Abstract
In all eukaryotes, a specialized enzyme, RNA polymerase I (Pol I), is dedicated to transcribe the 35S rRNA gene from a multicopy gene cluster, the ribosomal DNA (rDNA). In certain Saccharomyces cerevisiae mutants, 35S rRNA genes can be transcribed by RNA polymerase II (Pol II). In these mutants, rDNA silencing of Pol II transcription is impaired. It has been speculated that upstream activating factor (UAF), which binds to a specific DNA element within the Pol I promoter, plays a crucial role in forming chromatin structures responsible for polymerase specificity and silencing at the rDNA locus. We therefore performed an in-depth analysis of chromatin structure and composition in different mutant backgrounds. We demonstrate that chromatin architecture of the entire Pol I-transcribed region is substantially altered in the absence of UAF, allowing RNA polymerases II and III to access DNA elements flanking a Pol promoter-proximal Reb1 binding site. Furthermore, lack of UAF leads to the loss of Sir2 from rDNA, correlating with impaired Pol II silencing. This analysis of rDNA chromatin provides a molecular basis, explaining many phenotypes observed in previous genetic analyses.
Chromatin is the template of all processes involved in DNA metabolism in the eukaryotic cell. Accordingly, chromatin is a dynamic structure which changes in its composition and posttranslational modification, correlating with the functional state of a genomic locus (13, 25). It is important to understand how chromatin structure is established, maintained, and altered, thereby controlling the access to the genetic information, and to identify factors and molecular mechanisms involved in these processes.
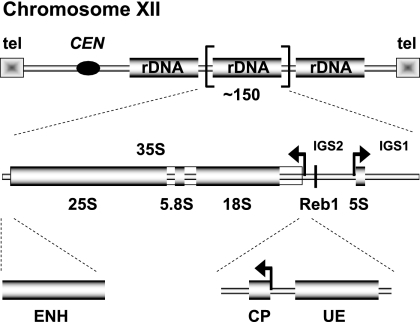
An excellent example to study the correlation between transcription and chromatin structure is the ribosomal DNA (rDNA) locus in Saccharomyces cerevisiae (hereafter called yeast). The rDNA is located on the right arm of chromosome XII and consists of 150 to 200 transcription units arranged head to tail in a tandem array (31, 33) (Fig. 1). Each of these repeated units is composed of the RNA polymerase I (Pol I)-transcribed 35S rRNA gene (precursor for the 18S, 5.8S, and 25S rRNAs) and two intergenic spacers (IGS1 and IGS2). IGS1 contains the transcriptional enhancer (ENH) for 35S rRNA transcription and is separated from IGS2 by the 5S rRNA gene transcribed by RNA polymerase III (Pol III) (Fig. 1). Interestingly, the 35S rRNA genes coexist in (at least) two different chromatin states in an actively dividing yeast cell (11). Actively transcribed 35S rRNA genes are largely devoid of histone molecules and bound by the HMG box protein Hmo1, whereas transcriptionally inactive 35S rRNA genes are nucleosomal (26).
FIG. 1.
Schematic representation of the yeast rDNA locus. The position of the rDNA repeat cluster on chromosome XII with respect to the centromere (CEN) and telomeres (tel) is shown. Each rDNA repeat consists of the Pol I-transcribed 35S rRNA gene (precursor for the 18S, 5.8S, and 25S rRNAs), the RNA Pol III-transcribed 5S rRNA gene, and two intergenic spacer regions, IGS1 and -2. Arrows mark the transcription start sites and direction. The positions of three regulatory DNA elements—enhancer (ENH), core promoter (CP), and upstream element (UE)—and of the Pol I promoter-proximal Reb1 binding site are indicated.
Efficient transcription initiation by Pol I requires the following four transcription factors, forming together with the RNA polymerase a preinitiation complex (PIC): upstream activating factor (UAF), core factor (CF), TATA-binding protein (TBP; yeast Spt15), and Rrn3 (31, 33). Another factor, Net1, a component of the regulator of nucleolar silencing and telophase exit (RENT) complex (see below), localizes at the Pol I promoter and stimulates Pol I transcription both in vitro and in vivo (36). Reb1, a ubiquitous DNA-binding protein factor, has two recognition sites in rDNA, within the ENH region and at the 5′ end of the Pol I promoter (27) (Fig. 1). Interestingly, both of the Reb1 recognition sites, but especially the promoter-proximal DNA element, contribute to efficient 35S rRNA gene transcription in vivo (23).
UAF is a multiprotein complex binding to the upstream element (UE) of the Pol I promoter (Fig. 1). The complex consists of the following six subunits: Rrn5, Rrn9, Rrn10, Uaf30, and the histones H3 and H4 (18, 19, 38). Uaf30 was demonstrated to be important for UAF recruitment to the UE (15), whereas the functions of the other factors (besides mediating specific protein-protein interactions [40]) are still unknown. It is generally accepted that UAF nucleates PIC formation. However, it is under discussion whether UAF, TBP, and CF can independently form a stable complex at the Pol I promoter (19) or if CF together with Rrn3-bound Pol I cycles on and off the promoter over the course of each initiation event (2, 3). In good agreement with in vitro data, UAF plays an important role for Pol I transcription in vivo, and in the absence of single UAF subunits, 35S rRNA production is severely impaired (19, 28, 38).
Apart from its role in stimulating Pol I transcriptional activity, UAF plays an important role in inhibiting transcription of 35S rRNA genes by RNA polymerase II (Pol II). Deletion of any of the genes coding for the subunit Rrn5, Rrn9, or Rrn10 prevents Pol I transcription of 35S rRNA genes, which are then transcribed by Pol II from a cryptic promoter upstream from the RNA Pol I initiation site (43). This polymerase switch (PSW) is accompanied by an increase in the copy number of rDNA repeats (32). Deletion of the gene coding for Uaf30 leads to transcription of 35S rRNA genes by both Pol I and Pol II, without significant changes in rDNA repeat copy number, but can also result in a PSW phenotype under certain conditions (38). The fact that a single factor controls promoter usage by different RNA polymerases is so far a unique feature of UAF, but the mechanism remains ill defined.
UAF is also important for silencing of Pol II reporter genes integrated into the rDNA locus (9) and the Pol II-dependent production of noncoding RNAs (ncRNAs) within IGS1 and IGS2 (7). Efficient silencing of Pol II transcription in rDNA further depends on Pol I transcription (7, 9) and on the histone deacetylase Sir2 together with Net1 and Cdc14 forming the RENT complex (4, 12, 39, 41). This has led to the model that UAF, perhaps together with other Pol I-associated factors, nucleates a special chromatin structure at the Pol I promoter, being repressive for 35S rRNA synthesis by Pol II and spreading to other rDNA regions by factors like RENT (9). However, an in-depth molecular characterization of the UAF-dependent rDNA chromatin structure is still missing.
In this study, we investigated rDNA chromatin structure and composition upon deletion of components of the Pol I transcription machinery. We found that deletion of UAF results in a reorganization of Pol I promoter chromatin. In the absence of UAF, flanking regions of the promoter-proximal Reb1 binding site become accessible for binding of Pol II and III and associated transcription factors and may be sites of transcription initiation in these strains. Furthermore, the integrity of the UAF complex is required for the association of Sir2 with the rDNA locus, which can explain defective silencing of Pol II transcription upon deletion of UAF components. We demonstrate that alterations in chromatin composition extend throughout the 35S rRNA coding sequence. Interestingly, none of the above-described changes can be provoked by short-term inactivation of Pol I transcription. Taken together, our analyses shed light on how UAF organizes rDNA chromatin, thereby determining RNA polymerase specificity at the Pol I promoter and rDNA silencing of Pol II transcription.
MATERIALS AND METHODS
Plasmids and yeast strains.
Complete lists of the oligonucleotides and yeast strains used in this study are presented in Tables 1 and 2.
TABLE 1.
