Abstract
Cells possess mechanisms that permit survival and recovery from stress, several of which regulate the phosphorylation of eukaryotic translation initiation factor 2α (eIF2α). We identified the human OGFOD1 protein as a novel stress granule component that regulates the phosphorylation of eIF2α and the resumption of translation in cells recovering from arsenite-induced stress. Coimmunoprecipitation studies revealed that OGFOD1 associates with a small subset of stress granule proteins (G3BP1, USP10, Caprin1, and YB-1) and the ribosome in both unstressed and stressed cells. Overexpression of OGFOD1 led to increased abundance of phosphorylated eIF2α, both in unstressed cells and in cells exposed to arsenite-induced stress, and to accelerated apoptosis during stress. Conversely, knockdown of OGFOD1 resulted in smaller amounts of phosphorylated eIF2α and a faster accumulation of polyribosomes in cells recovering from stress. Finally, OGFOD1 interacted with both eIF2α and the eIF2α kinase heme-regulated inhibitor (HRI), which was identified as a novel stress granule resident. These findings argue that OGFOD1 plays important proapoptotic roles in the regulation of translation and HRI-mediated phosphorylation of eIF2α in cells subjected to arsenite-induced stress.
Cells respond to stress by downregulation of global cellular translation, which is often accomplished by the phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) (16).
Phosphorylation of eIF2α and stalled protein synthesis lead to the formation of discrete cytoplasmic foci known as stress granules (14), which contain translationally repressed mRNA, translation initiation factors, such as eIF2α, and 40S ribosomal subunits. Several functions have been ascribed to stress granules, including the sorting of stalled translation complexes for mRNA decay, mRNA storage, and the return to translation (2). A variety of stress granule residents are thought to aid in these tasks. For example, Y-box 1 (YB-1), a DNA- and RNA-binding protein, is important for the regulation of transcription, translation, and DNA repair (7, 25); T-cell intracellular antigen 1 (TIA1) is an RNA-binding protein that shuttles between the nucleus, where it regulates splice site selection, and the cytoplasm, where it inhibits translation (8); and the canonical stress granule protein G3BP1 (Ras-GTPase-activating protein SH3-domain-binding protein 1) has been shown to function as a DNA and RNA helicase and also as a phosphorylation-dependent endoribonuclease (5, 27).
Of the stress granule proteins identified, eIF2α is among the best characterized. The translation initiation factor eIF2 is a three-subunit complex (with α, β, and γ subunits) that is a key regulatory target for cells undergoing stress. Most typically, negative regulation of eIF2 occurs when phosphorylation of the α subunit prevents GDP-GTP exchange on the γ subunit (11). In complex with phosphorylated eIF2α, eIF2γ remains bound to GDP and unable to bind initiator tRNA. Four different eIF2α kinases and two phosphatase complexes ensure that the appropriate amount of eIF2α phosphorylation is maintained in the cell (21). Inhibition of the eIF2α kinases has been shown to result in decreased survival of cells under stress, and inhibition of the phosphatases has been shown to have a cytoprotective effect (4, 10, 13, 17). In addition, misregulation of eIF2α phosphorylation has also been shown to result in increased cell death (20, 26). Thus, it appears that phosphorylation of eIF2α and inhibition of translation initiation are critical events in the survival of stress.
In our ongoing study to understand the regulation of mRNA expression by microRNAs in normal and stressed cells, we paid attention to recent studies that introduced two homologous yeast proteins, Schizosaccharomyces pombe Ofd1 [2-oxoglutarate and Fe(II) dioxygenase 1] and Saccharomyces cerevisiae Tpa1p (termination and polyadenylation 1), as novel regulators of gene expression. Interestingly, while Ofd1 is a transcriptional regulator (12), Tpa1p is required for efficient translation termination, maintenance of mRNA poly(A) tail length, and inhibition of mRNA decay (15). Because Tpa1p displays functions that are reminiscent of those of microRNAs in mammalian cells, we investigated the role of the human homolog of Ofd1 and Tpa1p, OGFOD1 [2-oxoglutarate and Fe(II)-dependent oxygenase domain containing 1], in the posttranscriptional regulation of gene expression.
We have identified human OGFOD1 as a novel stress granule component that interacts with a specific subset of stress granule proteins, the ribosome, the eIF2α kinase heme-regulated inhibitor (HRI), and eIF2α itself. Modulation of OGFOD1 abundance influenced the levels of phosphorylated eIF2α in unstressed cells and in cells recovering from arsenite-induced stress, resulting in altered translational recovery and cellular survival. Therefore, OGFOD1 is a novel stress granule protein which functions to link recovery from stress with translation regulation.
MATERIALS AND METHODS
Cell culture.
HeLa cells were maintained in Dulbecco modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 110 mg/liter sodium pyruvate, and 2 mM l-glutamine. For in vivo protease inhibition, cells were treated for 6 h at 37°C with EDTA-free Complete protease inhibitor (Roche) dissolved in growth medium.
Immunofluorescence staining.
HeLa cells were seeded in 8-well Lab-Tek II chamber slides 1 to 2 days before fixation to reach a confluence of 60 to 70%. Before fixation, stress granules were induced by adding 1 mM sodium arsenite (Riedel-deHaën) for 30 min at 37°C or by adding 10 μM thapsigargin (Santa Cruz Biotechnology) for 1 h at 37°C. Cells were fixed with 4% paraformaldehyde (PFA)-phosphate-buffered saline (PBS) for 20 min, washed three times with PBS, permeabilized for 10 min with 0.5% Triton X-100, washed three times with PBS, blocked with 1% gelatin from coldwater fish skin (G7765; Sigma) in PBS for 30 min, incubated for 1 h with primary antibody in 1% gelatin-PBS, washed twice in PBS, and incubated for 1 h with secondary antibodies in 1% gelatin-PBS. After 2 washes with PBS, nuclei were stained with Hoechst 33258 and slides were mounted with Fluoromount G (Southern Biotech). Antibodies are described in Table S1 in the supplemental material. Confocal pictures were taken with a Zeiss LSM 510 microscope.
Protein extraction and Western blot analysis.
Total protein was prepared using RIPA lysis buffer (150 mM NaCl, 1 mM EDTA, 100 mM Tris-HCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing EDTA-free Complete protease inhibitor cocktail (Roche). Cells were washed in cold PBS, harvested in cold PBS, resuspended in RIPA lysis buffer, and incubated on ice. Lysates were cleared by centrifugation prior to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transfer to Immobilon-P membranes (Millipore).
Standard enhanced chemiluminescence (ECL; GE) Western blotting techniques were used. Antibodies are described in Table S1 in the supplemental material. Unless noted otherwise, Western blot primary antibody incubations were carried out overnight at 4°C. All Western blot secondary antibody incubations were carried out for 1 h at room temperature. PBS containing 5% milk and 0.1% Tween 20 was used for blocking as well as for primary and secondary antibody incubation. Wash steps were performed in PBS containing 0.1% Tween 20. Tris-buffered saline (TBS) containing 0.1% Tween 20 was used in assays detecting cleaved poly(ADP-ribose) polymerase (PARP), cleaved caspase-9, or phosphorylated eIF2α.
