains several clusters of GC-rich DNA results in reduced activity of the E2F1 promoter (13). Therefore, we were intrigued by the observation that a number of Sp/KLF family members were induced by R5020 in our array; furthermore, oPOSSUM identified an enrichment of Sp1 sites in the promoters of PR-regulated genes.
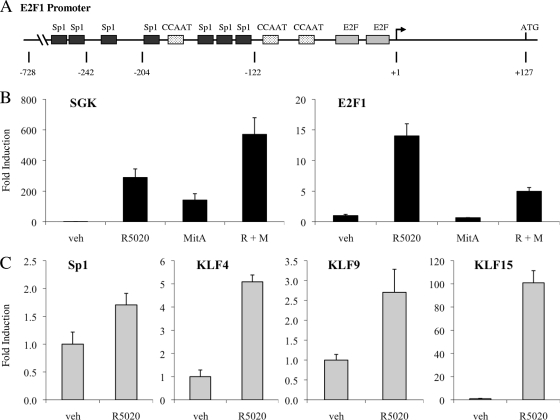
FIG. 5.
A member of the Sp/KLF family is involved in regulation of E2F1 transcription. (A) Schematic depicting regulatory elements within the E2F1 promoter (13). (B and C) Synchronized T47D:A18 cells were pretreated with vehicle (veh) or 200 nM mithramycin A (MitA) for 30 min and then treated with vehicle or 100 pM R5020 (R) for 18 h (B) or treated with vehicle or 100 pM R5020 for 18 h (C). Cells were lysed, and RNA was isolated and reverse transcribed. SGK, E2F1, SP1, KLF4, KLF9, and KLF15 mRNA levels were quantified using qPCR and normalized to the housekeeping gene 36B4. Results are expressed as mean fold induction over vehicle-treated cells ± SEM (n = 3).
To determine whether binding of an Sp/KLF family member to GC-rich DNA within the E2F1 promoter is important for progestin-dependent E2F1 induction, we pretreated T47D:A18 cells with mithramycin A, an antibiotic that binds to GC-rich DNA and blocks recruitment of transcription factors to these regions (20). Pretreatment with mithramycin A suppresses R5020-mediated induction of E2F1 transcription but does not decrease progestin-induced mRNA levels of the primary PR target gene SGK1, although basal levels of SGK1 mRNA did increase (Fig. 5B). Thus, we hypothesized that a transcription factor belonging to the Sp/KLF superfamily may be involved in PR-mediated induction of E2F1 expression.
Krüppel-like factor 15 (KLF15) is required for maximal induction of E2F1 expression by PR.
To further interrogate the potential involvement of an Sp/KLF family member in progestin regulation of E2F1 transcription, we utilized qPCR analysis to examine the expression of various Sp/KLF family members in synchronized T47D:A18 cells treated with 100 pM R5020 for 18 h. In fact, R5020 induces transcription of several Sp/KLF family members, including Sp1, KLF4, KLF9, and KLF15 (Fig. 5C). KLF15 was the most robustly induced Sp/KLF family member among those that we examined; furthermore, R5020 increased KLF15 mRNA levels rapidly within 2 h, which preceded PR-mediated induction of E2F1 expression (see Fig. S3 at http://mcdonnelllab.duhs.duke.edu). Additionally, qPCR studies with cycloheximide confirm that KLF15, unlike E2F1, does not require nascent protein synthesis for full expression and thus behaves more like a classic PR target gene (see Fig. S6 at http://mcdonnelllab.duhs.duke.edu). Therefore, we chose to evaluate the potential role of KLF15 in PR-mediated induction of E2F1 expression.
