Abstract
Alternative 5′ splice site selection is one of the major pathways resulting in mRNA diversification. Regulation of this type of alternative splicing depends on the presence of regulatory elements that activate or repress the use of competing splice sites, usually leading to the preferential use of the proximal splice site. However, the mechanisms involved in proximal splice site selection and the thermodynamic advantage realized by proximal splice sites are not well understood. Here, we have carried out a systematic analysis of alternative 5′ splice site usage using in vitro splicing assays. We show that observed rates of splicing correlate well with their U1 snRNA base pairing potential. Weak U1 snRNA interactions with the 5′ splice site were significantly rescued by the proximity of the downstream exon, demonstrating that the intron definition mode of splice site recognition is highly efficient. In the context of competing splice sites, the proximity to the downstream 3′ splice site was more influential in dictating splice site selection than the actual 5′ splice site/U1 snRNA base pairing potential. Surprisingly, the kinetic analysis also demonstrated that an upstream competing 5′ splice site enhances the rate of proximal splicing. These results reveal the discovery of a new splicing regulatory element, an upstream 5′ splice site functioning as a splicing enhancer.
The splicing of nuclear pre-mRNAs is a fundamental process required for the expression of most metazoan genes. It is carried out by the spliceosome, which recognizes splicing signals and catalyzes the removal of noncoding intronic sequences to assemble protein-coding sequences into mature mRNAs prior to export to the cytoplasm and translation (2). Of the approximately 25,000 genes carried by the human genome (16), more than 90% are believed to produce transcripts that are alternatively spliced, enriching the proteomic diversity of higher eukaryotic organisms (9, 29). Because regulation of this process can determine when and where a particular protein isoform is expressed, changes in alternative splicing patterns modulate many cellular activities.
Exon recognition and the regulation of alternative splicing are complex processes. Splicing enhancers and silencers, either exonic or intronic, occur frequently and influence alternative splicing. Furthermore, it has been established that the strength of splice sites (31), the intron/exon architecture (i.e., the exon or intron definition modes of splice site recognition) (8), local RNA secondary structure (13, 26), and the process of transcription by RNA polymerase II (19) influence splice site choice. Given the divergent sequence and architecture of genes, every exon is flanked by a unique pair of splice site signals and contains a unique group of splicing enhancers, silencers, and secondary structures. The sum of contributions from each of these elements then defines the overall recognition potential of an exon (11).
When analyzing pre-mRNA sequences, it quickly becomes evident that multiple or cryptic splices sites are abundant, especially 5′ splice sites (1). The mechanism as to which specific 5′ splice site is preferred over others must rely on selection mechanisms independent of whether the exon is alternative or constitutive. In a set of classical experiments, Reed and Maniatis (22) showed that the proximity between the 5′ and the 3′ splice sites determines, in part, the selection of competing splice sites. Using tandem repeats of either 5′ or 3′ splice sites, it was shown that the spliceosome exhibited a strong preference for pairing the most proximal splice sites. In the case of competing 5′ splice sites of equal U1 snRNA base pairing affinity, the most proximal splice site was preferentially chosen. While these experiments established a splice site selection phenomenon referred to as the proximity rule, mechanistic and thermodynamic insights into the preference for proximal splice sites have been minimal. Using a model system of tandem 5′ splice sites separated by 16 nucleotides, it was shown that the ability of U1 snRNA to base pair with competing splice sites is the major determinant of 5′ splice site selection, in agreement with previous models (24). However, recent computational analyses indicate that pairwise dependencies between 5′ splice site nucleotides are also significant in promoting splice site selection (23).
In this work, we have carried out a systematic analysis of 5′ splice site recognition using in vitro splicing assays. While the observed rates of splicing correlate largely with the binding potential between U1 snRNA and the 5′ splice site, we show that U1 snRNA interactions with the 5′ splice site were significantly aided by the proximity of the downstream exon. Tethered splicing activators upstream of weak 5′ splice sites activate splicing similarly and independent of whether splice sites are recognized within a single step (referred to as intron defined) or independent steps (referred to as outside intron definition). In the context of competing splice sites separated by 180 nucleotides, the proximal splice site was preferentially used even though the distal splice site had a much higher U1 snRNA binding potential. Interestingly, a splicing enhancer sandwiched between splice sites had opposing effects on the distal and proximal splice sites, activating the proximal splice site while repressing the distal splice site. Surprisingly, the kinetic analysis uncovered that a functional but unselected upstream 5′ splice site acts as bone fide splicing enhancer for proximal splice site selection.
MATERIALS AND METHODS
Pre-mRNAs.
DNA for regions of the first two exons (excluding the 5′ splice site of exon 2) of human β-globin was cloned downstream of the T7 promoter into SP73 plasmid. An MS2 hairpin was cloned approximately 80 nucleotides upstream of the original 5′ splice site. Thirteen different 5′ splice site cassettes were separately cloned into the original 5′ splice site region. Possible cryptic 5′ splice sites were inactivated. A splicing-neutral sequence was cloned between the MS2 hairpin and 5′ splice site cassette. The intronic lengths of these constructs were increased by adding 240 bp of neutral sequence originated from the intronic region of SMN6 to create long intron constructs. A GT-less exonic and 5′ splice site cassette region were cloned 80 bp upstream of the MS2 hairpin to create the 5′ splice site competition constructs with either a strong competitor, CAG|GUAAGUgU, or an abolished 5′ splice site with the sequence gua|cUucuaca (sequence 0 in Tables 1 to 3), where uppercase letters indicate matches with U1 snRNA and lowercase letters indicate mismatches with U1 snRNA. Potential cryptic 5′ splice sites (those containing a GT) were mutated within the exonic and intronic regions surrounding the 5′ splice site cassette of exon 1. Diagrams of the modified β-globin minigenes are depicted in the corresponding figures. To generate 5′ competition deletion constructs, the first-generation plasmids were digested with XbaI and Eco NI, blunted with T4 polymerase, and ligated with T4 ligase. This removed a 104-bp segment from the vector, decreasing the distance between the competing 5′ splice sites to 80 nucleotides (nt).