Northern blot analysis.
Total RNA was prepared using the TRIzol reagent (Invitrogen). RNA was resuspended in loading buffer (50% formamide, 6.7% formaldehyde, 1% MOPS-EDTA-sodium acetate [MESA] buffer), denatured at 65°C for 15 min, and separated in a 6.7% formaldehyde-1.25% agarose gel. Gels were run in MESA buffer (Sigma), with constant recirculation of the buffer. The RNA was transferred and UV cross-linked to a Zeta probe membrane (Bio-Rad).
OGFOD1 and actin Northern blots were performed using the ExpressHyb hybridization reagent (Clontech). OGFOD1 and actin probes were generated using the RadPrime DNA labeling system (Invitrogen) and [α-32P]dATP. Histone H2BC Northern blots were performed using an oligonucleotide probe as previously described (28). The histone H2BC probe had the sequence 5′-CACCTTGTACACGTACACAG.
Immunoprecipitations.
HeLa cell lysates were prepared in Net2 buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 0.05% NP-40 [Roche]) containing EDTA-free Complete protease inhibitor cocktail (Roche) and 10 mM ribonucleoside-vanadyl complexes (New England Biolabs) by sonication three times for 10 s each time, with 30-s intervals on ice (output 1, 50% duty cycle), using an S-250A sonifier (Branson Ultrasonics). Cleared sonicates were incubated with antibody-bound beads for 1 h at 4°C.
Immunoprecipitations followed by RNase A (Ambion/Applied Biosystems) treatment were performed as described above, except that ribonucleoside-vanadyl complexes were omitted from the sonication buffer. Briefly, following immunoprecipitation, beads were washed five times in Net2 buffer, incubated in 0.1 μg/μl RNase A for 15 min at room temperature, and washed two times in Net2 buffer. Untreated controls were performed in the presence of 10 mM ribonucleoside-vanadyl complexes.
Antibodies were prebound to 3.2 mg of protein A-Sepharose CL-4B (GE Lifesciences) in Net2 buffer overnight at 4°C. Antibodies are described in Table S1 in the supplemental material. Immunoprecipitating and coimmunoprecipitating proteins were detected by Western blot analysis.
OGFOD1 and G3BP1 overexpression.
The full-length human OGFOD1 cDNA (Image clone 5518545) was obtained from Invitrogen, amplified by PCR, and subcloned into pcDNA3, creating pcDNA3-OGFOD1. The plasmid pXL5-G3BP1, which carries the full-length G3BP1 cDNA, was obtained from OriGene (sc116551).
pcDNA3-OGFOD1 (2 μg) or pXL5-G3BP1 (1 μg) was transiently transfected into HeLa cells by use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions. Cells were harvested for Western blot analysis or prepared for staining at 72 h posttransfection.
OGFOD1, G3BP1, and USP10 RNA interference (RNAi).
Small interfering RNA (siRNA) duplexes (100 nM) were transiently transfected into HeLa cells by use of DharmaFECT 1 (Dharmacon/Thermo Scientific) according to the manufacturer's directions. Cells were harvested for Western blot analysis or polysome analysis or prepared for staining at 72 h posttransfection. Cells were harvested for Northern blot analysis at 48 h posttransfection.
Negative-control siRNA#2 was obtained from Dharmacon/Thermo Scientific and had the following sequences: NC#2 sense strand, 5′-GUAUCUCUUCAUAGCCUUAUU; and NC#2 antisense strand, 5′-UAAGGCUAUGAAGAGAUACUU. Negative-control siRNA#3 was also obtained from Dharmacon/Thermo Scientific and had the following sequences: NC#3 sense strand, 5′-AUGUAUUGGCCUGUAUUAGUU; and NC#3 antisense strand, 5′-CUAAUACAGGCCAAUACAUUU. The OGFOD1 siRNA duplex corresponded to siRNA#3 of the Dharmacon Smartpool and had the following sequences: OGFOD1 siRNA sense strand, 5′-CCACUGAUAUCACUGAAGAUU; and OGFOD1 siRNA antisense strand, 5′P-UCUUCAGUGAUAUCAGUGGUU. Other siRNA sequences were as follows: G3BP1 siRNA sense strand, 5′-CAGGAAGACUUGAGGACAUUU; G3BP1 siRNA antisense strand, 5′-AUGUCCUCAAGUCUUCCUGUU; USP10 siRNA sense strand, 5′-GCCUUUGAGCCCACAUAUAUUUU; and USP10 siRNA antisense strand, 5′-AAUAUAUGUGGGCUCAAAGGCUU.
Induction of stress and recovery from stress.
For induction of stress, cells were treated with 1 mM sodium arsenite for 30 min at 37°C. In recovery experiments, cells were first treated with arsenite as described above and then washed two times with normal growth medium, covered with normal growth medium, and incubated for 3 h at 37°C.
Polysome analysis.
Cells were covered with fresh medium 16 to 20 h prior to harvest. Following stress induction and recovery (as described above), cells were treated with 100 μg/ml cycloheximide for 3 min at 37°C. Cells were then washed two times on ice with ice-cold PBS containing 100 μg/ml cycloheximide. Each 10-cm dish of cells was harvested in 400 μl lysis buffer (300 mM NaCl, 15 mM Tris-HCl, pH 7.5, 15 mM MgCl2, 100 μg/ml cycloheximide, 1 mg/ml heparin) and incubated on ice for 15 min. Lysates were cleared at 8,400 × g at 4°C for 5 min. Cleared lysates were layered onto 10 to 50% sucrose gradients (300 mM NaCl, 15 mM Tris-HCl, pH 7.5, 15 mM MgCl2, 100 μg/ml cycloheximide, 1 mg/ml heparin) with a 60% sucrose cushion. Gradients were spun in an SW41 rotor at 35,000 rpm at 4°C for 2 h 45 min and then fractionated with an Isco Retriever II/UA-6 detector system.
RESULTS
Human OGFOD1 protein is a novel stress granule component.