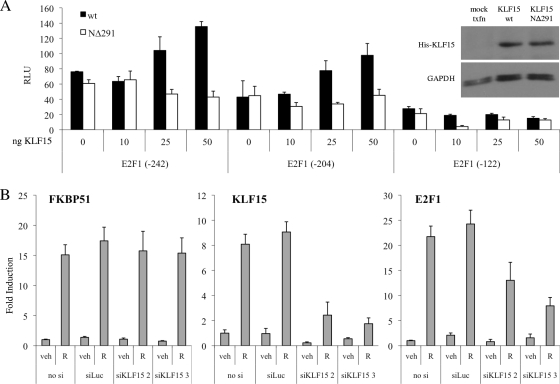
Using a position weight matrix previously described for KLF15 (22), the E2F1 promoter was scanned for putative KLF15-binding motifs using TESS. This analysis identified three putative KLF15-binding sites within the 82-bp GC-rich DNA region mentioned above (see Fig. S7 at http://mcdonnelllab.duhs.duke.edu). Unfortunately, KLF15 antibodies suitable for ChIP analysis are not yet available, and thus we could not directly examine whether KLF15 is recruited to these putative binding sites in the E2F1 promoter. As an alternative approach to probe the involvement of KLF15 in E2F1 gene regulation, we utilized luciferase assays to explore the connection between KLF15 and the E2F1 promoter. T47D:A18 cells were transiently transfected with a series of reporter gene constructs that contain successively smaller regions of the E2F1 promoter, in combination with increasing amounts of wild-type KLF15 or a KLF15 mutant that lacks the N-terminal DNA-binding domain (KLF15 NΔ291). Wild-type KLF15 increased activation of the longer E2F1 promoter fragments in a dose-dependent manner but was unable to activate the smallest promoter fragment (−122), which lacks the GC-rich DNA region containing the putative KLF15-binding sites (Fig. 6A). In contrast, addition of the mutant KLF15 NΔ291 construct did not affect activation of any E2F1 reporter constructs, indicating that the DNA-binding ability of KLF15 is required for induction of E2F1 activity.
FIG. 6.
KLF15 is necessary for maximal PR-mediated regulation of E2F1. (A) T47D:A18 cells were transiently cotransfected with various hE2F1-luc promoter fragment reporters along with increasing amounts of a vector expressing wild-type KLF15 or the KLF15 NΔ291 deletion mutant for 48 h and then were harvested and assayed for luciferase activity. Luciferase values were normalized to a β-galactosidase control. Data are the mean relative light units (RLU) ± SEM for one representative experiment performed in triplicate. Inset, Western blot control confirming equal expression of His-tagged KLF15 variants using GAPDH as a loading control. (B) T47D:A18 cells were transiently transfected with Stealth siRNAs targeting KLF15 (siKLF15 2 and 3) or a negative-control luciferase sequence (siLuc) at a final concentration of 100 nM for 48 h. Cells were synchronized by serum starvation for 24 h and then treated with vehicle (veh) or 100 pM R5020 (R) for 18 h. KLF15, E2F1, and FKBP51 mRNA levels were quantified using qPCR and normalized to the housekeeping gene 36B4. Results are expressed as mean fold induction over vehicle-treated cells ± SEM (n = 3).
To further implicate KLF15 in progestin regulation of E2F1 expression, we performed knockdown studies using two independent siRNAs targeting KLF15. Since we could not identify a reliable, working antibody that would detect KLF15 expression in T47D:A18 cells, we were unable to confirm knockdown of KLF15 at the protein level. However, qPCR analysis demonstrates that both siRNAs can inhibit basal and R5020-mediated induction of KLF15 mRNA levels to various extents, and even partial knockdown of KLF15 transcription had an inhibitory effect on R5020-mediated induction of E2F1 mRNA levels (Fig. 6B). In contrast, knockdown of KLF15 did not decrease the regulation of other classic PR target genes such as FKBP51. Taken together, these findings indicate that progestin-mediated induction of KLF15 is required for maximal induction of E2F1 expression by PR.