TABLE 1.
Splicing rates of 5′ splice sites correlate with U1 snRNP potential
| Construct |
Short intron |
Long intron |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | 5′ splice site sequencea | ΔGb | MaxEnt scorec | Avg rated |
Fold changed | MS2-RS-9G8 |
Fold change | ||
| − MS2-RS-9G8 | + MS2-RS-9G8 | − MS2-RS-9G8 | + MS2-RS-9G8 | ||||||
| 0 | gua|cUucuaca | >0 | −26.3 | * | * | * | * | * | * |
| 1 | CAa|GUcAagcc | 0 | −9.4 | 0.15 | 0.64 | 4.3 | ** | ** | ** |
| 2 | guG|GUucuaca | −0.9 | −8.2 | * | * | * | 0.007 | 0.04 | 6.4 |
| 3 | gAG|GUucuaca | −2.8 | −1.6 | 0.11 | 0.47 | 4.1 | 0.03 | 0.16 | 5.2 |
| 4 | CAG|GUucuaca | −3.6 | 0.7 | 0.10 | 0.36 | 3.5 | 0.07 | 0.20 | 2.7 |
| 5 | CuG|GUcuGUAU | −3.3 | 3.1 | 0.14 | 0.58 | 4.0 | 0.14 | 0.41 | 3.0 |
| 6 | guG|GUucGUAU | −3.3 | 3.3 | 0.25 | 0.85 | 3.4 | 0.08 | 0.35 | 4.7 |
| 7 | guG|GUucGUca | −1.6 | 3.3 | 0.69 | 1.13 | 1.6 | 0.11 | 0.23 | 2.1 |
| 8 | CAG|GUugGUAU | −8.5 | 8.1 | 0.37 | 1.62 | 4.4 | 0.24 | 0.54 | 2.3 |
| 9 | Cuc|GUAAGUca | −3.5 | 9.6 | 0.20 | 0.66 | 3.2 | ** | ** | ** |
| 10 | CAc|GUAAGUca | −4.0 | 10.2 | 0.34 | 1.33 | 3.9 | 0.15 | 0.41 | 2.8 |
| 11 | CAG|GUAAGUAg | −11.3 | 10.9 | 1.10 | 1.35 | 1.2 | 0.37 | 0.40 | 1.1 |
| 12 | CAG|GUAAGUca | −9.4 | 10.9 | 0.59 | 0.75 | 1.3 | 0.44 | 0.74 | 1.7 |
Uppercase letters indicate matches with U1 snRNA, and lowercase letters indicate mismatches with U1 snRNA.
ΔG values are based on nearest-neighbor contributions (32).
Rows are organized according to MaxEnt scores, which are values based on modeling the sequences of short sequence motifs that simultaneously account for nonadjacent as well as adjacent dependencies between positions. This method is based on the maximum-entropy principle and generalizes most previous probabilistic models of sequence motifs such as weight matrix models and nonhomogeneous Markov models (31).
The data show rate and fold activation comparisons of the single 5′ splice sites in the context of intron versus exon definition, with and without MS2-RS-9G8 recruitment. Average rates and fold activation were determined from multiple repeats with less than 30% difference between experiments. *, value not different from background. **, not tested in this context.
TABLE 3.
Rates of proximal 5′ splice site usage in the presence of a competing upstream 5′ splice site
| Construct |
Short intron |
Long intron |
||||||
|---|---|---|---|---|---|---|---|---|
| No. | 5′ splice site sequencea | MaxEnt scoreb | Avg ratec |
Fold changec | Avg rate |
Fold change | ||
| − MS2-RS-9G8 | + MS2-RS-9G8 | − MS2-RS-9G8 | + MS2-RS-9G8 | |||||
| 0 | gua|cUucuaca | −26.3 | * | * | * | * | * | * |
| 1 | CAa|GUcAagcc | −9.4 | 0.91 | 0.92 | 1.0 | 0.62 | 0.74 | 1.2 |
| 2 | guG|GUucuaca | −8.2 | 0.001 | 0.002 | 1.1 | 0.40 | 0.83 | 2.1 |
| 3 | gAG|GUucuaca | −1.6 | 0.17 | 0.45 | 2.6 | ** | ** | ** |
| 4 | CAG|GUucuaca | 0.7 | 0.40 | 0.58 | 1.5 | 0.21 | 0.34 | 1.6 |
| 5 | CuG|GUcuGUAU | 3.1 | 0.88 | 0.98 | 1.1 | 0.33 | 0.60 | 1.8 |
| 6 | guG|GUucGUAU | 3.3 | 1.0 | 1.1 | 1.1 | 0.41 | 0.77 | 1.9 |
| 7 | guG|GUucGUca | 3.3 | 0.92 | 0.98 | 1.1 | 0.50 | 0.84 | 1.7 |
| 8 | CAG|GUugGUAU | 8.1 | 1.1 | 1.1 | 1.0 | 0.71 | 0.81 | 1.1 |
| 9 | Cuc|GUAAGUca | 9.6 | 0.82 | 1.00 | 1.2 | ** | ** | ** |
| 10 | CAc|GUAAGUca | 10.2 | 0.84 | 0.64 | 0.8 | 0.84 | 0.56 | 0.7 |
| 11 | CAG|GUAAGUAg | 10.9 | 0.82 | 0.83 | 1.0 | 0.26 | 0.21 | 0.8 |
| 12 | CAG|GUAAGUca | 10.9 | 1.0 | 1.0 | 1.0 | ** | ** | ** |
| Distal | CAG|GUAAGUgU | 10.9 | 0.61 | 0.06 | −10.3 | ** | ** | ** |
Uppercase letters indicate matches with U1 snRNA, and lowercase letters indicate mismatches with U1 snRNA.