Yeast strains used in this study
| Strain | Parent strain | Genotypea | Reference or source |
|---|---|---|---|
| NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 | 29 | |
| NOY558 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn7::LEU2/pNOY103 | 20 | |
| NOY699 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn5::LEU2/pNOY103 | 19 | |
| NOY408-1a | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rpa135::LEU2/pNOY103 | 30 | |
| NOY604 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn3::HIS3/pNOY103 | 44 | |
| NOY703 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn9::HIS3/pNOY103 | 19 | |
| CG379 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 | 6 | |
| YCC95 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 rrn3-8 | 6 | |
| y617 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 HTB2-MNase-3×HA::KanMX6 | 26 |
| y618 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 UAF30-MNase-3×HA::KanMX6 | 26 |
| y621 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 HMO1-MNase-3×HA::KanMX6 | 26 |
| y624 | NOY505 | matα ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 RPA190-MNase-3×HA::KanMX6 | This study |
| y881 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 RRN7-MNase-3×HA::KanMX6 | 26 |
| y895 | NOY703 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn9::HIS3 rrn7::LEU2/pNOY103 | This study |
| y938 | YCC95 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 rrn3-8 RPA190-MNase-3×HA::KanMX6 | This study |
| y945 | CG397 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 RPA190-MNase-3×HA::KanMX6 | This study |
| y952 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 FOB1-MNase-3×HA::KanMX6 | This study |
| y954 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 HTZ1-MNase-3×HA::KanMX6 | This study |
| y1109 | NOY558 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn7::LEU2/pNOY103 UAF30-MNase-3×HA::KanMX6 | This study |
| y1120 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 | This study |
| y1121 | y881 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 RRN7-MNase-3×HA::KanMX6 | This study |
| y1143 | CG397 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 RRN11-MNase-3×HA::KanMX6 | This study |
| y1144 | YCC95 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 rrn3-8 RRN11-MNase-3×HA::KanMX6 | This study |
| y1145 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 HHO1-MNase-3×HA::KanMX6 | This study |
| y1148 | YCC95 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 rrn3-8 SPT15-MNase-3×HA::KanMX6 | This study |
| y1149 | CG397 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 SPT15-MNase-3×HA::KanMX6 | This study |
| y1151 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 RRN9-MNase-3×HA::KanMX6 | This study |
| y1171 | y617 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 HTB2-MNase-3×HA::KanMX6 | This study |
| y1172 | y621 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 HMO1-MNase-3×HA::KanMX6 | This study |
| y1174 | y624 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 RPA190-MNase-3×HA::KanMX6 | This study |
| y1177 | y952 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 FOB1-MNase-3×HA::KanMX6 | This study |
| y1184 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 REB1-MNase-3×HA::KanMX6 | This study |
| y1185 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-102 SPT15-MNase-3×HA::KanMX6 | 26 |
| y1196 | NOY699 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn5::LEU2/pNOY103 RRN7-MNase-3×HA::KanMX6 | This study |
| y1273 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 BRF1-MNase-3×HA::KanMX6 | This study |
| y1294 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 RPO31-MNase-3×HA::KanMX6 | This study |
| y1328 | y1184 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 REB1-MNase-3×HA::KanMX6 | This study |
| y1329 | y1185 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 SPT15-MNase-3×HA::KanMX6 | This study |
| y1330 | y1294 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 RPO31-MNase-3×HA::KanMX6 | This study |
| y1346 | y1185 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-102 sir2Δ::klURA3 Spt15-MNase-3×HA::KanMX6 | This study |
| y1450 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 SIR2-MNase-3×HA::KanMX6 | This study |
| y1453 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 NET1-MNase-3×HA::KanMX6 | This study |
| y1560 | CG397 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 RPO31-MNase-3×HA::KanMX6 | This study |
| y1566 | YCC95 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 rrn3-8 RPO31-MNase-3×HA::KanMX6 | This study |
| y1681 | NOY505 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 RPB3-MNase-3×HA::KanMX6 | This study |
| y1688 | y1681 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 RPB3-MNase-3×HA::KanMX6 | This study |
| y1689 | y1453 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 NET1-MNase-3×HA::KanMX6 | This study |
| y1690 | y1145 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 HHO1-MNase-3×HA::KanMX6 | This study |
| y1691 | y1450 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 SIR2-MNase-3×HA::KanMX6 | This study |
| y1692 | y1151 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 RRN9-MNase-3×HA::KanMX6 | This study |
| y1694 | y1273 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 BRF1-MNase-3×HA::KanMX6 | This study |
| y1695 | Y954 | mataade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 uaf30Δ::klURA3 HTZ1-MNase-3×HA::KanMX6 | This study |
| y1707 | YCC95 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 rrn3-8 NET1-MNase-3×HA::KanMX6 | This study |
| y1708 | YCC95 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 rrn3-8 RPB3-MNase-3×HA::KanMX6 | This study |
| y1712 | CG397 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 NET1-MNase-3×HA::KanMX6 | This study |
| y1713 | CG397 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 RPB3-MNase-3×HA::KanMX6 | This study |
| y940 | CG397 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 UAF30-MNase-3×HA::KanMX6 | This study |
| y1139 | CG397 | matα ade5 his7-2 leu2-112 trp1-289 ura3-52 RRN7-MNase-3×HA::KanMX6 | This study |
| y1926 | NOY408-1a | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rpa135::LEU2/pNOY103 RRN7-MNase-3×HA::KanMX6 | This study |
| y1927 | NOY408-1a | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rpa135::LEU2/pNOY103 UAF30-MNase-3×HA::KanMX6 | This study |
| y1928 | NOY604 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn3::HIS3/pNOY103 RRN7-MNase-3×HA::KanMX6 | This study |
| y1929 | NOY604 | matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn3::HIS3/pNOY103 UAF30-MNase-3×HA::KanMX6 | This study |
klURA3, Kluyveromyces lactis URA3.
TABLE 2.
Oligonucleotides used in this studya
| Oligonucleotide | Sequence | Purpose | Gene/locus |
|---|---|---|---|
| 843 | GATGGTACCCATCGTTTCAGATTCCGAGCAATCAGATACAAAGGGCATTTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | UAF30 |
| 844 | CTACCGCGGACAACACAAATTTCAACGCCTTGAAATTTTCATGATATCCTTGATATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | UAF30 |
| 935 | GATGGTACCAAGAAGAAGAAGGATAAGAAGAAGGACAAATCCAACTCTTCTATTTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | HMO1 |
| 936 | CTACCGCGGATTTTAGAAAGACAGTAGAGTAATAGTAACGAGTTTGTCCGTCCAATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | HMO1 |
| 941 | GATGGTACCGGTACGGGTTCATTTGATGTGTTAGCAAAGGTTCCAAATGCGGCTTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RPA190 |
| 942 | CTACCGCGGAAACTAATATTAAATCGTAATAATTATGGGACCTTTTGCCTGCTTATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RPA190 |
| 1016 | GATGGTACCGAAGGTACTAGGGCTGTTACCAAATACTCCTCCTCTACTCAAGCCTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | HTB2 |
| 1017 | CTACCGCGGTAATAAAAAGAAAACATGACTAAATCACAATACCTAGTGAGTGACATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | HTB2 |
| 1157 | GATGGTACCGACTGCATTTCAAGGATCAAAAATGCCTGTCTGCATAGGATGAATTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RRN7 |
| 1158 | CTACCGCGGAGTATGCATAGAAATAGCAATCCAGCGAGAATAATTTAAAAGGAGATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RRN7 |
| 1334 | GATGGTACCAGAGCTAGTTGATTATTTTAGCTCCAATATTTCAATGAAAACAGAAAATTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | REB1 |
| 1335 | CTACCGCGGCTATCAAACATTATTGAGTTTTTCGCTTTCACCAATTATATTTTCCGGAAATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | REB1 |
| 1342 | GATGGTACCCCAAGCTTTTGAAGCTATATACCCTGTGCTAAGTGAATTTAGAAAAATGTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | SPT15 |
| 1343 | CTACCGCGGAAATGGAACAAATAGAAAACCTTTTTTTCTTTTCGTCTACTCCTTCCCCAATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | SPT15 |
| 1358 | GATGGTACCTATAAATAAAGCATTATTATTGAAAGTGGAAAAAAAGGGAAGTAAGAAATCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | HTZ1 |
| 1359 | CTACCGCGGATACAGGAGCAGGGAGAATTACGGGAAATGGGAAAGAAAAACTATTCTTCATCGATGAATTCGAGCT | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | HTZ1 |
| 1492 | GATGGTACCCCCCTCCGGCATTATTAAACTAAACAAGAAGAAGGTCAAACTCTCCACGTCGTACGCTGCAGGTCGA | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | HHO1 |
| 1493 | CTACCGCGGTTTGATAGTATTGCTATCACCATTGACATTCTCGTTTGGATATTCACTTTATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | HHO1 |
| 1498 | GATGGTACCACGAGTTGGACGACTGCCTCATAGAACTGCCTAATGGGAACATATCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RRN9 |
| 1499 | CTACCGCGGATGAATATTTCTTAATGGAAAAAGGTAAAAAAAAGATTTTCTCATATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RRN9 |
| 1500 | GATGGTACCACGGGTTGGACGAAATGCATTACAGTGATGAAGACTCAAGTGAGTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RRN11 |
| 1501 | CTACCGCGGAAGTTTCCCTAGTTGAAACCAAGTTATTAAAGTTTACTAGTTTGTATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RRN11 |
| 1682 | GATGGTACCAAGTTTCTCCAAGAAGATTAATTACGACGCCATTGACGGTTTGTTTAGGTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | BRF1 |
| 1683 | CTACCGCGGCGTCTTTATTTCCGTTCCCTTTTTCCTTCCTAGGGTTGATTACCTAAACGATCGATGAATTCGAGCTC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | BRF1 |
| 1688 | GATGGTACCGCGATGTCTATTTGAAAGTCTCTCAAATGAGGCAGCTTTAAAAGCGAACTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RPO31 |
| 1689 | CTACCGCGGGTGGTAGAAAAATAATACAAATGCTATAAAAAAGTTTAAAAACGACTACTATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RPO31 |
| 1723 | GATGGTACCGAAGCCAAGTGGTGGATTTGCATCATTAATAAAAGATTTCAAGAAAAAATCGTACGCTGCAGGTCGA | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | NET1 |
| 1724 | CTACCGCGGTTTTTTTTACTAGCTTTCTGTGACGTGTATTCTACTGAGACTTTCTGGTAATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | NET1 |
| 1727 | GATGGTACCGGATAAGGGCGTGTATGTCGTTACATCAGATGAACATCCCAAAACCCTCTCGTACGCTGCAGGTCGA | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | SIR2 |
| 1728 | CTACCGCGGTGTAAATTGATATTAATTTGGCACTTTTAAATTATTAAATTGCCTTCTACATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | SIR2 |
| 1744 | GATGGTACCACAGGGTAATGGAGATCAAACAAGAGACTTTGGCACATCAATGGAATTGTCGTACGCTGCAGGTCGA | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | FOB1 |
| 1745 | CTACCGCGGTTTTTTTTTCACCTATGGTGACTCCTCCTTTCATTCTATCCTACATATTAATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | FOB1 |
| 2293 | GTTCCGCGGTGCATCTCAAATGGGTAATACTGGATCAGGAGGGTATGATAATGCTTGGTCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RPB3 |
| 2294 | GTTGGTACCTTTTCGGTTCGTTCACTTGTTTTTTTTCTTCTATTACGCCCACTTGAGAAATCGATGAATTCGAGCTCG | Primer used to obtain amplicon of K643 for genomic integration of MNase-3×HA::KanMX6 | RPB3 |
| 1509 | TTAACAAGTACTAAAGCGTTCGTTGACAGCTTTCTTTGCGTTGCCCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon from pBS1539 for deletion of the UAF30 gene | UAF30 |
| 1730 | TCGGTAGACACATTCAAACCATTTTTCCCTCATCGGCACATTAAAGCTGGCGTACGCTGCAGGTCGAC | Primer used to obtain amplicon from pBS1539 for deletion of the SIR2 gene | SIR2 |
| 817 | GAGGGACGGTTGAAAGTG | Primer used to obtain template for Southern probe prepn from genomic DNA | rDNA |
| 818 | ATACGCTTCAGAGACCCTAA | Primer used to obtain template for Southern probe prepn from genomic DNA | rDNA |
| 1161 | CAGGTTATGAAGATATGGTGCAA | Primer used to obtain template for Southern probe prepn from genomic DNA | rDNA |
| 1162 | AAAATGGCCTATCGGAATACA | Primer used to obtain template for Southern probe prepn from genomic DNA | rDNA |
| 2101 | GTAGTGCTCTGTGTGCTGCC | Primer used to obtain template for Southern probe prepn from genomic DNA | HMR |
| 2102 | GACGATTAAAAAGATGATCG | Primer used to obtain template for Southern probe prepn from genomic DNA | HMR |
| 1502 | AACAACGAAACGCCTTCATC | Primer used for PCR amplification of the klURA3 gene from pBS1539 to clone K937 | klURA3 |
| 1503 | AGGGAGCTCTACGACTCACTATAGGG | Primer used for PCR amplification of the klURA3 gene from pBS1539 to clone K937 | klURA3 |
| 710 | TGGAGCAAAGAAATCACCGC | Primer used for qPCR amplifying a region in 25S rDNA together with primer 711 | 25S |
| 711 | CCGCTGGATTATGGCTGAAC | Primer used for qPCR amplifying a region in 25S rDNA together with primer 710 | 25S |
| 920 | GCCATATCTACCAGAAAGCACC | Primer used for qPCR amplifying a region in 5S rDNA together with primer 921 | 5S |
| 921 | GATTGCAGCACCTGAGTTTCG | Primer used for qPCR amplifying a region in 5S rDNA together with primer 920 | 5S |
| 969 | TCATGGAGTACAAGTGTGAGGA | Primer used for qPCR amplifying rDNA promoter region together with primer 970 | Prom |
| 970 | TAACGAACGACAAGCCTACTC | Primer used for qPCR amplifying rDNA promoter region together with primer 969 | Prom |
| 2415 | GATCCCCGTCCAAGTTATGA | Primer used for qPCR amplifying a region at the 3′ end of the HMR-E together with primer 2416 | HMR |
| 2416 | ACCAGGAGTACCTGCGCTTA | Primer used for qPCR amplifying a region at the 3′ end of the HMR-E together with primer 2415 | HMR |
| 1348 | AGGGCTTTCACAAAGCTTCC | Primer used for qPCR amplifying enhancer element II together with primer 1349 | ENH |
| 1349 | TCCCCACTGTTCACTGTTCA | Primer used for qPCR amplifying enhancer element II together with primer 1348 | ENH |
| 712 | GAGTCCTTGTGGCTCTTGGC | Primer used for qPCR amplifying a region in 18S rDNA together with primer 713 | 18S |
| 713 | AATACTGATGCCCCCGACC | Primer used for qPCR amplifying a region in 18S rDNA together with primer 712 | 18S |
| 611 | AGGCGAAGAAAACCCACAAA | Primer used for qPCR amplifying a region in the NOC1 gene together with primer 612 | NOC1 |
| 612 | GTCGTCAGCATCCTCGTCAG | Primer used for qPCR amplifying a region in the NOC1 gene together with primer 611 | NOC1 |
klURA3, Kluyveromyces lactis URA3; qPCR, quantitative PCR; Prom, Pol I promoter.