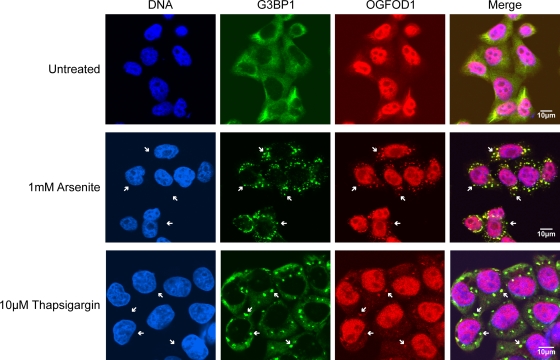
As an initial approach to examine whether human OGFOD1 has roles in translation or turnover of mRNAs in unstressed or stressed cells, we determined the subcellular localization of endogenous OGFOD1 in human HeLa cells by indirect immunofluorescence microscopy. As shown in Fig. 1, OGFOD1 is predominantly nuclear, with diffuse cytoplasmic staining, in unstressed cells. This is in contrast to the stress granule marker G3BP1, which is primarily cytoplasmic in unstressed cells (Fig. 1). After subjection of HeLa cells to arsenite-induced stress or thapsigargin-induced stress, each of which is known to induce the formation of cytoplasmic stress granules, a portion of OGFOD1 redistributed to discrete cytoplasmic foci that colocalized with G3BP1 (Fig. 1) and the stress granule marker TIA1 (see Fig. S1A in the supplemental material). These results indicate that OGFOD1 is a novel stress granule resident.
FIG. 1.
OGFOD1 colocalizes with G3BP1 in arsenite-induced and thapsigargin-induced stress granules. Endogenous OGFOD1 and G3BP1 were detected by indirect immunofluorescence in untreated HeLa cells, cells treated with 1 mM arsenite for 30 min at 37°C, or cells treated with 10 μM thapsigargin for 1 h at 37°C. An overlap of OGFOD1 and G3BP1 appears yellow in the merged image. White arrows point to stress granules.
OGFOD1 interacts with a subset of stress granule proteins in stressed and unstressed cells.
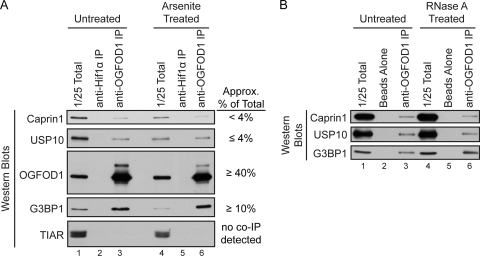
Next, we examined whether OGFOD1 interacts with other stress granule resident proteins. Because indirect immunofluorescence revealed that arsenite treatment induced the formation of stress granules in HeLa cells more robustly than thapsigargin treatment did, we chose to further investigate OGFOD1 during arsenite-induced stress. Coimmunoprecipitation analysis revealed that OGFOD1 interacted with a specific subset of stress granule residents, including G3BP1, USP10, Caprin1, and YB-1, in both unstressed and arsenite-stressed cells, in an RNA-independent manner (Fig. 2 and 3B). OGFOD1 did not coimmunoprecipitate with TIAR (Fig. 2A) and TIA1 (data not shown), which are abundant stress granule residents. Interestingly, a significant amount of cellular G3BP1 coimmunoprecipitated with OGFOD1, in contrast to Caprin1 and USP10 (Fig. 2A). USP10 and Caprin1 have previously been shown to bind directly to G3BP1; thus, it is possible that coimmunoprecipitation of USP10 and Caprin1 with OGFOD1 is mediated by the strong interaction of G3BP1 with OGFOD1.
FIG. 2.
OGFOD1 interacts with specific stress granule proteins in HeLa cells. (A) OGFOD1 coimmunoprecipitates specific stress granule proteins. Extracts from untreated or arsenite-treated HeLa cells were subjected to immunoprecipitation (IP) with an antibody against OGFOD1. Proteins were separated by SDS-PAGE and detected by Western blot analysis using the indicated antibodies. Immunoprecipitation of Hif1α served as a control for nonspecific interactions. (B) OGFOD1 coimmunoprecipitation of G3BP1, USP10, and Caprin1 is not RNase A sensitive. Extracts from untreated HeLa cells were subjected to immunoprecipitation with an antibody against OGFOD1 and then incubated with RNase A (RNase A treated). Proteins were separated by SDS-PAGE and detected by Western blot analysis using the indicated antibodies. Beads without added antibody served as a control for nonspecific interactions.
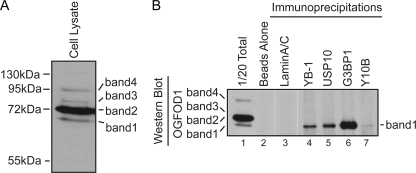
FIG. 3.
OGFOD1 band 1 interacts with stress granule proteins and the ribosome. (A) Detection of multiple forms of OGFOD1 in HeLa lysates by Western blot analysis. (B) OGFOD1 band 1 coimmunoprecipitates with stress granule proteins and the ribosome. Immunoprecipitations were performed on HeLa cell lysates, using antibodies against YB-1, USP10, G3BP1, and the large ribosomal subunit 5.8S rRNA (Y10B). Beads without added antibody and a lamin A/C immunoprecipitation were used as controls for nonspecific interactions.
OGFOD1 protein exists as multiple forms.
Western blot analyses (Fig. 2A and 3) indicated that the antibody directed against OGFOD1 detected four bands ranging from 68 kDa to 95 kDa, all larger than its predicted molecular mass of 63 kDa. Northern blot analysis revealed minor (2.3 kb) and major (3 kb) OGFOD1 mRNA species (see Fig. S2A in the supplemental material), which are both predicted to express open reading frames encoding 63-kDa proteins. The abundances of both mRNA species were dramatically reduced after treatment with siRNAs directed against OGFOD1, suggesting that they are both OGFOD1 mRNAs.
The abundances of all four protein bands decreased after siRNA treatment (see Fig. S2B in the supplemental material), arguing that bands 1 to 4 represent forms of OGFOD1. In addition, overexpression of the open reading frame of OGFOD1 in HeLa cells resulted in proportionally increased abundances of OGFOD1 bands 1 to 4 (see Fig. S2C in the supplemental material). Together, these results argue that bands 1 to 4 correspond to OGFOD1 forms that harbor various posttranslational modifications or, less likely, represent isoforms translated from minor mRNA splice variants.
To determine if OGFOD1 forms are modified by phosphorylation, we treated total protein extracts with calf intestinal phosphatase and analyzed OGFOD1 by Western blotting. Phosphatase treatment resulted in slightly reduced levels of OGFOD1 band 1 but did not result in the loss or increase of any other OGFOD1 band (data not shown). These findings suggest that migrations of the OGFOD1 bands are not substantially affected by posttranslational phosphorylation.
OGFOD1 band 1 interacts with stress granule proteins.
Next, we determined which of the OGFOD1 forms interacted with the stress granule proteins identified in Fig. 2. Immunoprecipitation of endogenous YB-1, USP10, and G3BP1 from unstressed HeLa cells followed by Western blot analysis using an OGFOD1-specific antibody revealed that the fastest-migrating species, OGFOD1 band 1, associated with all three stress granule residents but not with a control protein, nuclear resident lamin A/C (Fig. 3B, lanes 4 to 6). The Y10B antibody, which immunoprecipitates the large ribosomal subunit, also coimmunoprecipitated OGFOD1 band 1, although with a much lower efficiency than that with the stress granule proteins (Fig. 3B, lane 7). These results revealed that OGFOD1 band 1 interacts with specific stress granule residents and the ribosome in unstressed cells. Similar results were obtained in arsenite-stressed cells (data not shown).