DISCUSSION
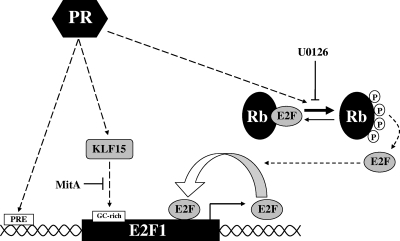
We show that PR is a component of several distinct pathways that function both directly and indirectly to positively upregulate E2F1 expression in breast cancer cells (Fig. 7). First, PR directly regulates E2F1 transcription by binding to proximal and distal enhancer sites located near E2F1. Second, progestin induces the hyperphosphorylation of Rb, which results in increased recruitment of E2F1 to its own promoter, thereby activating a positive feedback loop that further amplifies its transcription. Finally, PR induces expression of KLF15 and potentially other Sp/KLF family members, which bind to GC-rich regulatory regions within the E2F1 promoter and further activate transcription. Together, these pathways represent a complex multimodal regulatory system in which the combined actions of each component are required for maximal progestin-mediated upregulation of E2F1 transcription.
FIG. 7.
Model of multimodal regulation of E2F1 by progestins. Ligand-bound PR can bind to proximal and distal enhancer sites located near E2F1 and directly regulate E2F1 transcription. PR can also act indirectly through hyperphosphorylation of Rb and induction of KLF15 expression to achieve further progestin-mediated regulation of E2F1 expression in T47D cells.
In most breast cancer cell lines, estrogens are important for regulation of PR expression; however, the estrogen receptor (ER) has previously been shown to induce expression of E2F1, and we wanted to concentrate solely on PR-specific regulation of E2F1 expression. Therefore, we chose T47D cells as a model system for our studies because in this cell line, PR expression is uncoupled from ER signaling (14, 36). Given that progestins can stimulate proliferation of T47D cells in vitro and when propagated as xenografts in vivo, it was not unexpected to see that PR also modulates expression of E2F1, a transcription factor that controls cell cycle progression. However, we noted that E2F1 expression was also induced in response to progestins in BT483 breast cancer cells (Fig. 1D) and in ER-negative/PR-negative human mammary epithelial cells (HMECs) infected with a PR-B adenovirus (Fig. 2C), model systems where progestins do not stimulate proliferation. Importantly, the downstream biological effects of E2F1 are not limited to regulation of cell proliferation; indeed, E2F1 has been implicated in other critical processes such as DNA damage response, checkpoint control, and apoptosis (4). Defining the role(s) of these additional processes in PR biology is an area of continued exploration in our group. Additionally, the microarray analysis showed that treatment of T47D cells with R5020 stimulated the expression of E2F2 and E2F7; further studies are necessary to explore the roles of other E2F family members in PR signaling.
The initial purpose of our microarray study was to determine the overall involvement of the MAPK signaling pathway in PR regulation of target gene transcription. We were surprised to find that the expression levels of almost 80% of the 1,794 PR target genes identified in this analysis were affected by pretreatment with the MEK 1/2 inhibitor U0126 (Fig. 1A). Of course, since inhibition of MAPK reduces progestin-mediated upregulation of E2F1 expression (Fig. 1B and C), any PR target genes that are coregulated by this protein would be correspondingly affected. One explanation for the inhibitory effect of U0126 on progestin-mediated induction of E2F1 expression is the observation that MAPK inhibition partially suppressed PR-mediated hyperphosphorylation of Rb (Fig. 4C), which is necessary for release of E2F and activation of the positive feedback loop (Fig. 4A).
While the mechanism(s) by which progestins induce hyperphosphorylation of Rb has not been fully elucidated, it has been established that treatment of T47D cells with progestin leads to induction of cyclins D1 and E and increased activity of the cyclin D1/cdk4 complex (31, 33, 34), which has been implicated in phosphorylation of several sites on Rb (37). Previous studies have reported that progestin induction of cyclin D1 is dependent on rapid PR activation of the Src/MAPK pathway (2); therefore, we initially hypothesized that direct interactions between PR and Src family kinases might activate MAPK and contribute to progestin regulation of E2F1. However, we determined that R5020 effectively induces expression of E2F1 mRNA in cells expressing either wild-type PR-B or the mutant PR-BmPro (Fig. 2B), which cannot directly interact with c-Src or mediate rapid, nongenomic activation of Src/MAPK signaling.