Yeast strains y1926, y1927, y1928, and y1929 were created by crossing NOY408-1A and NOY604 with y940 and y1139, respectively. All other yeast strains expressing fusion proteins with a C-terminal micrococcal nuclease (MNase) marked by a triple hemagglutinin (HA) epitope were generated as described previously (26). For all strains used in this study, proper expression of the MNase fusion proteins was tested by Western blot analysis (data not shown). None of the MNase tags led to an obvious growth phenotype (data not shown). UAF30 and SIR2 deletion was performed essentially as described for deletion of HMO1 (26). Plasmid K937 was constructed by the insertion of a SmaI/SacI-restricted PCR fragment obtained from pBS1539 (34) using primers 1502 and 1503 into SmaI/SacI-restricted K643 (26). To establish UAF30 or SIR2 deletion strains, the URA3 gene from Kluyveromyces lactis (URA3) was amplified with primers 843 and 1509 or with primers 1728 and 1730 from plasmid K937. The amplified fragment was framed by 50 bp of DNA sequence homologous to the 5′ sequence upstream of the ATG and 50 bp of sequence homologous to the DNA downstream of the stop codon of the UAF30 or SIR2 open reading frame. The amplicon was directly used for transformation as described above. UAF30 deletion was confirmed by PCR. To obtain yeast strain y895, NOY703 was transformed with an XhoI/XbaI fragment of pNOY3166 (20). Positive clones were identified by PCR.
Yeast cultures, formaldehyde fixation, and isolation of nuclei.
In all experiments, yeast cells were cultured to exponential phase before formaldehyde fixation, and isolation of crude nuclei was performed as previously described (26). Growth medium used was yeast extract-peptone-galactose (YPG) in all experiments involving strains which rely on galactose-dependent expression of 35S rRNAs, whereas yeast extract-peptone-dextrose (YPD) was used in the remainder of the experiments. Growth temperature was 30°C, except for the experiments involving yeast strains carrying a temperature-sensitive allele of RRN3. In the latter case, cells were cultured at 24°C to exponential phase. Cells were split and either fixed with formaldehyde or further incubated in YPD at 37°C for another 120 min before formaldehyde cross-linking.
MNase digestion of yeast nuclei.
Nuclei isolated from formaldehyde cross-linked cells from 50 ml of an exponentially growing yeast culture were washed twice (1 min, 13,000 rpm, 4°C) with 500 μl of MNase buffer (15 mM Tris-HCl at pH 8, 50 mM NaCl, 1.4 mM CaCl2, 0.2 mM EDTA, 0.2 mM EGTA), suspended in 1 ml of MNase buffer, and split into five aliquots of 200 μl. The different aliquots were incubated in the absence or presence of different amounts of MNase (Sigma), as indicated in the figure legends, for 20 min at 37°C. Reactions were stopped by addition of an equal volume of IRN buffer (50 mM Tris-HCl at pH 8, 20 mM EDTA, 0.5 M NaCl), and DNA was analyzed as described for chromatin endogenous cleavage (ChEC) experiments (26).
ChEC and ChEC-psoralen analyses.
ChEC and ChEC-psoralen analyses were carried out using nuclei from formaldehyde cross-linked yeast cells as described previously (26).
Blot analysis and quantitation.
A list of probes used for blot hybridization is presented in Table 3. Southern blot analysis and quantitation were carried out as described previously (26), except that a FLA-3000 imaging system (Fujifilm) was used for data collection in some experiments. To obtain profiles of the MNase patterns after MNase digestion or ChEC, signal intensities over the indicated region of a lane were determined and divided by the respective total signal measured over the entire lane. To obtain slow-migrating band (s-band) or fast-migrating band (f-band) profiles from ChEC-psoralen analyses, signal intensities in each lane were normalized to the respective peak values and plotted against the distance of migration in the gel. Raw data were processed with the PeakFit software (Systat Software Inc.) using a Gaussian basis function (r2 values fit ≥0.98).
TABLE 3.
Description of Southern blot probes and targets
| Probe/target | Synthesis | Locus | Restriction enzyme | Fragment size (kb) |
|---|---|---|---|---|
| IGS2 | PCR from genomic DNA using primers 1161 and 1162 | rDNA | XcmI | 4.3 |
| rDNp | PCR from genomic DNA using primers 817 and 818 | rDNA | XcmI | 4.9 |
| rDNA | XcmI/SacII | 2 | ||
| GIT1 | PCR from genomic DNA using primers 2101 and 2102 | HMR | XcmI | 10.7 |
Chromatin immunoprecipitation (ChIP).
ChIP was performed mainly as described previously (14) in three independent experiments for each strain. Formaldehyde-fixed cells from 50 ml of an exponentially growing yeast culture were washed (1 min, 13,000 rpm, 4°C) with 1 ml of cold lysis buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] deoxycholate [DOC], 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM benzamidine) and suspended in 400 μl of lysis buffer. EGTA and EDTA used in the buffer suppress MNase activity. Glass beads (diameter, 0.75 to 1.0 mm; Roth) were added, and cells were disrupted on a VXR basic IKA Vibrax orbital shaker for 45 min with 2,000 to 2,200 rpm at 4°C. DNA was sonicated in a volume of 1 ml lysis buffer using a Branson Sonifier 250 to obtain an average DNA fragment size of 500 to 1,000 bp. Cell debris was removed by centrifugation (20 min, 13,000 rpm, 4°C). The chromatin extracts were split into three aliquots. A total of 40 μl of each aliquot served as an input control, and 250 μl of each aliquot was incubated for 90 min at 4°C with 1 μg of a monoclonal anti-HA antibody (3F10; Roche). Then, the lysates were incubated for 90 min at 4°C with 50 μl of protein G-Sepharose (Amersham) to enrich the MNase-HA3-tagged proteins bound by the antibody. After immunoprecipitation, the beads were washed three times with lysis buffer, twice with washing buffer I (50 mM HEPES [pH 7.5], 500 mM NaCl, 2 mM EDTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] DOC), and twice with washing buffer II (10 mM Tris-HCl [pH 8.0], 250 mM LiCl, 2 mM EDTA, 0.5% [vol/vol] Nonidet P-40, 0.5% [wt/vol] DOC), followed by a final washing step with TE buffer (10 mM Tri-HCl [pH 8.0], 1 mM EDTA). A total of 250 μl of buffer IRN (50 mM Tris-HCl [pH 8.0], 20 mM EDTA, 500 mM NaCl) was added to the immunoprecipitation (IP) beads and to the input samples. DNA was isolated as previously described for ChEC experiments (26). Both input and IP DNAs were suspended in 50 μl of TE buffer.
Relative DNA amounts present in input and IP DNAs were determined by quantitative PCR using SYBR green I dye (Roche) for DNA detection with a Rotor-Gene 3000 system (Corbett Life Science/Qiagen) and the comparative analysis software module. Primer pairs used for amplification are listed in Table 2. Input DNA was diluted 1:1,000 or 1:500, and IP DNA was diluted 1:200 or 1:20 prior to analysis. All samples were run in triplicate to ensure accuracy of the data.
RESULTS
Deletion of components of the basal Pol I transcription machinery leads to drastic changes in chromatin structure at the rDNA locus.