Abundance of OGFOD1 band 1 is regulated by G3BP1.
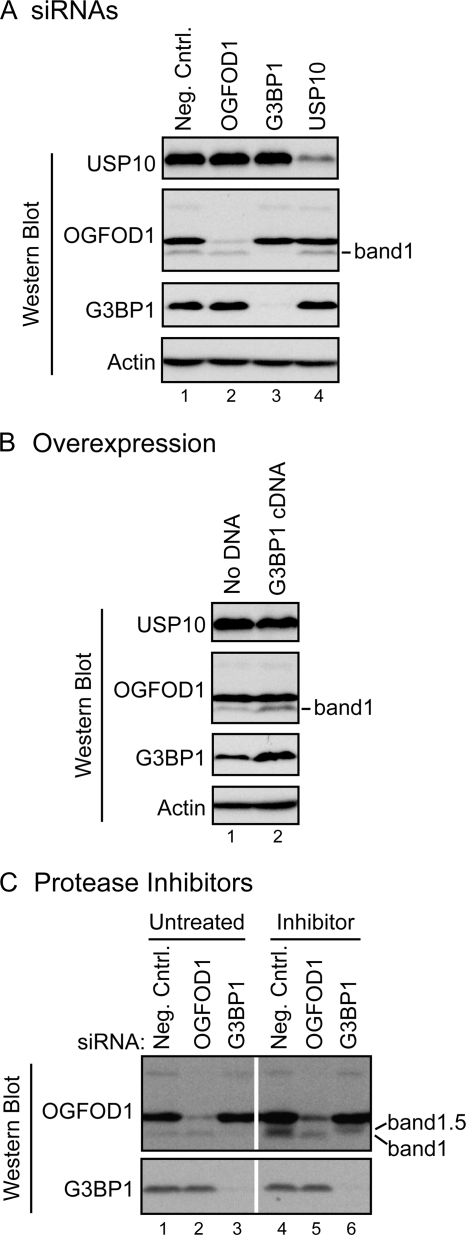
The interaction of OGFOD1 band 1 with distinct stress granule proteins was further characterized by siRNA-mediated gene knockdown of stress granule proteins. Figure 4A shows that G3BP1 siRNAs led to a dramatic reduction of G3BP1 protein abundance compared to the negative-control siRNAs. Surprisingly, knockdown of G3BP1 also resulted in selective loss of OGFOD1 band 1 (Fig. 4A, lane 3). In contrast, knockdown of OGFOD1 by RNAi did not diminish the abundance of either G3BP1 or USP10 (Fig. 4A, lane 2). Similarly, siRNA-mediated knockdown of USP10 did not result in altered abundances of OGFOD1 bands 1 to 4 or of G3BP1 (Fig. 4A, lane 4).
FIG. 4.
G3BP1 regulates OGFOD1 band 1 levels. (A) Depletion of G3BP1 by RNAi results in loss of OGFOD1 band 1. HeLa cells were treated with a negative-control siRNA (neg. cntrl.) and siRNAs against OGFOD1, G3BP1, or USP10 for 72 h prior to preparation of extracts. Endogenous proteins were detected by Western blot analysis. (B) Overexpression of G3BP1 leads to a larger amount of OGFOD1 band 1. HeLa cells were transiently transfected with a plasmid expressing a G3BP1 cDNA. Whole-cell protein extracts were prepared at 72 h posttransfection, separated by SDS-PAGE, and analyzed by Western blotting. (C) OGFOD1 band 1 is not degraded by serine and cysteine proteases during G3BP1 knockdown. HeLa cells were treated with a control siRNA (neg. cntrl.) and siRNAs against OGFOD1 and G3BP1 for 72 h. During the last 6 h of siRNA-mediated knockdown, cells were treated with normal medium (untreated) or medium containing a cocktail of serine and cysteine protease inhibitors (inhibitor). Endogenous proteins present in lysates were analyzed by Western blotting. Migration of novel OGFOD1 band 1.5 in the presence of protease inhibitors is indicated.
To test whether overexpression of G3BP1 could lead to increased abundance of OGFOD1 band 1, HeLa cells were transfected with a plasmid that expressed G3BP1. As shown in Fig. 4B, an increase in G3BP1 abundance correlated with an increased abundance of OGFOD1 band 1. In contrast, overexpression of G3BP1 did not affect the intracellular abundance of actin or USP10 (Fig. 4B). These results suggest that the generation or stability of OGFOD1 band 1 is affected by G3BP1.
To better understand the mechanism by which G3BP1 affects the abundance of OGFOD1 band 1 and to determine if proteases are involved in this process, cells were incubated with a cocktail of serine and cysteine protease inhibitors during the last 6 h of an siRNA-mediated knockdown of either OGFOD1 or G3BP1. While protease inhibitor treatment did not prevent G3BP1 siRNA-mediated loss of OGFOD1 band 1, it resulted in the appearance of a new form of OGFOD1, named band 1.5, that migrated between bands 1 and 2 (Fig. 4C, lane 6). These results suggest that G3BP1 may recruit a protease which processes OGFOD1 to generate band 1. It is less likely that G3BP1 is required to stabilize band 1 by protecting it from serine or cysteine proteases, because no rescue of band 1 was observed in the presence of protease inhibitors.
Presence of OGFOD1 is not required for stress granule formation.
To determine if OGFOD1 is required for the induction of stress granules, the appearance of stress granules was monitored when OGFOD1 abundance was depleted or enhanced. Although treatment with siRNAs directed against OGFOD1 mRNA dramatically reduced the abundance of OGFOD1 RNA and protein (see Fig. S2A and B in the supplemental material), stress granules containing G3BP1 readily formed when cells were treated with arsenite (see Fig. S3A in the supplemental material). In addition, overexpression of OGFOD1 in unstressed HeLa cells did not induce the formation of stress granules (see Fig. S3B in the supplemental material). These results suggest that OGFOD1 is not a stress granule nucleating protein, in contrast to G3BP1 and TIA1, which are both required for the formation of stress granules (1, 9).
OGFOD1 and G3BP1 modulate the phosphorylation status of eIF2α and the recovery of translation following arsenite-induced stress.