Rows are organized according to MaxEnt scores (31).
The data show rate and fold activation comparisons of each 5′ splice site in the context of intron versus exon definition, upstream 5′ splice site competition, and with and without MS2-RS-9G8 recruitment. Average rates and fold activation were determined from multiple repeats with less than 30% difference between experiments. *, value not different from background. **, not tested in this context.
All modified β-globin minigenes were digested with EcoRI. Presynthesized 32P-labeled RNA transcripts were synthesized with T7 RNA polymerase in the presence of 2 mM m7G(5′)ppp(5′) G cap analog at 37°C, resolved by 6% denaturing PAGE, and then purified.
In vitro splicing.
Reactions were carried out in 30% nuclear extract containing T7 polymerase-generated 32P-labeled RNA transcripts as the template, 1.0 mM ATP, 20 mM creatine phosphate, and 3.2 mM MgCl2, with or without saturating amounts of recombinant MS2-RS-9G8 (250 nM), as previously described (12). Reaction mixtures were incubated at 30°C for a determined time course as indicated in the figure legends. Following incubation, reaction mixtures were digested with proteinase K; phenol chloroform extracted; ethanol precipitated; resolved by either 6, 8, or 10% denaturing PAGE; and analyzed by PhosphorImager analysis (Bio-Rad) as previously described (12). The percent spliced is defined as [moles spliced product/(moles unspliced product + moles spliced product)]. To derive kinetic rate constants, the percent spliced over time was fit to a first-order rate description for product appearance. The background was determined individually for each lane. Average rates and fold activation were determined from multiple repeats with less than a 30% difference between experiments (r values for rate determinations were above 0.8).
Computational analysis of constitutive exons.
Five thousand constitutive exonic sequences with lengths of ≥100 nt were downloaded from the UCSC Genome Browser. Two exhaustive tables of 5′ splice sites were constructed: a table of weak splice sites with all possible 5′ splice sites with a MaxEnt score of <0 and a table of strong 5′ splice sites that contains all 5′ splice sites with a MaxEnt score of >7. Using a Python script, each sequence was searched for potential 5′ splice sites by sliding a 9-nt window from a position starting at 1nt downstream of the 3′ splice site to 100 nt away from the 3′ splice site. Each position was checked to see if it contained a strong or weak 5′ splice site from the hash tables. The total number of potential splice sites, either weak or strong, at each position was summed for the 5,000 exonic sequences, and these counts were binned in 10-nucleotide bins from 1 to 100 nt from the 3′ splice site. Each resulting bin was normalized to 5′ splice site frequencies observed for the first 10-nt bin closest to the 3′ splice site.
RESULTS
Thermodynamic contribution of ESEs to 5′ splice site activation.
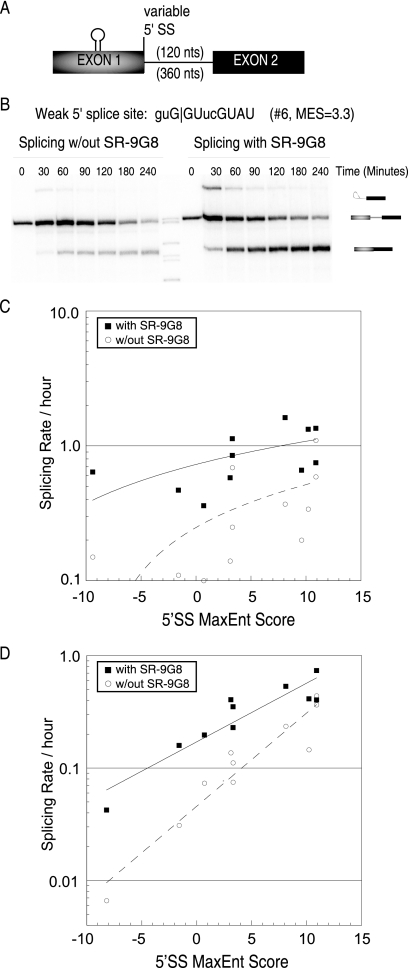
In Saccharomyces cerevisiae 5′ splice sites are well conserved compared to those in higher eukaryotes, where 5′ splice sites are degenerate. Although a consensus sequence for the 5′ splice site has been determined (Y−3A−2G−1/G+1U+2R+3A+4G+5U+6A+7U+8), many metazoan splice sites do not match this consensus. In fact, the splicing machinery often selects a weak 5′ splice site with fewer matches to the U1 snRNA consensus sequence over a stronger competing splice site (14, 18, 25, 28). Therefore, there must be motifs in the pre-mRNA such as exonic splicing enhancers (ESEs), which compensate for the weakness of a 5′ splice site. While many examples of ESE-dependent activation of weak splice sites have been described, it is currently unknown to what degree a U1 snRNA/5′ splice site consensus mismatch requires an ESE complex to compensate for the weak site. To investigate this dependency, we analyzed the splicing efficiency of β-globin-derived minigenes with variable intron-defined 5′ splice sites (Table 1) that are activated by an upstream ESE (Fig. 1A). In vitro splicing reactions in the presence or absence of MS2-RS-9G8 fusion protein demonstrated that the splicing rate of weaker 5′ splice sites (MaxEnt score of <4) increases up to 4-fold upon ESE activation (Fig. 1 and Table 1). In contrast, strong splice sites (MaxEnt score of >10) apparently do not significantly benefit from the presence of an activated ESE. Splicing under different conditions, such as with an altered percentage of nuclear extract or salt concentration, did not change the extent of ESE activation observed (data not shown). These results illustrate that an MS2-RS-9G8 ESE complex at the location used has a maximal activation potential of approximately 4-fold. As expected from previous observations (20), lowering the complementarity between U1 snRNA and the 5′ splice site reduced the rate of intron removal from maximal in vitro splicing of approximately 1 h−1 for a fully complementary splice site to 0.1 h−1 for a 5′ splice site of low complementarity (Fig. 1C). For the most part, the observed rates follow an overall trend expected from U1 snRNA binding potential, with stronger binding interactions resulting in higher rates of intron removal. Apparent deviations from linear regressions are likely to reflect the influence of additional factors that are currently not taken into account in models of splice site strength predictions (Fig. 1C and Table 1). Supporting this notion is the observation that the magnitude of changes in splicing rates is significantly different from expected energetic differences based on base pairing potential. For example, according to nearest-neighbor contributions (32), the difference in U1 snRNA binding potential between splice sites 3 and 11 is ∼1 × 106-fold, while the measured rates differ by only 10-fold.