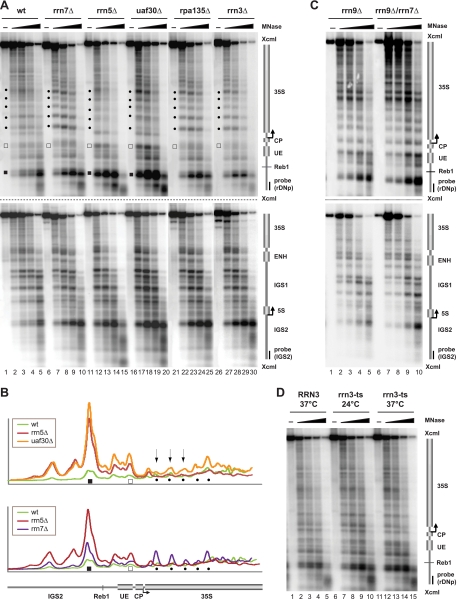
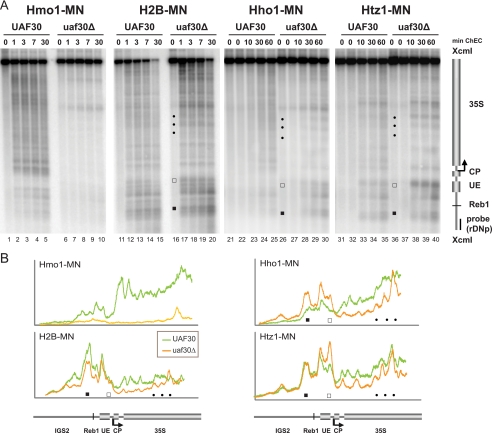
It has been suggested that the presence of UAF is required for establishment of a specific rDNA chromatin structure (9). Therefore, we examined if there are detectable changes in rDNA accessibility when UAF complex formation is impaired. We isolated nuclei from formaldehyde-cross-linked yeast cells deleted in either UAF30 (uaf30Δ) or RRN5 (rrn5Δ; PSW). Chromatin was digested with rising amounts of micrococcal nuclease (MNase). After DNA isolation and restriction enzyme digestion, we performed Southern blot analysis using the indirect end labeling technique, with a probe detecting the Pol I promoter region (Fig. 2A, top). The deletion of UAF30 and RRN5 led to substantial alterations in 35S rRNA gene accessibility for MNase compared to those of the same experiment performed with a wild-type strain (Fig. 2A, top, compare lanes 1 to 5 with lanes 11 to 20; Fig. 2B, top graph, profile analysis of lanes 3, 13, and 18 shown in Fig. 2A, top). We observed a decrease in protection against MNase digestion in the UAF deletion mutants around the promoter-proximal Reb1 binding site (Fig. 2A, top, and B, filled squares) and within the upstream element (Fig. 2A, top, and B, open squares). This suggests that UAF limits MNase access to these regions in the wild-type strain. This observation is in accordance with earlier findings studying the chromatin organization within IGS2 in the rrn5Δ strain (42).
FIG. 2.
Deletion of components of the basal Pol I transcription machinery leads to drastic changes in chromatin structure at the rDNA locus. (A) Yeast strains (NOY505, NOY408-1a, NOY558, NOY604, NOY699, and y1120) carrying a wild-type allele (wt) or deletions (Δ) in RRN3, RRN7, RRN5, RPA135, or UAF30 were cultured and treated with formaldehyde, as described in Materials and Methods. Crude nucleus preparations were incubated in the absence (−) or presence of 0.05, 0.15, 0.3, or 1 U of micrococcal nuclease (MNase), as detailed in Materials and Methods. DNA was isolated and analyzed using a Southern blot by indirect end labeling. An autoradiogram is shown. Different symbols highlight changes in MNase accessibility and are referred to in the text. A cartoon of the genomic region analyzed, including the positions and names of the probes used for indirect end labeling, is depicted on the right. (B) Profiles of DNA fragment patterns in individual samples obtained after digestion with 0.15 U of MNase as depicted in panel A. The genotype of the yeast strains analyzed is indicated in the legend of each graph. The relative signal intensity in the respective lanes was determined as described in Materials and Methods and plotted against the distance of migration in the gel. Different symbols highlight changes in MNase accessibility, as shown in panel A. A cartoon of the genomic region is indicated below the graphs. (C) Yeast strains (NOY703 and y895) carrying deletions in RRN9 and RRN7 were analyzed as described in panel A, except that 0.25, 0.5, 1, and 2 units of MNase were used for digestion. (D) Yeast strains (CG379 and Ycc95) carrying a wild-type copy or a temperature-sensitive allele (ts) of RRN3 were cultured at the permissive (24°C) and the restrictive (37°C) temperatures, as described in Materials and Methods. Chromatin was analyzed as described in panel A.
The structural changes extended into the 35S rRNA coding region (Fig. 2A, top, and B, filled dots). The observed alterations were more pronounced in the rrn5Δ strain than those in the uaf30Δ mutant. In fact, the MNase digestion pattern observed in the uaf30Δ mutant was somewhat reminiscent of the digestion pattern in the wild type (Fig. 2B, arrows). This is consistent with a heterogeneous population of Pol I- and Pol II-transcribed 35S rRNA gene chromatin structures in this strain (38). Instead, the MNase digestion patterns of rrn5Δ and rrn9Δ strains were indistinguishable (Fig. 2A, lanes 11 to 15, and C, lanes 1 to 5). This agrees with the observation that in these genetic backgrounds, 35S rRNA genes are no longer transcribed by Pol I (43).
We next analyzed if deletion of the genes coding for other PIC components, the core factor subunit Rrn7 (rrn7Δ), the essential Pol I transcription factor Rrn3 (rrn3Δ), or the Pol I subunit Rpa135 (rpa135Δ) influences rDNA chromatin structure. In these strains, transcription of chromosomal 35S rRNA genes is abolished (32), and the cells survive because the 35S rRNA is synthesized by Pol II from a multicopy plasmid under the control of the GAL7 promoter (30). These strains can be cultured only in galactose-containing media. Therefore, all the strains analyzed in the experiment presented in Fig. 2A were cultured in YPG. We also confirmed that growth in YPD does not alter rDNA accessibility for MNase in the wild-type strain or the uaf30Δ strain, since they were routinely grown in YPD in most of the subsequent experiments (data not shown).
rDNA accessibility for MNase was nearly identical in the rrn7Δ, rrn3Δ, and rpa135Δ strains but different from the accessibility for MNase in the wild-type strain or in the UAF deletion mutants (Fig. 2A, top, compare lanes 1 to 5 or lanes 11 to 20 with lanes 6 to 10 and lanes 21 to 30; Fig. 2B, bottom graph, profile analysis of lanes 3, 8, and 13 shown in Fig. 2A, top). We observed only a moderate increase in accessibility at the regions flanking the Pol I promoter-proximal Reb1 binding site and within the upstream element in the rrn7Δ, rrn3Δ, or rpa135Δ strain. Consistent with this result, the UAF complex is still bound to the promoter in these mutants (3) (see Fig. 5B). The regularly spaced rDNA fragments observed further downstream in the 35S rRNA coding sequence might indicate protection by translationally phased nucleosomes in the rrn7Δ, rrn3Δ, and rpa135Δ strains (Fig. 2A, top, and B, filled dots). Interestingly, when analyzing a mutant strain in which both the core factor gene RRN7 and the UAF gene RRN9 have been deleted, MNase produces an rDNA cleavage pattern identical to those observed in the rrn5Δ and rrn9Δ strains (compare Fig. 2A to C, lanes 11 to 15). Thus, the presence of UAF is required to establish the specific rDNA chromatin structure observed in the rrn7Δ strain (and presumably in the rrn3Δ and rpa135Δ strains). Along these lines, a PSW phenotype can be established in core factor deletion strains only upon deletion of UAF genes (32).
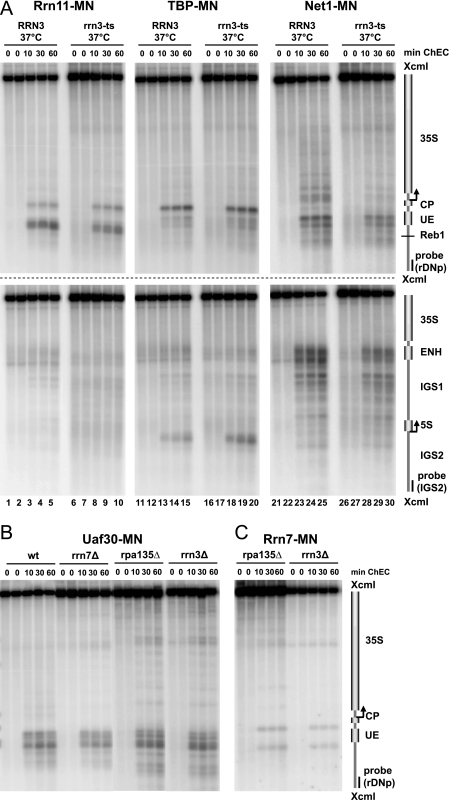
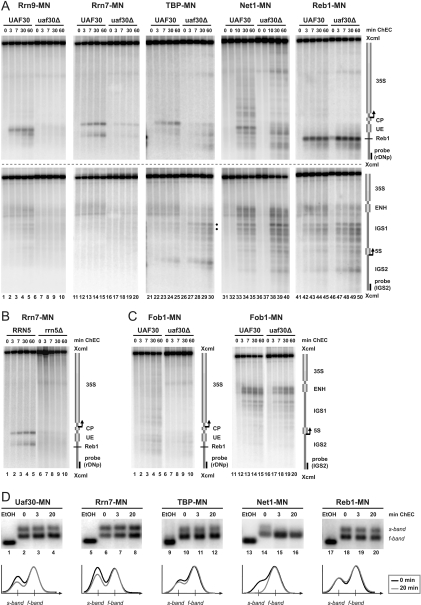
FIG. 5.
UAF and CF bind to the Pol I promoter in the absence of Pol I or Rrn3 in vivo. (A) Yeast strains (y1143, y1144, y1148, y1149, y1707, and y1712) carrying a wild-type copy or a temperature-sensitive (ts) allele of RRN3 and expressing the indicated MNase fusion proteins (MN) were treated as described in the legend to Fig. 2D. Crude nuclei were subjected to ChEC analysis as described in the legend to Fig. 3A. (B, C) Yeast strains (y618, y1109, y1926, y1927, y1928, and y1929) carrying a wild-type allele (wt) or deletions (Δ) in RRN3, RRN7, RPA135, or UAF30 and expressing the indicated MNase fusion proteins (MN) were treated and subjected to ChEC analysis as described in the legend to Fig. 3A.
Pol I transcription is either strongly impaired or abolished in the mutant strains analyzed above. Thus, we wanted to investigate whether interference with Pol I transcription at the 35S rRNA genes is sufficient to establish an alternative rDNA chromatin structure. We compared MNase digestion patterns of rDNA chromatin from a strain carrying a temperature-sensitive (TS) allele of the Pol I-specific transcription factor Rrn3 (rrn3) and the isogenic RRN3 wild-type strain (RRN3). After the shift to the restrictive temperature, Pol I can no longer (re)initiate transcription and will eventually leave the 35S rRNA gene locus (26) (see Fig. 6C). We could not observe any significant differences in rDNA cleavage patterns in chromatin isolated from the rrn3 TS strain cultured at 24°C or at the restrictive temperature (37°C) or from the RRN3 wild-type strain cultured at 37°C (Fig. 2D, compare lanes 1 to 5 with lanes 6 to 15). We conclude that the rDNA chromatin state established when 35S rRNA genes are actively transcribed by Pol I is stable even after Pol I has left the locus.
FIG. 6.