However, other studies have proposed an alternative mechanism for rapid activation of MAPK signaling by progestins, whereby PR interacts with unliganded ER, which in turn activates the Src/MAPK signaling pathway (1, 19). Furthermore, a recent study reported that progestin induction of cyclin D1 requires both the DNA-binding domains of PR, which allow PR to bind directly to distal regions of the cyclin D1 promoter, and the two ER-interacting domains (ERID) of PR, which allow PR to interact with ER to achieve rapid activation of Src/MAPK (27). Additional studies are necessary to determine whether PR activation of MAPK through this alternative, ER-dependent pathway and subsequent induction of cyclin D1 is the mechanism leading to progestin-mediated hyperphosphorylation of Rb, and subsequent induction of the positive feedback loop that amplifies E2F1 expression. Interestingly, we noted that the magnitude of PR-mediated induction of E2F1 expression in ER-negative cell lines, such as T47D:C42 cells (Fig. 2B) or HMECs (Fig. 2C), was not as great as that achieved by progestins in ER-positive cell lines, such as T47D:A18 cells (Fig. 1B) or BT483 cells (Fig. 1D). The significance of this observation is currently under investigation.
Bioinformatic analyses revealed a 277-gene subset of progestin-regulated transcripts that was enriched for E2F-binding sites (Fig. 1A); this subset includes classic E2F1 target genes such as those for CDC6, cyclin E, and CDK2. However, it is currently unclear whether the effects of progestins on these genes and others are mediated solely by secondary E2F1 actions or whether PR also directly regulates their transcriptional activity. Analyses with Patser showed that 99 progestin-regulated genes contain both putative PREs and E2F1-binding sites within their promoters (see Fig. S8 at http://mcdonnelllab.duhs.duke.edu), and this may indicate a trend of coregulation of target genes by direct actions of PR and E2F1. Interestingly, since the expression of as many as 277 R5020-regulated genes may be modulated by E2F1, a target of PR-B but not PR-A (Fig. 2A and B), it is possible that regulation of E2F1 by the PR-B isoform could be an important factor that contributes to the vastly different profiles of PR-A and PR-B as transcriptional regulators.
Similarly, several pieces of data suggest a trend of coregulation of target genes by PR and members of the Sp/KLF superfamily. For instance, pretreatment with mithramycin A affected R5020-mediated induction of many downstream PR target genes that we examined; moreover, we observed that knockdown of KLF15 inhibited R5020 induction of several PR target genes (data not shown). Bioinformatic analyses using Patser revealed that out of the 1,794 PR target genes detected in our microarray study, the promoters of 1,372 genes contain putative GC-rich binding sites for Sp/KLF family members (see Fig. S8 at http://mcdonnelllab.duhs.duke.edu). Studies are currently ongoing to determine whether cooperation between PR and KLF15 and/or other SP/KLF family members in the regulation of gene transcription constitutes a more global model of PR function.
While the extent to which PR engages in multimodal regulation of target genes remains to be determined, the data we have generated in this study indicate that the ability of PR to induce the expression of E2F and Sp/KLF family members and their resulting impact on gene expression provides a mechanism to explain secondary, cycloheximide-sensitive responses to progestins. In general, the indirect secondary responses that are stimulated by progestins have been less studied than primary transcriptional responses; however, this area of PR signaling deserves more attention, since the regulation of target gene expression by PR-stimulated transcription factors can dramatically influence the overall transcriptional program set into motion by progestins. In the context of PR regulation of E2F1 transcription, secondary factors such as E2F1 and KLF15 act to reinforce progestin-mediated induction of E2F1 expression, but E2F and Sp/KLF family members may act to suppress PR actions on other target genes.