FIG. 1.
Enhancer dependency of variable 5′ splice sites for efficient intron-defined splicing. (A) Diagram of modified β-globin minigene templates with variable 5′ splice sites (Table 1), an MS2 hairpin ESE activated by the addition of MS2-RS-9G8, and either a 120-nt or a 360-nt intron. (B) Autoradiogram showing a time course analysis of splicing with a weak 5′ splice site (guG|GUucGUAU; MaxEnt Score = 3.3 [see Table 1 for description]) without enhancer (left) compared to that with the same splice site activated by MS2-RS-9G8 recruitment (right). Pre-mRNA transcript, spliced product, and intermediate lariat bands are indicated. (C and D) Comparison of the splicing rates determined from short intron (C) or long intron (D) pre-mRNAs according to splice site strength (MaxEnt Score range, −9.4 to 10.9). Average rates were determined from multiple repeats with less than 30% difference between experiments. Further comparison of 5′ splice site sequences, strength (MaxEnt Score), and splicing rates can be found in Table 1.
To determine the influence of enhancer activation in the context of 5′ splice sites outside the intron definition context, i.e., splice sites that are defined independently from each other, we carried out similar rate analyses using isogenic pre-mRNA substrates with introns longer than the previously determined transition length between intron and exon definition (∼250 nt) (8). As was observed for intron-defined splice site recognition, reducing the complementarity between U1 snRNA and the exon-defined 5′ splice site lowered the rate of intron removal by more than 10-fold (Table 1; Fig. 1D). Significantly, in the presence of MS2-RS-9G8 fusion protein, the rate of intron removal increased for all tested pre-mRNAs without an apparent limit, as was observed in the intron-definition mode (Table 1). In agreement with previous studies (8), splicing of short introns is more efficient than the removal of larger introns (Table 1). These observations support the notion that the close proximity of a pairing-competent splice site compensates for weak splicing signals. In summary, the current methods predicting splice site strength correlate well with experimentally determined rates of splicing (Fig. 1; Table 1).
Proximal 5′ splice site preference.
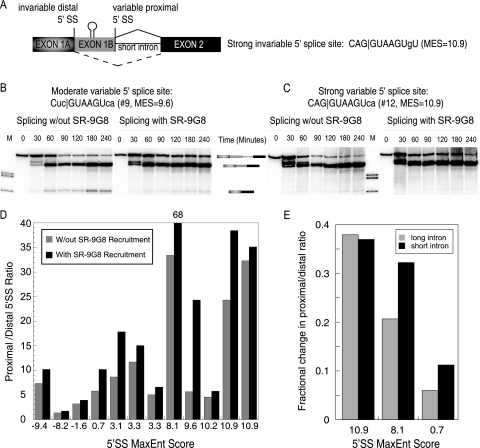
The proximity rule was formulated based on experimental observations using duplicated splice sites and refers to the preferential pairing between the most proximal 5′ and 3′ splice sites (22). Although established several years ago, its mechanism and thermodynamic contribution to splice site choice have yet to be investigated. To evaluate the proximity rule, we designed a series of 5′ splice site competition substrates that contain a strong, invariable distal splice site (CAG|GUAAGUgU) and one of 13 variable proximal splice sites (Fig. 2A; Tables 2 and 3). In addition, a splicing activator can be tethered between the competing splice sites through binding to an MS2 hairpin. Using this experimental setup, we intended to measure the degree to which splice site selection would shift from the proximal to the distal splice site by decreasing the strength of the proximal splice site with respect to the distal splice site. As expected from the proximity rule, the proximal splice site was chosen over the distal splice site if the distal and proximal splice sites were of similar strength (Fig. 2C). Surprisingly, proximal splicing was still preferred even if the proximal splice site was significantly weaker than the distal splice site (Fig. 2B and Table 2). In fact, our results show that under the conditions tested, proximal splicing was always the preferred pathway, although a shift in the proximal/distal ratio can be correlated to proximal 5′ splice site strength (Fig. 2D and Table 2). Furthermore, placement of a splicing activator complex between the competing splice sites increased the rate of proximal splicing, most noticeably in the context of significant distal splicing (Fig. 2B and Table 2). To test whether the observed preference for proximal splicing is influenced by intron length, we repeated the kinetic evaluations as described above with pre-mRNA substrates containing longer introns. Although the increase in intronic distance slightly increases the ratio toward the distal splice site (Table 2), the proximity rule still remains the stronger element in our experimental system. This trend suggests that increased intron length reduces the influence of the proximity effect. We conclude that the proximity of paired splice sites can have a profound influence on splice site choice, especially within the context of short introns.