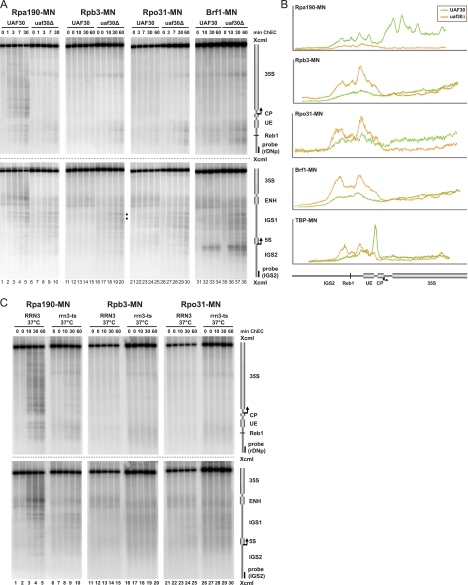
Pol II and III gain access to the Pol I promoter region in the UAF30 deletion mutant. (A) Yeast strains (y624, y1174, y1273, y1294, y1330, y1681, y1688, and y1694) carrying a wild-type allele or a deletion (Δ) in UAF30 and expressing the indicated MNase fusion proteins (MN) were treated and subjected to ChEC analysis as described in the legend to Fig. 3A. The positions of two Rpb3-MNase-mediated cuts at a bidirectional Pol II promoter within IGS1 are marked by filled dots. (B) Profiles of DNA fragment patterns in individual samples obtained after 30 min (Rpa190-MNase-expressing strains) or 60 min (other strains) of ChEC. Profile analysis was performed as described in the legend to Fig. 2B. (C) Yeast strains (y938, y945, y1560, y1566, y1708, and y1713) carrying a wild-type copy or a temperature-sensitive (ts) allele of RRN3 and expressing the indicated MNase fusion proteins (MN) were treated as described in the legend to Fig. 2D. Crude nuclei were subjected to ChEC analysis as described in the legend to Fig. 3A.
Hybridization of the above-described blots with a probe detecting the 5S rRNA gene and IGS1 region did not reveal significant changes in MNase digestion patterns for any of the strains (Fig. 2A and C, bottom, and data not shown). This has also been reported for MNase accessibility within IGS2 in the rrn7Δ and rrn5Δ strains analyzed in this study (42). Thus, the observed differences in the accessibility of rDNA chromatin in the respective genetic backgrounds are confined to the 35S rRNA gene. Taken together, this analysis demonstrates that various defined chromatin structures with characteristic differences in MNase accessibilities can be observed in rRNA genes, depending on the presence of UAF and correlating with the transcriptional state of the genes.
UAF30 deletion leads to an altered arrangement of structural chromatin components at the 35S rRNA genes.
Since less than 10% of the 35S rRNA genes are transcribed by Pol I in the uaf30Δ mutant (15) and the chromatin structure closely resembled those of a PSW strain (Fig. 2A and B), we performed most of our subsequent analyses in this genetic background. The rationale behind this decision was that the uaf30Δ mutant displayed a relatively mild growth defect (doubling time of 210 min in YPD) (data not shown) compared to growth of the PSW strains (doubling time of 7 h in YPD) (data not shown). Furthermore, the copy number of rDNA repeats was increased at most by a factor of 1.5 in all of the uaf30Δ strains analyzed in this study (see also reference 15) compared to a 2- to 4-fold increase in the repeat copy numbers obtained for the PSW strains (32) (data not shown).
We analyzed the association of several structural chromatin components with rDNA. To this end, we employed chromatin endogenous cleavage (ChEC) (35), which we have used in the past to investigate rDNA chromatin composition (26). A factor of interest is expressed from its endogenous locus as a fusion protein with a C-terminal MNase. Cells are grown in the desired conditions and then treated with formaldehyde to cross-link the respective factor to its DNA/chromatin binding site. Crude nuclei are prepared, and the MNase is activated by the addition of calcium. DNA is isolated and subjected to indirect end labeling Southern blot analysis to map cleavage events with high precision at any chromosomal location.
The ChEC patterns produced by MNase fusion proteins Hmo1, H2B, the yeast histone H1 homologue, Hho1, and the histone variant Htz1 expressed in UAF30 wild-type and uaf30Δ strains are shown in Fig. 3A. Hmo1-MNase cleavage within the 35S rRNA coding region was barely detectable in the uaf30Δ mutant (Fig. 3A, lanes 1 to 10). We observed qualitative changes in the cleavage patterns of the histone fusion proteins in agreement with the MNase accessibility pattern of chromatin from the uaf30Δ mutant presented in Fig. 2 (compare Fig. 2A, lanes 1 to 5 and lanes 16 to 20, with Fig. 3A, lanes 11 to 40; compare also the profile analyses presented in Fig. 2B and 3B). Thus, the reorganization of structural chromatin constituents in the uaf30Δ strain correlates well with the changes in MNase sensitivity of the rDNA.
FIG. 3.
UAF30 deletion alters the arrangement of structural chromatin components at the 35S rRNA genes. (A) Yeast strains (y617, y621, y954, y1145, y1171, y1172, y1690, and y1695) carrying a wild-type allele or a deletion (Δ) in UAF30 and expressing the indicated MNase fusion proteins (MN) were cultured and treated with formaldehyde as described in Materials and Methods. Crude nucleus preparations were subjected to chromatin endogenous cleavage (ChEC) in the absence or presence of calcium for the indicated times, as detailed in Materials and Methods. DNA was analyzed as described in the legend to Fig. 2A. Different symbols highlight changes in MNase accessibility, as described in the legend to Fig. 2A. (B) Profiles of DNA fragment patterns in individual samples obtained after 30 min (H2B-MN- and Hmo1-MN-expressing strains) or 60 min (other strains) of ChEC. Profile analysis was performed as described in the legend to Fig. 2B. Different symbols highlight changes in MNase accessibility as described in the legend to Fig. 2A.
UAF30 deletion leads to compositional and structural changes within Pol I promoter chromatin.
Recent data showed that Uaf30 is required for efficient association of UAF and CF with the Pol I promoter in vivo (15). We wanted to analyze structural and compositional changes occurring in Pol I promoter chromatin upon UAF30 deletion in more detail. ChEC experiments with strains expressing MNase fusion proteins of UAF subunit Rrn9, CF subunit Rrn7, Spt15 (the yeast TATA-binding protein [TBP]), the RENT subunit Net1, and Reb1 were carried out in both UAF30 wild-type and uaf30Δ strains. In these and most of the following experiments, we measured not only qualitative but also semiquantitative changes in cleavage events mediated by the respective MNase fusion proteins. The latter was achieved by analyzing very similar amounts of rDNA when comparing the UAF30 and uaf30Δ strains. This allows the direct translation of relative band intensities of MNase cleavage products into relative cleavage efficiencies.
Cleavage by UAF and CF components fused to MNase in the UAF30 wild-type strain occurred at their known binding sites at the UE and the core promoter (CP), respectively (26). Association of these factors with the promoter region was reduced (Rrn7-MN) or barely detectable (Rrn9-MN) in the uaf30Δ mutant (Fig. 4A, top, lanes 1 to 20). This is in accordance with previous chromatin immunoprecipitation (ChIP) experiments (15). Moreover, in an rrn5Δ strain expressing RRN7 as a MNase fusion protein, the association of CF with the promoter region was no longer detectable (Fig. 4B). This is consistent with the observation that in this strain, 35S rRNA is no longer synthesized by Pol I (43). TBP-MNase cut between the UE and the CP in the UAF30 wild-type strain. In the uaf30Δ strain, cleavage at the same site was diminished, but additional cuts occurred within the UE and upstream of the promoter-proximal Reb1 binding site (Fig. 4A, top, lanes 21 to 30). Consistently, these areas became more accessible for exogenously added MNase in the uaf30Δ strain (Fig. 2A, lanes 16 to 20). Net1-MNase made multiple cuts clustering around the Pol I promoter in the UAF30 wild-type strain extending into the 35S rRNA coding region (Fig. 4A, top, lanes 31 to 35). This cleavage pattern closely resembled those of other RENT components Sir2 and Cdc14 when expressed as MNase fusion proteins (see Fig. 7A, lanes 1 to 5; also data not shown). Similar to the change in the cleavage pattern for TBP-MNase in the uaf30Δ strain, we observed reduced Net1-MNase-mediated cleavage surrounding the Reb1 binding site in this genetic background (Fig. 4A, top, lanes 36 to 40). In contrast, Reb1-MNase-mediated cleavage at the Pol I promoter remained unaltered in the uaf30Δ strain (Fig. 4A, top, lanes 41 to 50).
FIG. 4.
UAF30 deletion leads to compositional and structural changes within Pol I promoter chromatin. (A to C) Yeast strains (y881, y1121, y1151, y1184, y1185, y1328, y1329, y1453, y1689, y1692, y881, y1196, y952, and y1177) carrying a wild-type allele or a deletion (Δ) in UAF30 and expressing the indicated MNase fusion proteins (MN) were treated and subjected to ChEC analysis as described in the legend to Fig. 3A. The positions of two TBP-MNase-mediated cuts at a bidirectional Pol II promoter within IGS1 are marked by filled dots. (D) Yeast strains (y618, y881, y1184, y1185, and y1453) expressing the indicated MNase fusion proteins (MN) were treated and subjected to ChEC analysis as described in the legend to Fig. 3A before psoralen cross-linking was performed. DNA was isolated, digested with XcmI and SacII, and analyzed in a Southern blot, with probe rDNp detecting a 2-kb fragment-encompassing promoter and 35S coding regions of the rRNA gene. Graphs at the bottom depict s- and f-band profiles for samples before or after 20 min of ChEC (for details, see Materials and Methods).
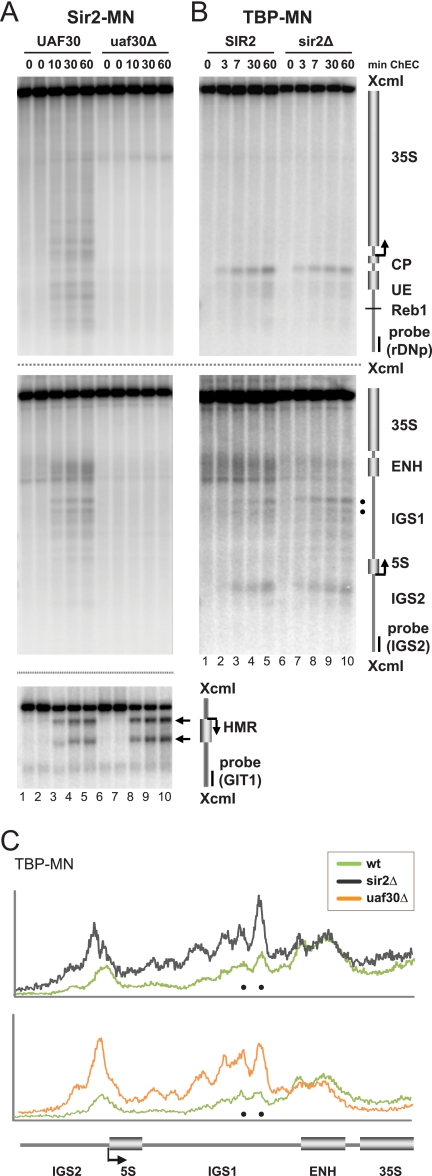
FIG. 7.