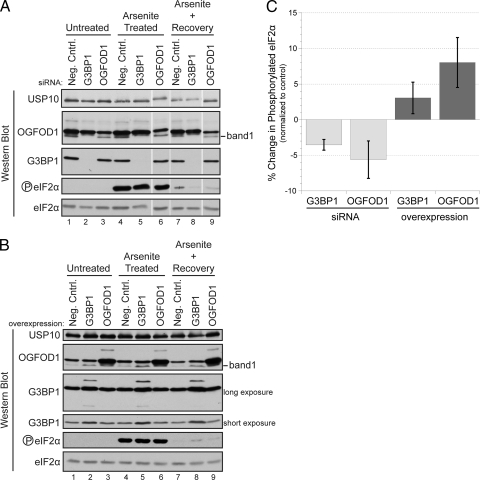
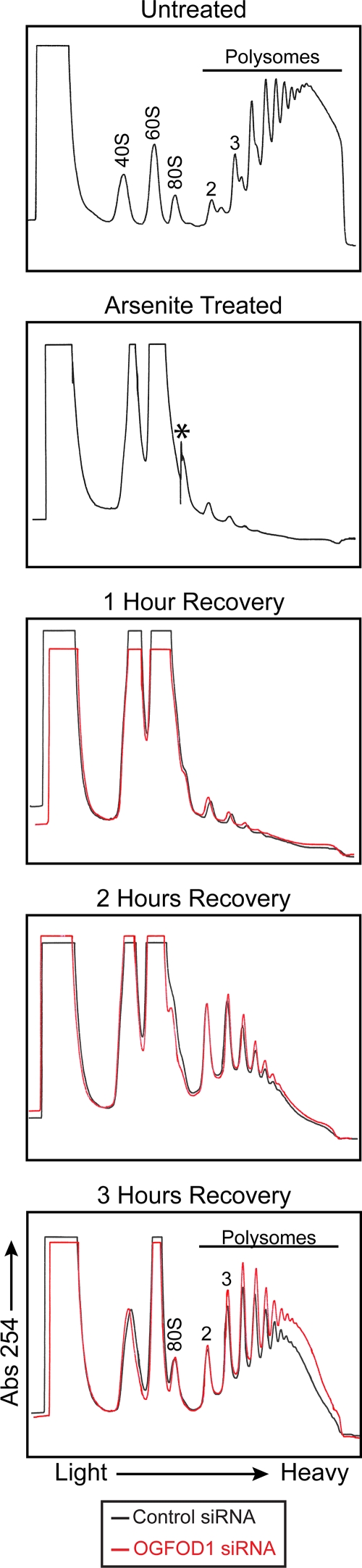
Arsenite-induced stress enhances the formation of stress granules (Fig. 1), increases the phosphorylation of eIF2α, and inhibits translation. We determined the amount of time required for dissolution of stress granules that formed when HeLa cells were treated with 1 mM arsenite for 30 min. Indirect immunofluorescence of G3BP1 and TIAR revealed that after 3 h of recovery, most stress granules marked by these two proteins had resolved and phosphorylation of eIF2α was significantly reversed (Fig. 5A, lane 7, and B, lane 7; see Fig. S1B in the supplemental material). In addition, Fig. 6 shows that a 30-min treatment with arsenite resulted in a rapid loss of polysomes with concomitant accumulation of 40S and 60S ribosomal subunits. New polysomes formed slowly after 3 h of recovery, and approximately 50% of the polysomes were reformed at that time compared to those in unstressed cells. The 3-h recovery time point was used in the subsequent experiments, because translation efficiency had significantly recovered and cellular morphologies appeared normal.
FIG. 5.
OGFOD1 regulates the phosphorylation of eIF2α during recovery from arsenite-induced stress. (A) Abundance of phosphorylated eIF2α decreases upon knockdown of OGFOD1 or G3BP1. Cells were treated with a negative-control siRNA (neg. cntrl.) and siRNAs against G3BP1 and OGFOD1 72 h prior to harvest. Extracts were prepared from unstressed cells (untreated), arsenite-treated cells, and cells recovering from arsenite-induced stress. Proteins were analyzed by Western blotting. (B) Abundance of phosphorylated eIF2α increases after overexpression of OGFOD1 or G3BP1. Extracts were prepared as described above from HeLa cells transiently transfected with an empty DNA vector (neg. cntrl.) or with plasmids expressing G3BP1 and OGFOD1 cDNAs and subjected to Western blot analysis. (C) Abundance of phosphorylated eIF2α in cells recovering from arsenite-induced stress changes upon knockdown or overexpression of OGFOD1 and G3BP1. Levels of phosphorylated eIF2α and total eIF2α were quantitated for three independent experiments, using Image J software. Representative Western blots are shown in panels A and B. Percent changes in phosphorylated eIF2α were calculated relative to total eIF2α levels and normalized to the negative control. Error bars represent the standard errors of the means.
FIG. 6.
OGFOD1 regulates formation of polysomes during recovery from arsenite-induced stress. Examination of polysome formation during stress recovery was performed by sucrose gradient sedimentation analysis. HeLa cells were treated with a negative-control siRNA (black line) or with an siRNA against OGFOD1 (red line) for a total of 72 h. Cells were treated with 1 mM arsenite for 30 min at 37°C and allowed to recover for the indicated times. Sucrose gradient analyses were performed as described in Materials and Methods. Absorbance peaks that correspond to the ribosomal 40S and 60S subunits as well as to 80S monosomes and polysomes are indicated. The Isco UA-6 detector was set to a sensitivity of 0.5 for all gradients except the arsenite-treated sample, which was collected at a sensitivity of 1.0. The asterisk indicates a user-initiated change in the UA-6 baseline setting so that all absorbance data could be retained.
To examine the role of G3BP1 and OGFOD1 in modulating the phosphorylation status of eIF2α, the abundance of phosphorylated eIF2α was examined in cells after siRNA-mediated gene knockdown of G3BP1 or OGFOD1. Phosphorylation of eIF2α was strongly induced in cells after arsenite treatment (Fig. 5A, lanes 4 to 6). After 3 h of recovery, low levels of phosphorylated eIF2α were still observed in cells treated with the negative-control siRNA (Fig. 5A, lane 7). In comparison, the abundance of phosphorylated eIF2α was further reduced in cells treated with siRNAs against G3BP1 and OGFOD1 (Fig. 5A, lanes 8 and 9; data are quantitated in Fig. 5C). Because the amount of total eIF2α did not change under these conditions (Fig. 5A, lanes 1 to 9), the reduced levels of phosphorylated eIF2α support a role for OGFOD1 and G3BP1 in modulating the phosphorylation status of eIF2α.
To further substantiate this hypothesis, OGFOD1 and G3BP1 were overexpressed in unstressed cells, cells stressed with arsenite, and cells recovering from stress. After 3 h of recovery from stress, larger amounts of phosphorylated eIF2α still persisted in cells overexpressing G3BP1 or OGFOD1 (Fig. 5B, lanes 7 to 9; data are quantitated in Fig. 5C). The accumulation of the phosphorylated form of eIF2α was not due to increased amounts of eIF2α in the cell (Fig. 5B), suggesting that overexpression of OGFOD1 and G3BP1 resulted in the activation of eIF2α kinases or the inhibition of eIF2α phosphatases.