FIG. 2.
Proximal 5′ splice sites are preferred over a strong distal 5′ splice site. (A) Diagram of a modified β-globin minigene template with an upstream invariable 5′ splice site (CAG|GUAAGUgU), a downstream variable 5′ splice sites, and a short intron. (B and C) Representative autoradiograms showing a time course of the in vitro splicing assay. (B) Gel analysis of splicing with a moderately strong 5′ splice site (Cuc|GUAAGUca; MaxEnt Score = 9.6) without enhancer (left) compared to that with the same splice site activated by MS2-RS-9G8 recruitment (right). (C) Same as panel B except the splice site is stronger (CAG|GUAAGUca; MaxEnt Score = 10.9). Pre-mRNA transcript and proximally and distally spliced product bands are indicated. (D) Comparison of the proximal-to-distal 5′ splice site ratio according to splice site strength with and without MS2-RS-9G8 recruitment. Further comparison of 5′splice site sequences, strength, and splicing rates can be found in Tables 2 and 3. (E) Fractional changes in the proximal-to-distal splice site selection ratio as a consequence of moving competing 5′ splice sites into closer proximity. The bar graph illustrates that reducing the distance between competing 5′ splice sites decreases proximal splice site selection in the context of short or long introns.
TABLE 2.
In the context of competing splice sites, proximal splice site selection is the predominant splice pattern
| Construct |
Short intron |
Long Intron |
||||||
|---|---|---|---|---|---|---|---|---|
| No. | 5′ splice site sequencea | MaxEnt scoreb | Avg ratioc |
Fold changec | Avg ratio |
Fold Change | ||
| − MS2-RS-9G8 | + MS2-RS-9G8 | − MS2-RS-9G8 | + MS2-RS-9G8 | |||||
| 0 | gua|cUucuaca | −26.3 | * | * | * | * | * | * |
| 1 | CAa|GUcAagcc | −9.4 | 7.4 | 10 | 1.4 | 2.6 | 3.7 | 1.4 |
| 2 | guG|GUucuaca | −8.2 | 1.4 | 1.7 | 1.2 | 5.3 | 8.8 | 1.7 |
| 3 | gAG|GUucuaca | −1.6 | 3.2 | 3.9 | 1.2 | ** | ** | ** |
| 4 | CAG|GUucuaca | 0.7 | 5.8 | 10 | 1.7 | ** | ** | ** |
| 5 | CuG|GUcuGUAU | 3.1 | 8.7 | 18 | 2.1 | 4.9 | 9.4 | 1.9 |
| 6 | guG|GUucGUAU | 3.3 | 12 | 15 | 1.2 | ** | ** | ** |
| 7 | guG|GUucGUca | 3.3 | 5.1 | 6.6 | 1.3 | 2.7 | 5.3 | 2.0 |
| 8 | CAG|GUugGUAU | 8.1 | 33 | 68 | 2.1 | 11 | 13 | 1.2 |
| 9 | Cuc|GUAAGUca | 9.6 | 5.7 | 24 | 4.2 | ** | ** | ** |
| 10 | CAc|GUAAGUca | 10.2 | 4.6 | 5.8 | 1.3 | 5.9 | 4.9 | 0.8 |
| 11 | CAG|GUAAGUAg | 10.9 | 32 | 35 | 1.1 | 7.9 | 9.4 | 1.2 |
| 12 | CAG|GUAAGUca | 10.9 | 24 | 38 | 1.6 | ** | ** | ** |
| Distal | CAG|GUAAGUgU | 10.9 | NA | NA | NA | NA | NA | NA |
Uppercase letters indicate matches with U1 snRNA, and lowercase letters indicate mismatches with U1 snRNA.
Rows are organized according to MaxEnt scores (31).
Average ratios of proximal/distal usage and fold activation were determined from multiple repeats with less than 30% difference between experiments. *, value not different from background. **, not tested in this context. NA, not applicable.
One contributing factor to proximal splice site selection could be the distance between the competing 5′ splice sites. In the constructs that we used, the proximal and distal splice sites are separated by 180 nt, thus potentially promoting preferential proximal splicing. To address whether the distance between competing splice sites modulates proximal splice site selection, we removed approximately 100 nt from a subset of the cis-competition constructs and determined the relative selection of proximal and distal 5′ splice sites. The results clearly demonstrate that the closer proximity of competing 5′ splice sites changes the balance of 5′ splice site usage in favor of distal splicing regardless of intron length (Fig. 2E). We conclude that the relative distance between competing splice sites influences their selection. The closer together splice sites are situated, the more influential is their intrinsic U1 snRNP binding affinity.
Selective enhancement by splicing enhancers.