Deletion of UAF30 leads to loss of Sir2 from the rDNA locus. (A, B) Yeast strains (y1450, y1691, y1185, and y1346) carrying a wild-type allele or a deletion (Δ) in UAF30 or SIR2 and expressing the indicated MNase fusion proteins (MN) were treated and subjected to ChEC analysis as described in the legend to Fig. 3A. The positions of two TBP-MNase-mediated cuts at a bidirectional Pol II promoter within IGS1 are marked by filled dots. In panel A, bottom, the mem-brane was also hybridized with probe GIT1 detecting the HMR locus. (C) Profiles of DNA fragment patterns in individual samples obtained after 60 min of ChEC. Profile analysis was performed as described in the legend to Fig. 2B.
The same membranes were hybridized with a probe detecting the 5S rRNA gene and IGS1. As observed earlier for multiple Pol I promoter-bound factors (26), Rrn9-MNase produced weak cuts within IGS1 and the ENH region in a strain expressing a UAF30 wild-type allele (Fig. 4A, bottom, lanes 1 to 5). These cleavage events were not observed in the uaf30Δ strain. TBP-MNase made a distinct cut at the 5′ end of the 5S rRNA gene in both UAF30 wild-type and uaf30Δ strains (Fig. 4A, bottom, lanes 21 to 30). This is consistent with its function as a subunit of the Pol III transcription factor TFIIIB (5). Furthermore, we observed enhanced cleavage by TBP-MNase in the uaf30Δ strain within IGS1 (Fig. 4A, bottom, compare lanes 21 to 25 with lanes 26 to 30, filled dots). This region was previously identified as a bidirectional Pol II promoter (22). Strong cleavage by Net1-MNase and Reb1-MNase was observed in IGS1 and at the ENH region upstream of the 3′ end of the 35S rRNA gene in the UAF30 and uaf30Δ strains (Fig. 4A, bottom, lanes 31 to 50). We conclude that UAF30 deletion does not influence association of these factors with IGS1.
Net1 association with this region has been shown to depend on Fob1 (16), a protein binding to DNA elements within the ENH region, blocking the progression of replication forks toward the 3′ end of 35S rRNA genes (21). Consistent with our observations in Net1-MNase-expressing strains, cleavage of the ENH region by a Fob1-MNase fusion protein is unaltered in the uaf30Δ strain (Fig. 4C, compare lanes 11 to 15 with lanes 16 to 20). A weak interaction of Fob1 with the Pol I promoter has previously been reported (16). Accordingly, we observe Fob1-MNase-mediated cleavage in this region in the UAF30 background, with a pattern similar to that observed upon cleavage by Net1-MNase (compare Fig. 4C, lanes 1 to 5, with 4A, top, lanes 31 to 35). Upon deletion of UAF30, Fob1-MNase-mediated cleavage at the Pol I promoter was reduced, and the digestion pattern was altered. This is again in good correlation with the changes observed in Net1-MNase expressing uaf30Δ cells (compare Fig. 4C, lanes 6 to 10, with 4A, top, lanes 36 to 40). It should be noted that Reb1-MNase-mediated cleavage at its recognition site within the ENH region was weak compared to the cleavage at the promoter-proximal binding site (Fig. 4A, lanes 41 to 50). This observation agrees with recent ChIP data (17) and raises questions about a potential role of Reb1 in efficient Pol I termination in vivo (24).
Together, these data confirm that Uaf30 is required for efficient recruitment of other UAF components and CF to the Pol I promoter. In the absence of Uaf30, TBP and Net1 association with the Pol I promoter changes qualitatively and quantitatively, whereas Reb1 binding is unaltered. Additionally, UAF30 deletion increases TBP interaction with the IGS1 region.
In contrast to Rrn9, Rrn7, TBP, and Net1, association of Reb1 with rDNA was not altered by deletion of UAF30. Reb1 also behaved differently than the other factors when we examined the respective association with either transcriptionally inactive or actively transcribed 35S rRNA genes. Psoralen cross-linking can be used to distinguish between the two rDNA chromatin states (10). In these analyses, restriction fragments derived from either actively transcribed or transcriptionally inactive nucleosomal 35S rRNA genes have a different mobility, producing a slow-migrating “s-band” or a fast-migrating “f-band” in native agarose gel electrophoresis, respectively (10). When ChEC and psoralen cross-linking are combined, preferential s- or f-band degradation by the MNase fusion proteins can be measured (26). We performed ChEC-psoralen analyses with wild-type cells expressing Uaf30-, Rrn7-, TBP-, Net1-, and Reb1-MNase fusion proteins. We observed that Uaf30-, Rrn7-, TBP-, and Net1-MNase preferentially degraded the s-band derived from actively transcribed 35S rRNA genes (Fig. 4D, lanes 1 to 16, and note graphs below for quantitation of the relative signal intensities). This supports a role for Net1 in stimulating Pol I transcription at the Pol I promoter (36). In contrast, Reb1-MNase-mediated s- and f-band degradation followed similar kinetics (Fig. 4D, lanes 17 to 20, and note graph below for quantitation of the relative signal intensities). This suggests that Reb1 binds upstream of both transcriptionally inactive and actively transcribed 35S rRNA genes. It is noteworthy that from the ChEC-psoralen analyses, it cannot be excluded that some of the other factors examined also associate with f-band chromatin in addition to their preferential interaction with s-band chromatin.
UAF and CF bind to the Pol I promoter in the absence of Pol I or Rrn3 in vivo.
The observed changes in transcription factor binding in the uaf30Δ strains could be a consequence of impaired Pol I transcription. We therefore performed ChEC experiments in the RRN3 wild-type and the rrn3 TS strains expressing either the CF subunit Rrn11, TBP, or Net1 as MNase fusion proteins. Cleavage by the respective MNase fusion proteins did not reveal significant differences in transcription factor association after shutdown of Pol I transcription at 37°C in the rrn3 TS strain (Fig. 5A). ChEC experiments with the RRN3 wild-type and rrn3 TS strains expressing either UAF30 or RRN10 as MNase fusion proteins yielded similar results (data not shown). Thus, although Pol I left the 35S rRNA gene promoter at the restrictive temperature in the rrn3 TS strain (26) (Fig. 6C), UAF, TBP, and CF remained bound to the Pol I promoter. This suggests that CF does not leave the promoter together with Rrn3-Pol I in vivo, as has been previously observed in in vitro experiments (2).
We further investigated whether UAF and CF could be detected in various other strains carrying deletions in genes of the Pol I PIC. ChEC experiments were performed in an rrn7Δ strain and a corresponding wild-type strain expressing UAF30 as MNase fusion protein and in rpa135Δ and rrn3Δ strains expressing either UAF30 or RRN7 as MNase fusion proteins. We observed cleavage by Uaf30-MNase at the Pol I promoter region in the wild-type strain and in all three mutant strains (Fig. 5B). Furthermore, the typical cleavage pattern for Rrn7-MN flanking the UE was detected in the rpa135Δ and rrn3Δ strains (Fig. 5C). We conclude that UAF binding to the UE does not require CF, Pol I, or Rrn3. Moreover, UAF and CF can bind to the Pol I promoter region even in the absence of the Rrn3-Pol I complex, although the association with the 35S promoter region might be reduced at least in the rpa135Δ background (compare Fig. 4A, lanes 11 to 15, to 5C).
Pol II and III gain access to the Pol I promoter region in the UAF30 deletion mutant.
Previous data revealed that uaf30Δ strains use both Pol I and Pol II for 35S rRNA synthesis with most of the transcripts produced by Pol I and with only ∼10% of transcription carried out by Pol II (38). Interestingly, the Pol II initiation site localized upstream of the Pol I initiation site, as shown by primer extension experiments (43). Since we observed binding of TBP upstream of the Pol I promoter in the uaf30Δ strain, we investigated if we could detect association of Pol II with this region.
We performed ChEC experiments in UAF30 wild-type and uaf30Δ strains expressing MNase fusion proteins of the Pol I subunit Rpa190, the Pol II subunit Rpb3, and as a control, the Pol III subunit Rpo31. As observed earlier (26), Rpa190-MNase produced a distinct cleavage pattern spreading along the entire 35S rRNA coding region in the UAF30 wild-type strain (Fig. 6A, top and bottom, lanes 1 to 5). In contrast, in the uaf30Δ strain, Pol I association with the transcribed region was strongly reduced (Fig. 6A, lanes 6 to 10, and B, profile analysis of lanes 5 and 10 in Fig. 6A, top). This is consistent with recent ChIP data and can be explained by the fact that only about 6% of the rRNA genes are actively transcribed in this genetic background (15). This association might be at the limit of detection in ChIP and ChEC experiments. For UAF30 wild-type strains expressing Rpb3-MNase and Rpo31-MNase, we did not detect cleavage within the investigated rDNA region (Fig. 6A, top, lanes 11 to 15 and lanes 21 to 25). In uaf30Δ strains, however, both fusion proteins produced a relative increase in cuts flanking the promoter-proximal Reb1 binding site (Fig. 6A, top, lanes 16 to 20 and lanes 26 to 30).
Whereas the detection of Rpb3-MNase-mediated cleavage could be expected in the uaf30Δ strain, Rpo31-MNase-mediated cleavage came as a surprise. We therefore performed a ChEC experiment with a strain expressing a MNase fusion protein of Brf1, another component of the Pol III transcription factor TFIIIB. We observed enhanced cleavage in the uaf30Δ strain that was very similar to that obtained after ChEC with Rpb3-, Rpo31-, or TBP-MNase (Fig. 6A, top, lanes 31 to 38; Fig. 6B, profile analysis of lanes 15, 20, 25, 30, 34, and 38 in Fig. 6A, top, and of lanes 25 and 30 in Fig. 4A, top). This indicates that in the absence of UAF, Pol II and Pol III may gain access to the Pol I promoter.