Finally, induction of KLF15 expression by PR has ramifications that extend beyond its role in progestin-mediated regulation of E2F1. KLF15 is a recently discovered transcription factor, and the transcriptional mechanisms that regulate KLF15 promoter activity are poorly understood; however, several recent studies support a role for NRs in regulation of KLF15 expression. In ovariectomized mice, treatment with estradiol and progesterone upregulates KLF15 expression in the uterine epithelium (24). In addition, dexamethasone treatment induces KLF15 expression in chondrocytes (12), and both corticosterone and the glucocorticoid receptor-specific agonist cortivazol upregulate KLF15 expression in cardiomyocytes (39). Furthermore, little is known about the biological function(s) of KLF15 in the breast. In our qPCR analysis of breast cancer cells, we observed that basal transcription of KLF15 was low; in contrast, KLF15 is highly expressed in the liver, kidney, heart, and skeletal muscle (35). Studies involving KLF15 in other tissues have revealed an emerging role for KLF15 in regulation of metabolic processes such as glucose homeostasis (9) and lipid accumulation (5). It is clear that further studies are warranted to determine how progestin-mediated activation of KLF15 signaling may affect metabolic signaling processes in the breast.
In conclusion, although E2F1 transcription is affected by the direct interaction of PR with the regulatory regions near E2F1, we also established that maximal induction of E2F1 expression by progestins requires the actions of additional transcription factors, such as E2F1 and KLF15, on the E2F1 promoter. The same may be true for a much larger subset of PR target genes. In fact, we suspect that PR often acts in concert with these and other secondary factors to coregulate target gene expression, depending on the cell- or tissue-specific context. These results suggest a paradigm for multimodal PR gene regulation that entails cooperation between direct and indirect pathways of PR signaling to achieve the desired downstream transcriptional cascade.
Acknowledgments
This work was supported by DOD grant W81XWH-06-1-0745 (H.E.W.) and by NIH grants DK048807 (D.P.M.); EY13499 (D.C.O.); and DK074967, CA089393, and CA080111 (M.B.).
We thank the members of the McDonnell laboratory for critical review of the manuscript and Ganesan Sathya for construction of the pcDNA3-hPR-B plasmid. We are grateful to Dean Edwards at Baylor College of Medicine in Houston, TX, for PR antibodies and T47D:C42 cell lines; to Craig Jordan at the Fox Chase Cancer Center in Philadelphia, PA, for T47D:A18 cells; and to Kathryn Horwitz at University of Colorado in Denver, CO, for hPR-Bcys. We also thank the NIEHS Microarray Core for technical assistance related to the gene expression microarray work.
Footnotes
Published ahead of print on 1 February 2010.
REFERENCES
- 1.Ballare, C., M. Uhrig, T. Bechtold, E. Sancho, M. Di Domenico, A. Migliaccio, F. Auricchio, and M. Beato. 2003. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol. Cell. Biol. 23:1994-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boonyaratanakornkit, V., E. McGowan, L. Sherman, M. A. Mancini, B. J. Cheskis, and D. P. Edwards. 2007. The role of extranuclear signaling actions of progesterone receptor in mediating progesterone regulation of gene expression and the cell cycle. Mol. Endocrinol. 21:359-375. [DOI] [PubMed] [Google Scholar]
- 3.Boonyaratanakornkit, V., M. P. Scott, V. Ribon, L. Sherman, S. M. Anderson, J. L. Maller, W. T. Miller, and D. P. Edwards. 2001. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol. Cell 8:269-280. [DOI] [PubMed] [Google Scholar]
- 4.DeGregori, J., and D. G. Johnson. 2006. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6:739-748. [DOI] [PubMed] [Google Scholar]