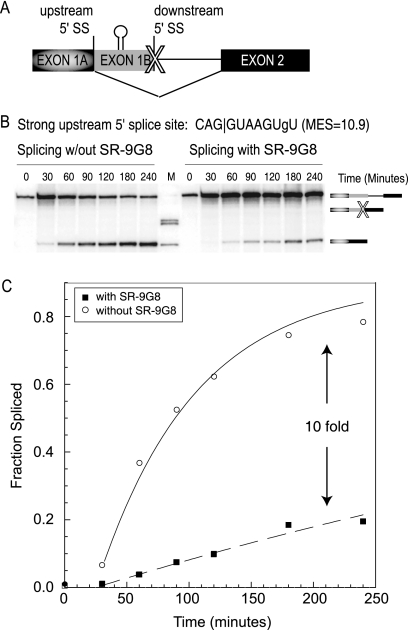
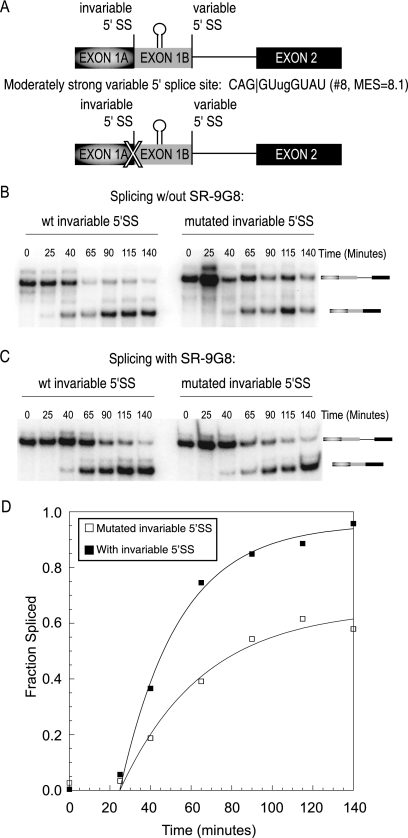
Exonic splicing enhancers are usually characterized by their enhancement of spliceosomal recruitment to regulated splice sites. Recently, it was demonstrated that splicing enhancers could act as barriers to prevent exon skipping as well (4, 15, 21). This effect is also observed for the cis-competition substrates with weak proximal splice sites (Fig. 2B; Table 2). Upon the addition of MS2-RS-9G8, distal splicing is significantly reduced, while proximal splicing is as strong as in the absence of the activator. When analyzing stronger proximal 5′ splice sites, the distally spliced band is less visible, and the inhibitory effect on distal splicing is hardly noticeable (Fig. 2C). In principle, a loss of distal splicing could be the indirect consequence of increased proximal splicing or the result of direct inhibition of distal splicing. To differentiate between these potential models, we analyzed intron removal kinetics of a pre-mRNA substrate where the proximal splice site has been mutated (Fig. 3A). The data show that the rate of distal splicing is reduced 10-fold in the presence of MS2-RS-9G8 enhancer complexes tethered downstream of the distal splice site (Fig. 3B and C). These results demonstrate that splicing enhancer complexes located between competing 5′ splice sites can act to increase the recognition of the proximal splice site while at the same time inhibiting distal splicing. Conceptually, these results are analogous to those of Ibrahim et al., which demonstrated that splicing enhancers located between competing 3′ splice sites activate the upstream and proximal 3′ splice site while inhibiting distal 3′ splice site selection (15).
FIG. 3.
MS2-RS-9G8 Recruitment inhibits upstream 5′ splice site. (A) Diagram of the β-globin minigene template with an abolished downstream 5′ splice site. The X indicates the abolished proximal splice site. (B) Representative autoradiogram showing a time course of the in vitro splicing assay. Gel analysis of splicing with the strong upstream 5′ splice site (CAG|GUAAGUgU; MaxEnt Score = 10.9) without MS2-RS-9G8 recruitment (left) compared to that with the same splice site inhibited by MS2-RS-9G8 recruitment (right) is shown. Pre-mRNA transcript and distally spliced product bands are indicated. (C) Graph comparison of the fraction spliced during the time course of the experiment.
Competing upstream 5′ splice sites function as enhancers of proximal splice site selection.
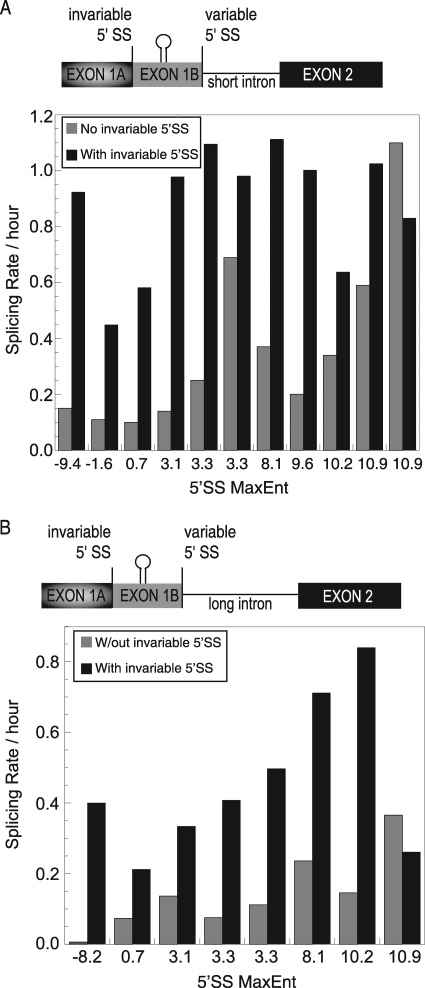
Alternative 5′ splice sites compete for a common downstream 3′ splice site. As such, the use of one 5′ splice site is expected to inhibit the use of the alternative splice site. The rate analysis described above allowed assessment of whether intron removal in the context of competing splice sites is carried out with altered kinetics compared to the noncompetitive scenario. Surprisingly, rate comparisons demonstrate that the presence of an upstream 5′ splice site significantly activates splicing of the various proximal 5′ splice sites regardless of intron length (Fig. 4). This rate increase could be the consequence of the additional nucleotide sequences that are associated with the 5′ competition constructs (the cis-competition substrates contain an extra 80 nucleotides at their 5′ end). Alternatively, this rate increase could be triggered by the presence of a functional upstream 5′ splice site. To differentiate between these possibilities, the upstream 5′ splice site sequence was abolished and splicing rates were determined (Fig. 5A). Compared to the cis-competition substrate, intron removal kinetics for the upstream splice site mutant substrate was reduced by 3-fold, consistent with the interpretation that an upstream functional splice site can act as an enhancer of proximal splicing (Fig. 5B and D). This enhancement in splicing kinetics was not apparent when a splicing enhancer complex was activated between the competing splice sites, because the canonical enhancer on its own already triggered maximal splicing efficiency (Fig. 5C). We conclude that the presence of a competing upstream 5′ splice site can function as a splicing enhancer to the downstream 5′ splice site.