Because knockdown of OGFOD1 resulted in reduced amounts of phosphorylated eIF2α during recovery from stress, and since the abundance of phosphorylated eIF2α directly correlates with repression of translation, we hypothesized that under these conditions, reduced levels of OGFOD1 would result in enhanced recovery of translation. To test this notion, cells were treated with either a control siRNA or an siRNA against OGFOD1 for 72 h, stressed with arsenite for 30 min, and then allowed to recover. Lysates were prepared and polysomal profiles analyzed by sucrose gradient sedimentation analysis. Figure 6 shows that knockdown of OGFOD1 strikingly enhanced polysome formation during recovery from stress. These results indicate that knockdown of OGFOD1 results in an enhanced recovery of translation as cells recover from stress. In contrast, overexpression of OGFOD1 under similar conditions resulted in decreased polysome levels compared to that in the negative control, suggesting that recovery of translation is slowed under these conditions (data not shown). Taken together, these findings argue that OGFOD1 functions as a rheostat to regulate the recovery from stress via modulation of eIF2α phosphorylation.
OGFOD1 promotes apoptosis in response to stress.
Phosphorylation of eIF2α and formation of stress granules have been shown to significantly impact the ability of a cell to survive stress (3, 6, 26). We have demonstrated that OGFOD1 and G3BP1 are stress granule proteins that regulate the abundance of phosphorylated eIF2α and that OGFOD1 regulates the recovery of translation as cells recover from arsenite-induced stress (Fig. 5 and 6).
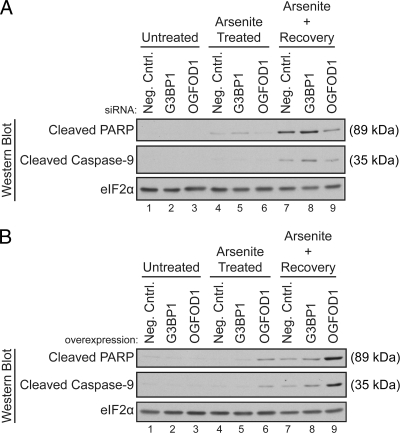
To determine if OGFOD1 plays a role in cellular survival after stress, we monitored the activation of caspase-9 and the cleavage of PARP in unstressed cells, arsenite-stressed cells, and cells recovering from stress. Knockdown of OGFOD1 resulted in decreased levels of cleaved caspase-9 and cleaved PARP, whereas knockdown of G3BP1 led to a slight increase in caspase-9 and to PARP cleavage (Fig. 7A). Overexpression of OGFOD1 resulted in a strong accumulation of activated caspase-9 and cleaved PARP in both stressed cells and cells recovering from stress (Fig. 7B). Taken together, these results indicate that OGFOD1 plays a proapoptotic role in cells exposed to arsenite.
FIG. 7.
OGFOD1 regulates survival of arsenite-induced stress. (A) Knockdown of OGFOD1 reduces accumulation of the 89-kDa PARP cleavage product in cells recovering from arsenite-induced stress. Cells were treated with a negative-control siRNA (neg. cntrl.) and siRNAs against G3BP1 or OGFOD1 72 h prior to harvest. Extracts were prepared from unstressed cells (untreated), arsenite-treated cells, and cells recovering from arsenite-induced stress. Proteins were analyzed by Western blotting. (B) Overexpression of OGFOD1 increases accumulation of the 35-kDa caspase-9 and 89-kDa PARP cleavage products in cells exposed to arsenite. Extracts were prepared as described above from HeLa cells transiently transfected with an empty DNA vector (neg. cntrl.) or with plasmids expressing G3BP1 and OGFOD1 cDNAs and subjected to Western blot analysis.
OGFOD1 interacts with HRI and eIF2α.
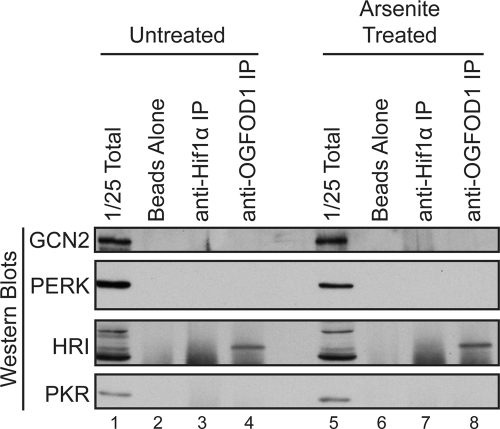
We hypothesized that OGFOD1 regulates the phosphorylation status of eIF2α via activation of an eIF2α kinase or inhibition of an eIF2α phosphatase. Thus, we performed immunoprecipitation analyses in both unstressed and arsenite-stressed cells to examine whether OGFOD1 interacts with any of the four known eIF2α kinases. Results from these experiments revealed that OGFOD1 interacts with a specific form of the eIF2α kinase HRI but not with GCN2, PERK, or PKR (Fig. 8, lanes 4 and 8). These findings argue that OGFOD1 may regulate the phosphorylation of eIF2α and the recovery of translation via interaction with HRI.
FIG. 8.
OGFOD1 interacts with eIF2α kinase HRI. Extracts from untreated or arsenite-treated HeLa cells were subjected to immunoprecipitation with an antibody against OGFOD1. Proteins were separated by SDS-PAGE and detected by Western blot analysis using the indicated antibodies. Beads without added antibody and Hif1α immunoprecipitation served as controls for nonspecific interactions.
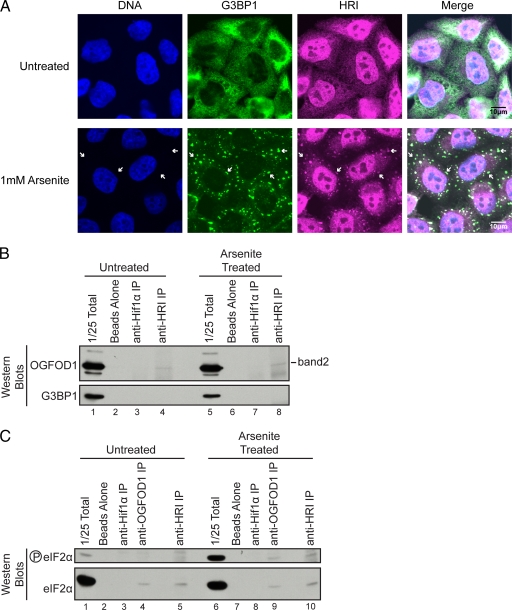
HRI has previously been shown to mediate the formation of stress granules and the survival of arsenite-induced stress in mice (18, 20). Based on the interaction of OGFOD1 with HRI, we sought to determine if HRI itself is a stress granule resident. Indirect immunofluorescence of endogenous HRI revealed that, similar to OGFOD1, HRI in unstressed cells is predominantly nuclear, with diffuse cytoplasmic staining (Fig. 9). Upon arsenite-induced stress, a portion of HRI relocalized to discrete cytoplasmic foci containing the stress granule marker G3BP1 (Fig. 9A).
FIG. 9.