- 5.Du, X., R. L. Rosenfield, and K. Qin. 2009. KLF15 is a transcriptional regulator of the human 17beta-hydroxysteroid dehydrogenase type 5 gene. A potential link between regulation of testosterone production and fat stores in women. J. Clin. Endocrinol Metab. 94:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DuSell, C. D., M. Umetani, P. W. Shaul, D. J. Mangelsdorf, and D. P. McDonnell. 2008. 27-Hydroxycholesterol is an endogenous selective estrogen receptor modulator. Mol. Endocrinol. 22:65-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giangrande, P. H., E. A. Kimbrel, D. P. Edwards, and D. P. McDonnell. 2000. The opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor binding. Mol. Cell. Biol. 20:3102-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giangrande, P. H., G. Pollio, and D. P. McDonnell. 1997. Mapping and characterization of the functional domains responsible for the differential activity of the A and B isoforms of the human progesterone receptor. J. Biol. Chem. 272:32889-32900. [DOI] [PubMed] [Google Scholar]
- 9.Gray, S., B. Wang, Y. Orihuela, E. G. Hong, S. Fisch, S. Haldar, G. W. Cline, J. K. Kim, O. D. Peroni, B. B. Kahn, and M. K. Jain. 2007. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab. 5:305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hertz, G. Z., and G. D. Stormo. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15:563-577. [DOI] [PubMed] [Google Scholar]
- 11.Ho Sui, S. J., J. R. Mortimer, D. J. Arenillas, J. Brumm, C. J. Walsh, B. P. Kennedy, and W. W. Wasserman. 2005. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 33:3154-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James, C. G., V. Ulici, J. Tuckermann, T. M. Underhill, and F. Beier. 2007. Expression profiling of dexamethasone-treated primary chondrocytes identifies targets of glucocorticoid signalling in endochondral bone development. BMC Genomics 8:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, D. G., K. Ohtani, and J. R. Nevins. 1994. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 8:1514-1525. [DOI] [PubMed] [Google Scholar]
- 14.Keydar, I., L. Chen, S. Karby, F. R. Weiss, J. Delarea, M. Radu, S. Chaitcik, and H. J. Brenner. 1979. Establishment and characterization of a cell line of human breast carcinoma origin. Eur. J. Cancer 15:659-670. [DOI] [PubMed] [Google Scholar]
- 15.Li, X., D. M. Lonard, and B. W. O'Malley. 2004. A contemporary understanding of progesterone receptor function. Mech. Ageing Dev. 125:669-678. [DOI] [PubMed] [Google Scholar]
- 16.Lomberk, G., and R. Urrutia. 2005. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 392:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundberg, A. S., and R. A. Weinberg. 1998. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol. 18:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magklara, A., and C. L. Smith. 2009. A composite intronic element directs dynamic binding of the progesterone receptor and GATA-2. Mol. Endocrinol. 23:61-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Migliaccio, A., D. Piccolo, G. Castoria, M. Di Domenico, A. Bilancio, M. Lombardi, W. Gong, M. Beato, and F. Auricchio. 1998. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. EMBO J. 17:2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, D. M., D. A. Polansky, S. D. Thomas, R. Ray, V. W. Campbell, J. Sanchez, and C. A. Koller. 1987. Mithramycin selectively inhibits transcription of G-C containing DNA. Am. J. Med. Sci. 294:388-394. [DOI] [PubMed] [Google Scholar]
- 21.Murphy, C. S., J. J. Pink, and V. C. Jordan. 1990. Characterization of a receptor-negative, hormone-nonresponsive clone derived from a T47D human breast cancer cell line kept under estrogen-free conditions. Cancer Res. 50:7285-7292. [PubMed] [Google Scholar]
- 22.Otteson, D. C., H. Lai, Y. Liu, and D. J. Zack. 2005. Zinc-finger domains of the transcriptional repressor KLF15 bind multiple sites in rhodopsin and IRBP promoters including the CRS-1 and G-rich repressor elements. BMC Mol. Biol. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otteson, D. C., Y. Liu, H. Lai, C. Wang, S. Gray, M. K. Jain, and D. J. Zack. 2004. Kruppel-like factor 15, a zinc-finger transcriptional regulator, represses the rhodopsin and interphotoreceptor retinoid-binding protein promoters. Invest. Ophthalmol. Vis Sci. 45:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan, H., L. Zhu, Y. Deng, and J. W. Pollard. 2006. Microarray analysis of uterine epithelial gene expression during the implantation window in the mouse. Endocrinology 147:4904-4916. [DOI] [PubMed] [Google Scholar]