FIG. 4.
Upstream 5′ splice sites activate the downstream variable 5′ splice site. (A) The graph compares splicing rates of various short intron-proximal 5′ splice sites with and without upstream activation according to splice site strength. (B) The graph compares splicing rates of various long intron-proximal 5′ splice sites with and without upstream activation according to splice site strength.
FIG. 5.
A competing upstream 5′ splice sites increases proximal splicing efficiency. (A) Diagrams of β-globin minigene templates with functional upstream and downstream 5′ splice sites (upper) compared to the isogenic construct with an abolished upstream 5′ splice site (lower). The X indicates the abolished splice site. (B) Representative autoradiogram showing a time course experiment in the absence of MS2-RS-9G8. The wild-type (wt) upstream 5′ splice site (left) is compared to the abolished upstream 5′ splice site (right). Pre-mRNA transcript and proximally spliced product bands are indicated on the right. (C) Representative autoradiogram showing an identical time course experiment in the presence of MS2-RS-9G8. (D) Quantitation of the data shown in panel B in the absence of MS2-RS-9G8.
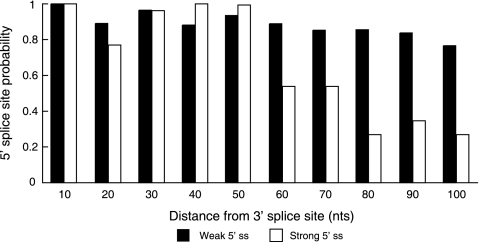
These observations suggest that a strong upstream 5′ splice site not only competes with but also enhances the downstream 5′ splice site. Currently, it is unclear which of these two competing effects dominates to promote the final outcome of a splicing reaction. Given our observation that the selection between closer splice sites is more dependent on their respective U1 snRNP binding potentials (i.e., the upstream 5′ splice site is a stronger competitor for the downstream 3′ splice site), it is possible that the upstream 5′ splice site enhancer function is distance dependent. To test this hypothesis and to gain information regarding its potential biological significance, we performed a computational analysis evaluating the distribution of potential 5′ splice sites across 5,000 constitutive exons. Interestingly, strong 5′ splice sites are enriched in these exons within 50 nt of the exon's 3′ splice site (Fig. 6). In contrast, weak 5′ splice sites are evenly distributed across the whole exon. These analyses demonstrate that strong 5′ splice sites that are not utilized as competing splice sites are overrepresented within the 5′ portions of constitutive exons, potentially functioning within these locations as enhancers for the downstream 5′ splice site. One explanation for this striking alternative 5′ splice site enrichment may be exon size limits. Human internal exons are typically larger than 75 nt, in support of the notion that cross-exon recognition is most efficient across an optimal exon length of approximately 120 nt. These considerations suggest that exon size optimization or constraints of the spliceosome may dictate whether an alternative 5′ splice site competes with or enhances downstream 5′ splice site selection.
FIG. 6.
Distribution of strong and weak potential 5′ splice sites along constitutive exons. The presence of potential 5′ splice sites in 5,000 constitutive exons was measured using the MaxEnt scoring scheme (31), where a 5′ MaxEnt score of >7 represents a strong 5′ splice site and a MaxEnt score of <0 represents a weak 5′ splice site. The y axis shows the two classes of potential 5′ splice sites binned in 10-nt bins. Each bin was normalized to 5′ splice site frequencies observed for the first 10-nt bin closest to the 3′ splice site. The x axis represents the distance of 5′ splice site bins from the 3′ splice site.
DISCUSSION
The experiments presented here describe a systematic approach to evaluate the combinatorial regulation of 5′ splice site activation. Through analysis of variable proximal splice sites, we show that 5′ splice site selection frequencies correlate well with their U1 snRNA base pairing potential. In agreement with previous results, recognition of splice sites flanking short introns (i.e., intron definition mode) is much more efficient than long intron removal. Interestingly, the proximity of competing 5′ splice sites to a pairing 3′ splice site proved to be much more influential in establishing splicing patterns than the actual 5′ splice site/U1 snRNA base pairing potential. Unexpectedly, our kinetic evaluation revealed the discovery of two new splicing regulatory functions, i.e., SR protein recruitment as a bidirectional silencer/enhancer and an upstream 5′ splice site functioning as a splicing enhancer.
5′ splice site strength and ESE activation.