OGFOD1 and HRI interact with eIF2α. (A) HRI colocalizes with G3BP1 in arsenite-induced stress granules. Endogenous HRI and G3BP1 were detected by indirect immunofluorescence in untreated HeLa cells or in cells treated with 1 mM arsenite for 30 min at 37°C. An overlap of HRI and G3BP1 appears white in the merged image. White arrows point to stress granules. (B) OGFOD1 band 2 coimmunoprecipitates with HRI. Extracts from untreated or arsenite-treated HeLa cells were subjected to immunoprecipitation with an antibody against HRI. Beads without added antibody and Hif1α immunoprecipitation were used as controls for nonspecific interactions. Proteins were separated by SDS-PAGE and detected by Western blot analysis using the indicated antibodies. (C) OGFOD1 and HRI coimmunoprecipitate eIF2α and phosphorylated eIF2α. Extracts from untreated or arsenite-treated HeLa cells were subjected to immunoprecipitation with antibodies against OGFOD1 and HRI. Beads without added antibody and Hif1α immunoprecipitation were used as controls for nonspecific interactions. Proteins were separated by SDS-PAGE and detected by Western blot analysis using the indicated antibodies.
To further substantiate the interaction between OGFOD1 and HRI and to determine which form of OGFOD1 interacts with HRI, we performed coimmunoprecipitation analyses using an HRI-specific antibody. Results from these experiments revealed that HRI interacted with OGFOD1 band 2, but not with G3BP1, in both unstressed and arsenite-stressed cells (Fig. 9B, lanes 4 and 8). In support of these findings, we also noted that G3BP1 was unable to coimmunoprecipitate HRI (data not shown). These results reveal that different forms of OGFOD1 interact with HRI and G3BP1 and that G3BP1 may not play a direct role in the regulation of eIF2α phosphorylation.
Finally, because OGFOD1 influences the phosphorylation status of eIF2α and interacts with HRI, we sought to determine if OGFOD1 interacts with eIF2α itself. Coimmunoprecipitation analyses revealed that OGFOD1 and HRI coimmunoprecipitated both nonphosphorylated and phosphorylated eIF2α (Fig. 9C, lanes 4, 5, 9, and 10). In combination, these results suggest that OGFOD1 plays a central role in the regulation of translation and HRI-mediated phosphorylation of eIF2α in response to arsenite-induced stress.
DISCUSSION
The founding member of the OGFOD1 family, S. cerevisiae Tpa1p, has been shown to regulate mRNA deadenylation, mRNA stability, and translation termination (15). Unlike Tpa1p, human OGFOD1 does not interact with translation termination factors or the poly(A) binding protein (data not shown). We report here that human OGFOD1 relocalizes to cytoplasmic stress granules after arsenite-induced stress and modulates translational recovery by regulating the phosphorylation status of eIF2α.
An extensive analysis of OGFOD1 interactions in unstressed and stressed cells revealed that it interacts with a subset of stress granule proteins that includes G3BP1, Caprin1, USP10, and YB-1 but does not include TIA1, TIAR, FMRP, SMN, PABP, and eIF4G (Fig. 2 and 3B; data not shown). Furthermore, a specific form of OGFOD1 was found to mediate these interactions. It is not clear if all four proteins are found in a single large macromolecular complex, but based on the previous identification of Caprin1 and USP10 as G3BP1 binding partners and the strong interaction between G3BP1 and OGFOD1 band 1, it is likely that OGFOD1, G3BP1, Caprin1, and USP10 exist as a macromolecular complex.
OGFOD1 exists as multiple forms in the cell that are likely generated as a combined result of both modifying and cleavage activities. The presence of so many different forms suggests that OGFOD1 may be highly regulated at the posttranslational level. Although immunofluorescence revealed that there is a strong disparity between cytoplasmic and nuclear localization of total OGFOD1 (Fig. 1), results from subcellular fractionation experiments suggested that the relative ratios of bands 1 to 4 are maintained in both compartments (data not shown). While we have shown that OGFOD1 band 1 interacts with a number of different proteins and the ribosome, the interaction between band 1 and G3BP1 appears to be the most significant. Based on coimmunoprecipitation studies, it appears that a considerable portion of G3BP1 interacts with OGFOD1 and that nearly all of OGFOD1 band 1 is associated with G3BP1 (Fig. 2 and 3B). The interaction between OGFOD1 band 1 and G3BP1 likely has biological significance, because modulating the level of G3BP1 influences the abundance of OGFOD1 band 1 (Fig. 4). For example, siRNA-mediated knockdown of G3BP1 resulted in the loss of both G3BP1 and OGFOD1 band 1. In fact, the most effective way to knock down OGFOD1 band 1 is via the knockdown of G3BP1.
Based on the results discussed above, it is possible that G3BP1 binding either stabilizes OGFOD1 band 1 or is responsible for the generation of band 1. Results from experiments using protease inhibitors suggested that G3BP1 may recruit a serine or cysteine protease to a larger form of OGFOD1, with rapid cleavage resulting in the formation of OGFOD1 band 1. It will be important to determine how band 1 and each of the other OGFOD1 forms is generated so that their abundances can be modulated and their specific biological roles determined.
Very little is known about the roles of stress granule residents in aiding cells to recover from stress. In mammalian cells, one of the best-characterized facets of the stress response is the phosphorylation of eIF2α. Phosphorylation of eIF2α occurs as a response to stress and results in stalled translation initiation (16). This process has been shown to be critical for cells to survive a number of different stresses, including hypoxia, endoplasmic reticulum stress, and oxidative stress (17, 20). In fact, inhibition of the dephosphorylation of eIF2α has been shown to enhance cellular survival from stress (4, 13). Constitutive expression of an eIF2α phosphorylation mimic and persistent translational repression, on the other hand, have also been linked to decreased cell survival (19, 26).
We have identified OGFOD1 and G3BP1 as modulators of the eIF2α-mediated stress response. Overexpression of OGFOD1 results in an increased abundance of phosphorylated eIF2α in unstressed cells (see Fig. S2C in the supplemental material). Overexpression of Caprin1, an OGFOD1- and G3BP1-interacting protein, has also been shown to result in increased abundance of phosphorylated eIF2α in unstressed cells (24). These results suggest that it may be a general function of OGFOD1 and its associated factors to regulate the abundance of phosphorylated eIF2α in unstressed cells.
OGFOD1 and G3BP1 also play a significant role in maintaining the abundance of phosphorylated eIF2α when cells recover from stress. Knockdown of OGFOD1 or G3BP1 resulted in reduced amounts of phosphorylated eIF2α during recovery from stress (Fig. 5A). While the changes in eIF2α phosphorylation were subtle, they were reproducible (Fig. 5C) and correlated with polysomal profiles observed during recovery from stress (Fig. 6). It is known that subtle changes in the phosphorylation status of eIF2α may have dramatic consequences on the activity of the translation apparatus (22, 23). Phosphorylation of eIF2α is inhibitory to translation; thus, the observed reduction in phosphorylation of eIF2α after OGFOD1 knockdown is functionally relevant.