- 25.Proietti, C., M. Salatino, C. Rosemblit, R. Carnevale, A. Pecci, A. R. Kornblihtt, A. A. Molinolo, I. Frahm, E. H. Charreau, R. Schillaci, and P. V. Elizalde. 2005. Progestins induce transcriptional activation of signal transducer and activator of transcription 3 (Stat3) via a Jak- and Src-dependent mechanism in breast cancer cells. Mol. Cell. Biol. 25:4826-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu, M., and C. A. Lange. 2003. MAP kinases couple multiple functions of human progesterone receptors: degradation, transcriptional synergy, and nuclear association. J. Steroid Biochem. Mol. Biol. 85:147-157. [DOI] [PubMed] [Google Scholar]
- 27.Quiles, I., L. Millan-Arino, A. Subtil-Rodriguez, B. Minana, N. Spinedi, C. Ballare, M. Beato, and A. Jordan. 2009. Mutational analysis of progesterone receptor functional domains in stable cell lines delineates sets of genes regulated by different mechanisms. Mol. Endocrinol. 23:809-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitoh, M., M. Ohmichi, K. Takahashi, J. Kawagoe, T. Ohta, M. Doshida, T. Takahashi, H. Igarashi, A. Mori-Abe, B. Du, S. Tsutsumi, and H. Kurachi. 2005. Medroxyprogesterone acetate induces cell proliferation through up-regulation of cyclin D1 expression via phosphatidylinositol 3-kinase/Akt/nuclear factor-kappaB cascade in human breast cancer cells. Endocrinology 146:4917-4925. [DOI] [PubMed] [Google Scholar]
- 29.Schug, J., and G. C. Overton. 1997. Modeling transcription factor binding sites with Gibbs sampling and minimum description length encoding. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5:268-271. [PubMed] [Google Scholar]
- 30.Shen, T., K. B. Horwitz, and C. A. Lange. 2001. Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol. Cell. Biol. 21:6122-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skildum, A., E. Faivre, and C. A. Lange. 2005. Progesterone receptors induce cell cycle progression via activation of mitogen-activated protein kinases. Mol. Endocrinol. 19:327-339. [DOI] [PubMed] [Google Scholar]
- 32.Stepanova, M., F. Lin, and V. C. Lin. 2006. Establishing a statistic model for recognition of steroid hormone response elements. Comput. Biol. Chem. 30:339-347. [DOI] [PubMed] [Google Scholar]
- 33.Sutherland, R. L., O. W. Prall, C. K. Watts, and E. A. Musgrove. 1998. Estrogen and progestin regulation of cell cycle progression. J. Mammary Gland Biol. Neoplasia 3:63-72. [DOI] [PubMed] [Google Scholar]
- 34.Thuneke, I., H. M. Schulte, and A. M. Bamberger. 2000. Biphasic effect of medroxyprogesterone-acetate (MPA) treatment on proliferation and cyclin D1 gene transcription in T47D breast cancer cells. Breast Cancer Res. Treat. 63:243-248. [DOI] [PubMed] [Google Scholar]
- 35.Uchida, S., S. Sasaki, and F. Marumo. 2001. Isolation of a novel zinc finger repressor that regulates the kidney-specific CLC-K1 promoter. Kidney Int. 60:416-421. [DOI] [PubMed] [Google Scholar]
- 36.Vegeto, E., M. G. Cocciolo, F. Raspagliesi, A. Piffanelli, R. Fontanelli, and A. Maggi. 1990. Regulation of progesterone receptor gene expression. Cancer Res. 50:5291-5295. [PubMed] [Google Scholar]
- 37.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 38.Wu, R. C., J. Qin, P. Yi, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2004. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol. Cell 15:937-949. [DOI] [PubMed] [Google Scholar]
- 39.Yoshikawa, N., M. Nagasaki, M. Sano, S. Tokudome, K. Ueno, N. Shimizu, S. Imoto, S. Miyano, M. Suematsu, K. Fukuda, C. Morimoto, and H. Tanaka. 2009. Ligand-based gene expression profiling reveals novel roles of glucocorticoid receptor in cardiac metabolism. Am. J. Physiol. Endocrinol. Metab. 296:E1363-E1373. [DOI] [PubMed] [Google Scholar]