We next investigated whether the binding of Pol II and Pol III at the Pol I promoter region is a consequence of impaired Pol I transcription in the uaf30Δ strain. ChEC experiments were carried out in the RRN3 and rrn3 TS strains, expressing Rpa190-, Rpb3-, or Rpo31-MNase. Expectedly, Rpa190-MNase cleavage in the rrn3 TS strain could no longer be detected within the 35S rRNA coding region at 37°C (Fig. 6C, lanes 6 to 10). The same has been observed for another Pol I subunit, Rpa43, in similar experiments (26). In addition, Rpb3- and Rpo31-MNase fusion proteins did not cleave at the Pol I promoter under this condition (Fig. 6C, lanes 16 to 20 and 26 to 30).
To complete the ChEC analyses in the Rpa190-, Rpb3-, and Rpo31-MNase-expressing strains, membranes were hybridized with a probe detecting the 5S rRNA gene and IGS1 region. As expected, we found Rpo31- and Brf1-MNase-mediated cleavage at the 5S rRNA gene in the wild-type and the uaf30Δ strains (Fig. 6A, bottom, lanes 21 to 38). Rpo31-MNase-mediated cleavage in the 5S rRNA gene was rather weak (being even stronger in IGS1), but we obtained similar results for several other Pol III subunits (data not shown). Interestingly, we detected enhanced cleavage by Rpb3-MNase in the uaf30Δ strain at the bidirectional Pol II promoter within IGS1 (Fig. 6A, bottom, lanes 11 to 20, filled dots). This correlates with increased cleavage by TBP-MNase in this genetic background (Fig. 4A, bottom, lanes 21 to 30, filled dots). This result again suggests that UAF30 deletion decreases rDNA silencing of Pol II transcription within this region.
Deletion of UAF30 leads to loss of Sir2 from the rDNA locus.
Since the Sir2 gene is required for efficient silencing of Pol II transcription at the rDNA locus (4, 7, 12, 39), we investigated the Sir2 association with rDNA in ChEC experiments in the UAF30 wild-type and uaf30Δ strains. Sir2 is part of the RENT complex, associating with both the Pol I promoter and enhancer regions of the 35S rRNA gene (16). Accordingly, we observed Sir2-MNase cleavage at the Pol I promoter and within the IGS1 and ENH region in the UAF30 wild-type strain (Fig. 7A, top and middle, lanes 1 to 5). The overall cleavage pattern closely resembled the Net1-MNase cleavage pattern (Fig. 4A). In the uaf30Δ strain, association of Sir2 with both the Pol I promoter and IGS1/ENH regions was no longer detectable (Fig. 7A, top and middle, lanes 6 to 10), whereas cleavage by Sir2-MNase at the HMR locus was not affected (Fig. 7A, bottom, lanes 1 to 10).
We speculated that the absence of Sir2 in the uaf30Δ strain might account for increased cleavage by TBP- and Rpb3-MNase within IGS1 (Fig. 4A and 6A). We therefore performed a ChEC experiment in SIR2 wild-type and sir2Δ strains, expressing TBP-MNase. Whereas TBP-MNase-mediated cleavage at the Pol I promoter was unaltered in the sir2Δ strain (Fig. 7B, top, compare lanes 1 to 5 with lanes 6 to 10), we observed enhanced cleavage within IGS1, very similar to the TBP-MNase-mediated cleavage in the uaf30Δ strain (compare the filled dots shown in Fig. 7B, bottom, lanes 6 to 10, with those shown in Fig. 4A, bottom, lanes 26 to 30; Fig. 7B, profile analysis of lanes 25 and 30 shown in Fig. 4A, bottom, and of lanes 5 and 10 in Fig. 7B, bottom).
Thus, UAF30 is required for Sir2 recruitment to the Pol I promoter and the IGS1/ENH region. Whereas the latter is likely the reason for enrichment of TBP and Pol II within the IGS1 rDNA region, the loss of Sir2 does not lead to detectable changes in chromatin composition and structure at the Pol I promoter (Fig. 7B and data not shown). In good agreement, it has been reported that SIR2 deletion is insufficient to cause 35S rRNA gene transcription by Pol II (32).
We performed ChIP experiments to confirm that Sir2 interaction with rDNA is impaired upon UAF30 deletion. For immunoprecipitation, we used a triple HA tag, which is fused to the C termini of all MNase fusion proteins. In extracts of UAF30 cells, Sir2 efficiently coprecipitated DNA fragments of the Pol I promoter and ENH regions, whereas fragments of the 18S and 5S rDNA regions were not enriched (Fig. 8A, bars in dark gray, graph on the left). In contrast, coprecipitation of the Pol I promoter and ENH regions was at background levels in extracts of uaf30Δ cells (Fig. 8A, bars in light gray, graph on the left). Consistent with the ChEC experiments, robust precipitation of a DNA fragment in close proximity to the HMR-E element was observed in extracts of UAF30 wild-type and deletion strains, whereas no enrichment was detected for a DNA fragment of the single-copy NOC1 locus (Fig. 8A, graph on the right). It should be noted that the fraction of input DNA of the HMR fragment coprecipitating with Sir2 in extracts from UAF30 cells was five times higher than the fraction of input DNA of the fragments from Pol I promoter and ENH regions (Fig. 8A, compare scales of y axes in the respective graphs). This is again in agreement with ChEC experiments, where we observed strong Sir2-MNase-mediated degradation of the full-length XcmI fragment encompassing the HMR locus (Fig. 7A, bottom) but only moderate degradation of the two different full-length XcmI fragments of the rDNA locus (Fig. 7A, top). Taken together, ChIP confirms that Sir2 is lost from the rDNA locus upon deletion of UAF30 but can still efficiently bind to the silent mating-type locus. Furthermore, it appears that only a subpopulation of the rDNA is bound by Sir2.
FIG. 8.
ChIP analysis confirms results obtained in ChEC experiments. (A to D) Yeast strains (y1184, y1185, y1328, y1329, y1450, y1453, y1689, and y1691) carrying a wild-type allele (wt) or a deletion (Δ) in UAF30 and expressing the proteins indicated in the upper left corner of each panel as fusion proteins with a C-terminal triple HA tag were subjected to ChIP experiments as described in Materials and Methods. The amounts of specific DNA fragments present in the input and retained on the beads were determined by quantitative PCR with primer pairs listed in Table 2. The bar graphs depict percentages of total input DNA retained after ChIP of the respective triple HA-tagged proteins. Error bars represent the standard deviations from three independent ChIP experiments, each of which was analyzed in triplicate quantitative PCRs. A cartoon representing the rDNA locus, depicting the locations of the different rDNA regions amplified in the quantitative PCRs, is shown at the top. prom, Pol I promoter.
We additionally performed ChIP experiments with Net1 (Fig. 8B). As for Sir2, Net1 precipitated preferentially DNA fragments of the Pol I promoter and ENH regions in extracts from UAF30 cells. (Fig. 8B, bars in dark gray). In extracts of uaf30Δ cells, we observed a reduction in coprecipitation of the Pol I promoter fragment, but not the ENH fragment (Fig. 8B, bars in light gray). The reduced precipitation of Pol I promoter fragment in extracts of uaf30Δ cells correlated with reduced chromatin endogenous cleavage by Net1-MNase in this genetic background (Fig. 4A, top, lanes 36 to 40). The smaller amount of input Pol I promoter fragment recovered after ChIP might also be in part because Net1 localizes upstream of its original binding sites in UAF30 cells (Fig. 4A, top, lanes 36 to 40) and therefore upstream of the Pol I promoter region amplified by the primer pair used for detection in quantitative PCR. In good agreement with the ChEC data, we got very similar results with regard to the precipitation of the Pol I promoter fragment in a TBP ChIP experiment (Fig. 8C). The TBP ChIP also confirmed the result obtained by ChEC that TBP interaction with the 5S rRNA gene promoter is not impaired upon UAF30 deletion (Fig. 4A, bottom, lanes 21 to 30). Finally, ChIP revealed that Reb1 binding at its Pol I promoter-proximal recognition site is unaltered in a uaf30Δ strain (Fig. 8D), a conclusion which was independently drawn from ChEC experiments (Fig. 4A, top, lanes 41 to 50).
DISCUSSION
We found drastic structural changes in a large chromatin domain, encompassing the yeast 35S rRNA gene, upon deletion of components of the RNA Pol I transcription factor UAF (Fig. 2A). Additionally, an alternative rDNA chromatin state was found in deletion mutants of CF components, a subunit of Pol I, or RRN3 (Fig. 2A). Interestingly, establishment of the latter chromatin state was also dependent on the presence of UAF (Fig. 2C). This argues that UAF organizes rDNA chromatin downstream of its binding site consistent with an earlier model of UAF function (9).
The absence or presence of UAF determines which RNA polymerase transcribes the 35S rRNA genes (43). It is thus conceivable that the observed chromatin alterations within the 35S rRNA coding region are the consequence of transcription by different RNA polymerase systems. However, we show that conditional shutdown of Pol I transcription in an rrn3 TS strain does not lead to a rapid chromatin transition (Fig. 2D and 5A). Therefore, rDNA chromatin states are at least transiently stable. This is in agreement with recent data demonstrating that while Pol I leaves the rDNA at the restrictive temperature in an rrn3 TS strain, Hmo1—a constituent of actively transcribed 35S rRNA genes—remains associated with this locus (26).
As observed for Hmo1, UAF and CF remain bound to the Pol I promoter upon inactivation of RNA Pol I transcription in the rrn3 TS strain (Fig. 5A). This argues against the release of CF from the promoter after each initiation cycle, as it has been proposed by in vitro experiments (2). Furthermore, CF is also bound to the Pol I promoter in strains carrying deletions in the genes coding for Rrn3 and Rpa135 (Fig. 5C). Our results demonstrate that partial PIC assembly by UAF and CF can occur in the absence of Pol I (or Rrn3) in vivo. An earlier study had investigated Pol I promoter PIC assembly by comparative in vivo footprinting in the same wild-type strain and in several of the deletion strains we used in our study (3). It was suggested that CF binding depends on the binding of Rrn3 and Pol I. While we were investigating the physical association of CF with the Pol I promoter, Bordi et al. inferred the absence or presence of this factor indirectly from the DNA protection pattern (3). Thus, it cannot be excluded that the assigned CF footprint was indeed a footprint of a fully assembled PIC, including Rrn3 and Pol I. However, since Pol I promoter cleavage by Rrn7-MNase was rather weak (especially in the rpa135Δ strain), it could be that partial PIC formation in the deletion strains is inefficient and therefore escapes detection in in vivo footprinting analyses. Despite this discrepancy, high-resolution analyses of Pol I promoter elements performed earlier showed identical DNA protection patterns in CF, RRN3, and Pol I deletion strains (3). These results accord well with our low-resolution MNase cleavage patterns, being indistinguishable in the three genetic backgrounds (Fig. 2A). Finally, it was concluded that UAF binds the UE in the absence of CF, Rrn3, or Pol I (3), which is consistent with our observations (Fig. 5B). In summary, our results support a model of Keys and coworkers, with UAF recruiting CF and TBP to the Pol I promoter, which serves as a platform for transcription initiation by the Rrn3-Pol I complex in vivo (19).