In support of these findings, we also demonstrated that overexpression of OGFOD1 or G3BP1 resulted in increased amounts of phosphorylated eIF2α during the recovery from stress (Fig. 5B). As might be expected from an increase in the level of phosphorylated eIF2α, OGFOD1 overexpression correlated with a decrease in polysomes during the recovery from stress (data not shown). Previous work has revealed that similar aberrant accumulation of phosphorylated eIF2α and prolonged translational repression correlate with decreased cell survival (19, 26). In accordance with these findings, we have demonstrated that overexpression of OGFOD1 also correlates with decreased cell survival (Fig. 7B). The increased activation of the initiator caspase-9 suggests that cells overexpressing OGFOD1 are less well suited to survive arsenite-induced stress.
To determine the mechanism by which the OGFOD1 complex regulates eIF2α phosphorylation, we sought to identify the eIF2α kinases that interact with OGFOD1. Results from these experiments indicated that OGFOD1 band 2 interacted with HRI and that both HRI and OGFOD1 interacted with eIF2α, independent of its phosphorylation state. Based on our finding that OGFOD1 interacts with HRI and previous work demonstrating that HRI mediates the cellular response to arsenite in mice, we suggest that OGFOD1 modulates phosphorylation of eIF2α and the recovery of translation following arsenite-induced stress via regulation of HRI activity (20).
OGFOD1 bears a domain with strong similarity to the prolyl-hydroxylase domain of the HIFα-modifying proteins PHD1 to PHD3. The presence of this domain suggests that the mechanism by which OGFOD1 regulates eIF2α phosphorylation may involve hydroxylation of a specific proline residue within HRI, or potentially within eIF2α. Because OGFOD1 associates with both HRI and eIF2α in unstressed and stressed cells, it is likely that the mechanism by which OGFOD1 controls the eIF2α regulatory cascade will reveal novel modes of translation control under stressed and homeostatic conditions.
Supplementary Material
Acknowledgments
We are grateful to Karla Kirkegaard and Christopher J. Potter for critical readings of the manuscript. We also thank Joan A. Steitz for the kind gift of the Y10B antibody and Duncan Brown for the design of the USP10 siRNA.
This work was supported by National Institutes of Health grants R01 AI069000 and R01 CA140445 (to P.S.). K.A.W. was supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation. S.S. was supported by Deutsche Forschungsgemeinschaft postdoctoral fellowship SCHU 2294/2-1.
Footnotes
Published ahead of print on 12 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430-436. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2008. Stress granules: the tao of RNA triage. Trends Biochem. Sci. 33:141-150. [DOI] [PubMed] [Google Scholar]
- 3.Arimoto, K., H. Fukuda, S. Imajoh-Ohmi, H. Saito, and M. Takekawa. 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10:1324-1332. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, M., K. F. Bryant, C. Jousse, K. Long, H. P. Harding, D. Scheuner, R. J. Kaufman, D. Ma, D. M. Coen, D. Ron, and J. Yuan. 2005. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science 307:935-939. [DOI] [PubMed] [Google Scholar]
- 5.Costa, M., A. Ochem, A. Staub, and A. Falaschi. 1999. Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the ras transduction pathway. Nucleic Acids Res. 27:817-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisinger-Mathason, T. S., J. Andrade, A. L. Groehler, D. E. Clark, T. L. Muratore-Schroeder, L. Pasic, J. A. Smith, J. Shabanowitz, D. F. Hunt, I. G. Macara, and D. A. Lannigan. 2008. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell 31:722-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evdokimova, V., P. Ruzanov, M. S. Anglesio, A. V. Sorokin, L. P. Ovchinnikov, J. Buckley, T. J. Triche, N. Sonenberg, and P. H. Sorensen. 2006. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol. Cell. Biol. 26:277-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forch, P., and J. Valcarcel. 2001. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis 6:463-468. [DOI] [PubMed] [Google Scholar]
- 9.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han, A. P., C. Yu, L. Lu, Y. Fujiwara, C. Browne, G. Chin, M. Fleming, P. Leboulch, S. H. Orkin, and J. J. Chen. 2001. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20:6909-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinnebusch, A. G., T. E. Dever, and K. Asano. 2007. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae, p. 225-268. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Hughes, B. T., and P. J. Espenshade. 2008. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 27:1491-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jousse, C., S. Oyadomari, I. Novoa, P. Lu, Y. Zhang, H. P. Harding, and D. Ron. 2003. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163:767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeling, K. M., J. Salas-Marco, L. Z. Osherovich, and D. M. Bedwell. 2006. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:5237-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimball, S. R. 1999. Eukaryotic initiation factor eIF2. Int. J. Biochem. Cell Biol. 31:25-29. [DOI] [PubMed] [Google Scholar]
- 17.Koumenis, C., C. Naczki, M. Koritzinsky, S. Rastani, A. Diehl, N. Sonenberg, A. Koromilas, and B. G. Wouters. 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol. Cell. Biol. 22:7405-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, L., A. P. Han, and J. J. Chen. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 21:7971-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, P. D., C. Jousse, S. J. Marciniak, Y. Zhang, I. Novoa, D. Scheuner, R. J. Kaufman, D. Ron, and H. P. Harding. 2004. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McEwen, E., N. Kedersha, B. Song, D. Scheuner, N. Gilks, A. Han, J. J. Chen, P. Anderson, and R. J. Kaufman. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925-16933. [DOI] [PubMed] [Google Scholar]
- 21.Ron, D., and H. P. Harding. 2007. eIF2α phosphorylation in cellular stress responses and disease, p. 345-368. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Rowlands, A. G., K. S. Montine, E. C. Henshaw, and R. Panniers. 1988. Physiological stresses inhibit guanine-nucleotide-exchange factor in Ehrlich cells. Eur. J. Biochem. 175:93-99. [DOI] [PubMed] [Google Scholar]
- 23.Scorsone, K. A., R. Panniers, A. G. Rowlands, and E. C. Henshaw. 1987. Phosphorylation of eukaryotic initiation factor 2 during physiological stresses which affect protein synthesis. J. Biol. Chem. 262:14538-14543. [PubMed] [Google Scholar]
- 24.Solomon, S., Y. Xu, B. Wang, M. D. David, P. Schubert, D. Kennedy, and J. W. Schrader. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27:2324-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorokin, A. V., A. A. Selyutina, M. A. Skabkin, S. G. Guryanov, I. V. Nazimov, C. Richard, J. Th'ng, J. Yau, P. H. Sorensen, L. P. Ovchinnikov, and V. Evdokimova. 2005. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 24:3602-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava, S. P., K. U. Kumar, and R. J. Kaufman. 1998. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273:2416-2423. [DOI] [PubMed] [Google Scholar]
- 27.Tourriere, H., I. E. Gallouzi, K. Chebli, J. P. Capony, J. Mouaikel, P. van der Geer, and J. Tazi. 2001. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell. Biol. 21:7747-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wehner, K. A., J. E. Gallagher, and S. J. Baserga. 2002. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 22:7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.