Pol II initiates 35S rRNA transcription from a cryptic promoter upstream of the Pol I transcription start site in UAF deletion strains (43). Accordingly, we find TBP translocation from its binding site within the Pol I PIC to regions upstream, flanking a promoter-proximal Reb1 recognition sequence in uaf30Δ strains (Fig. 4A). We observe Pol II binding at the same positions (Fig. 6A). In addition, Reb1 binding to the promoter-proximal recognition sequence is strong and unaltered in the uaf30Δ strains (Fig. 4A). It has been demonstrated that Reb1 binding establishes nucleosome-free flanking regions, which are available for interaction with DNA-binding factors (8). It is thus plausible that UAF prevents establishment of these nucleosome-free regions in a wild-type strain, thereby restricting the access of Pol II to the Pol I promoter. Interestingly, Pol III and the Pol III transcription factor Brf1 also bind to these sites in uaf30Δ strains (Fig. 6A and data not shown). Since Pol III terminates at thymidine (T) runs with more than 5 consecutive T residues in the coding strand (1), we consider it unlikely that the enzyme is capable of producing a full-length 35S rRNA containing multiple potential termination signals. Nevertheless, Pol III could be responsible for the synthesis of residual (perhaps prematurely terminated) 35S rRNA transcripts still detected in PSW strains upon conditional inactivation of Pol II (43).
We find that Sir2 association with rDNA is abolished in the uaf30Δ strain and a PSW strain lacking RRN5 (Fig. 7A and 8A and data not shown). The loss of the Sir2-rDNA interaction in UAF deletion strains might be responsible for impaired Pol II silencing and for histone hyperacetylation at two Pol II promoters driving ncRNA synthesis within IGS1 and -2 (7, 9). Importantly, Sir2 levels are not limiting in UAF deletion strains, because the protein still binds to other genomic target sites (Fig. 7A and 8A and data not shown). Furthermore, SIR2 overexpression cannot rescue the Pol II silencing defect in the rDNA of PSW strains (9). Thus, it is possible that UAF is directly involved in Sir2 recruitment to rDNA.
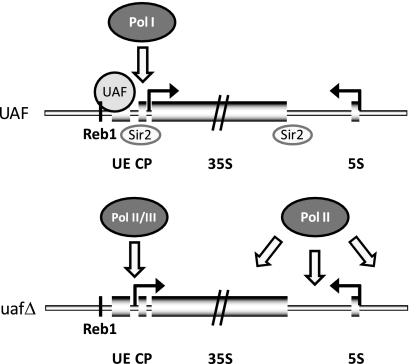
Taken together, this work is a detailed analysis of changes in rDNA chromatin structure occurring upon deletion of components of the Pol I transcription machinery. We present evidence showing how UAF organizes chromatin over a large chromosomal domain by controlling access of different RNA polymerases to the Pol I promoter and determining Pol II silencing in rDNA (Fig. 9). Our results provide a solid molecular basis for earlier observations (9) suggesting that a single factor like UAF can be a mediator between transcriptional activity and the chromatin structure of a genomic locus.
FIG. 9.
UAF organizes the rDNA chromatin structure. In a wild-type strain, UAF acts locally at the Pol I promoter by stimulating Pol I transcription and restricting access of other polymerases to a cryptic promoter located upstream of the Pol I initiation site. Protection of the cryptic promoter is lost upon deletion of UAF components (uafΔ), and Pol II and III gain access to this region. UAF is further required for efficient recruitment of Sir2 to rDNA and therefore for silencing of Pol II transcription. Thus, the specific transcriptional state of the 35S rRNA genes may determine the chromatin structure at the entire transcribed domain.
Acknowledgments
We thank Masayasu Nomura and colleagues for providing plasmids and strains and for their inspiring work on rDNA chromatin. We are grateful to Philipp Milkereit and Herbert Tschochner for constant discussions and helpful comments on the manuscript.
This work was supported by a grant in the context of the DFG research unit FOR 1068 (to J.G.). H.G. and M.W. received fellowships from the Studienstiftung des Deutschen Volkes and the Elitenetzwerk Bayern, respectively.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Allison, D. S., and B. D. Hall. 1985. Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 4:2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aprikian, P., B. Moorefield, and R. H. Reeder. 2001. New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol. 21:4847-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordi, L., F. Cioci, and G. Camilloni. 2001. In vivo binding and hierarchy of assembly of the yeast RNA polymerase I transcription factors. Mol. Biol. Cell 12:753-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255-269. [DOI] [PubMed] [Google Scholar]
- 5.Buratowski, S., and H. Zhou. 1992. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71:221-230. [DOI] [PubMed] [Google Scholar]
- 6.Cadwell, C., H. J. Yoon, Y. Zebarjadian, and J. Carbon. 1997. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 17:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesarini, E., F. R. Mariotti, F. Cioci, and G. Camilloni. 2010. RNA polymerase I transcription silences noncoding RNAs at the ribosomal DNA locus in Saccharomyces cerevisiae. Eukaryot. Cell 9:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chasman, D. I., N. F. Lue, A. R. Buchman, J. W. LaPointe, Y. Lorch, and R. D. Kornberg. 1990. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev. 4:503-514. [DOI] [PubMed] [Google Scholar]
- 9.Cioci, F., L. Vu, K. Eliason, M. Oakes, I. N. Siddiqi, and M. Nomura. 2003. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell 12:135-145. [DOI] [PubMed] [Google Scholar]
- 10.Conconi, A., R. M. Widmer, T. Koller, and J. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57:753-761. [DOI] [PubMed] [Google Scholar]
- 11.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritze, C. E., K. Verschueren, R. Strich, and R. Easton Esposito. 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16:6495-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hager, G. L., J. G. McNally, and T. Misteli. 2009. Transcription dynamics. Mol. Cell 35:741-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecht, A., and M. Grunstein. 1999. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 304:399-414. [DOI] [PubMed] [Google Scholar]
- 15.Hontz, R. D., S. L. French, M. L. Oakes, P. Tongaonkar, M. Nomura, A. L. Beyer, and J. S. Smith. 2008. Transcription of multiple yeast ribosomal DNA genes requires targeting of UAF to the promoter by Uaf30. Mol. Cell. Biol. 28:6709-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, J., and D. Moazed. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17:2162-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawauchi, J., H. Mischo, P. Braglia, A. Rondon, and N. J. Proudfoot. 2008. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 22:1082-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keener, J., J. A. Dodd, D. Lalo, and M. Nomura. 1997. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 94:13458-13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keys, D. A., B. S. Lee, J. A. Dodd, T. T. Nguyen, L. Vu, E. Fantino, L. M. Burson, Y. Nogi, and M. Nomura. 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10:887-903. [DOI] [PubMed] [Google Scholar]
- 20.Keys, D. A., L. Vu, J. S. Steffan, J. A. Dodd, R. T. Yamamoto, Y. Nogi, and M. Nomura. 1994. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 8:2349-2362. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, T., and T. Horiuchi. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1:465-474. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, T., and A. R. D. Ganley. 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309:1581-1584. [DOI] [PubMed] [Google Scholar]
- 23.Kulkens T., C. A. van der Sande, A. F. Dekker, H. van Heerikhuizen, and R. J. Planta. 1992. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 11:4665-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang, W. H., B. E. Morrow, Q. Ju, J. R. Warner, and R. H. Reeder. 1994. A model for transcription termination by RNA polymerase I. Cell 79:527-534. [DOI] [PubMed] [Google Scholar]
- 25.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128:707-719. [DOI] [PubMed] [Google Scholar]
- 26.Merz, K., M. Hondele, H. Goetze, K. Gmelch, U. Stoeckl, and J. Griesenbeck. 2008. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 22:1190-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrow, B. E., S. P. Johnson, and J. R. Warner. 1989. Proteins that bind to the yeast rDNA enhancer. J. Biol. Chem. 264:9061-9068. [PubMed] [Google Scholar]
- 28.Nogi, Y., L. Vu, and M. Nomura. 1991. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 88:7026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nogi, Y., R. Yano, J. Dodd, C. Carles, and M. Nomura. 1993. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol. Cell. Biol. 13:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nogi, Y., R. Yano, and M. Nomura. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 88:3962-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nomura, M. 2001. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 66:555-565. [DOI] [PubMed] [Google Scholar]
- 32.Oakes, M., I. Siddiqi, L. Vu, J. Aris, and M. Nomura. 1999. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol. 19:8559-8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planta, R. J. 1997. Regulation of ribosome synthesis in yeast. Yeast 13:1505-1518. [DOI] [PubMed] [Google Scholar]
- 34.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Séraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 35.Schmid, M., T. Durussel, and U. K. Laemmli. 2004. ChIC and ChEC; genomic mapping of chromatin proteins. Mol. Cell 16:147-157. [DOI] [PubMed] [Google Scholar]
- 36.Shou, W., K. M. Sakamoto, J. Keener, K. W. Morimoto, E. E. Traverso, R. Azzam, G. J. Hoppe, R. M. Feldman, J. DeModena, D. Moazed, H. Charbonneau, M. Nomura, and R. J. Deshaies. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8:45-55. [DOI] [PubMed] [Google Scholar]
- 37.Reference deleted.
- 38.Siddiqi, I. N., J. A. Dodd, L. Vu, K. Eliason, M. L. Oakes, J. Keener, R. Moore, M. K. Young, and M. Nomura. 2001. Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J. 20:4512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 40.Steffan, J. S., D. A. Keys, J. A. Dodd, and M. Nomura. 1996. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 10:2551-2563. [DOI] [PubMed] [Google Scholar]
- 41.Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies, A. D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245-256. [DOI] [PubMed] [Google Scholar]
- 42.Vogelauer, M., F. Cioci, and G. Camilloni. 1998. DNA protein-interactions at the Saccharomyces cerevisiae 35 S rRNA promoter and in its surrounding region. J. Mol. Biol. 275:197-209. [DOI] [PubMed] [Google Scholar]
- 43.Vu, L., I. Siddiqi, B. S. Lee, C. A. Josaitis, and M. Nomura. 1999. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 96:4390-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto, R. T., Y. Nogi, J. A. Dodd, and M. Nomura. 1996. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 15:3964-3973. [PMC free article] [PubMed] [Google Scholar]