Using a systematic approach, we investigated to what degree a splicing enhancer complex can activate a downstream 5′ splice site. We designed a series of 5′ splice site test substrates with variable complementarity to U1 snRNA either in the intron definition context or in the exon definition context. As anticipated from previous work, the predicted strength of interaction between the 5′ splice site and the U1 snRNA, using either free energy of binding (32) or MaxEnt scoring (31), does correlate reasonably well with the splicing efficiencies observed (Table 1). This gives merit to the value of predicting splicing efficiency through U1 snRNA binding to the 5′ splice site. However, the magnitude of the observed changes in splicing rates do not correlate linearly with expected differences based on base pairing potential (the difference in U1 snRNA binding potential between splice sites 3 and 11 is ∼1 × 106-fold, while the measured rates differ only by 10-fold), supporting the notion that the affinity of U1 snRNP to 5′ splice sites is also augmented by protein/RNA interactions (7). In the presence of the splicing enhancer complex MS2-RS-9G8, splicing rates increased up to 4-fold in the intron definition context. In general, stronger splice sites spliced efficiently even in the absence of enhancer complexes at close to maximal in vitro splicing rates (12).
When the splicings of pre-mRNAs harboring differently sized introns were compared, a significant intron length dependency was observed, demonstrating that a greater distance from the downstream 3′ splice site results in significantly lower splicing kinetics. Thus, it is tempting to speculate that the selection of intron-defined splice sites, i.e., splice sites recognized within one kinetic step (8), may require different sets or types of regulatory elements than splice sites that are recognized independently from each other, i.e., splice sites that are outside the intron definition mode. Given spatial constraints, intron-defined alternative splicing may depend much more on splicing signals within the immediate surroundings, while exhibiting less dependence on more distant splicing signals.
Proximity effect.
Under the experimental conditions used, the “proximity effect” proved to be more influential in dictating splice site choice than anticipated (Fig. 2 and Table 2). The results suggest that the “proximity effect” should carry significant weight when predicting alternative splice site preference. What is unclear is to what degree the distance between competing splice sites controls the influence of the “proximity effect”. Clearly, moving the competing splice sites closer to each other resulted in a significant shift in distal splice site selection (Fig. 2E). Furthermore, recent work showed that the use of competing splice sites within very close proximity (in this case 16 nucleotides) is more dependent on the U1 snRNA binding potential than what was observed here (24). One attractive explanation for these distance effects might be that longer sequences separating competing splice sites have a higher probability of harboring splicing enhancers, thereby further increasing the selection of the proximal 5′ splice site. Intron length also influences splice site selection, as our data are consistent with the idea that longer introns dilute the influence of the proximity effect.
Directional inhibition through SR protein binding.
Previous work has shown that SR proteins can interfere with splicing (5, 15, 27). Our results obtained from the 5′ splice site competition experiments also indicate that an RS domain enhancer can induce splicing interference, albeit at a different location along the pre-mRNA. These results are reminiscent of observations demonstrating that the exonic or intronic location of SR proteins induces activation or repression (15, 17). Therefore, it is evident that splicing enhancer elements that recruit SR proteins direct spliceosome activity according to its pre-mRNA context. The data presented here suggest that these enhancer complexes exhibit dual functionality and bidirectionality in which pre-mRNA-bound SR proteins preferentially recruit the spliceosome to the downstream 5′ splice site and inhibit its recruitment to the upstream 5′ splice site. Recent computational work gives further relevance to the dual functionality of splicing elements. The role of splicing regulatory elements in the determination of exon/intron boundaries conforms to the more complex integrated splicing code recently presented and agrees with the results of unpublished fluorescence-activated screens (30). Especially noticeable is the context dependence of enhancer and silencer elements (10). How the observed apparent directionality is achieved is currently unclear. However, one can envision mechanisms whereby the interaction between SR proteins and the spliceosome result in preferential deposition of spliceosomal components downstream of the enhancer sequence, thereby sequestering the spliceosome from the competing upstream 5′ splice site.
Upstream 5′ splice site sequence functions as enhancer.
Using trans- and cis-splicing assays, it has been demonstrated that a downstream 5′ splice site could enhance the efficiency of intron removal (3, 6). This enhancement was SR protein dependent, suggesting that the presence of SR proteins mediates interactions between U1 snRNP and U2 snRNP (3). Our kinetic analysis of competing 5′ splice sites resulted in the unexpected discovery that the presence of a competing upstream 5′ splice site increased the rate of proximal splicing. Even though the U1 snRNP binding potential of proximal 5′ splice sites was significantly lower than that of the distal splice site, mainly proximal splicing was observed. However, the presence of the upstream 5′ splice site surprisingly accelerated proximal splicing. These results demonstrate that an upstream 5′ splice site can function as an enhancer of splicing without being used as a significant target for the splicing reaction itself. It is unclear how such an enhancement could be achieved. Perhaps an upstream 5′ splice site initially facilitates the recruitment of additional spliceosomal components. However, during the establishment of splicing patterns, the proximity of the competing 5′ splice sites to the 3′ splice site dominates the selection process in establishing committed splice site pairing. As argued by our computational analysis of constitutive exons (Fig. 6), such scenarios could be envisioned if a competing 5′ splice site is situated too close to an upstream 3′ splice site, essentially rendering upstream 5′ splice selection improbable due to exon size restrictions. Alternatively, even though the sequence of the distal 5′ splice site looks like a functional splice site, it may not act like one due to incomplete signals or adjacent inhibitory elements. Independent of these considerations, the utilization of the distal splice site in the context of the abolished proximal 5′ splice site argues strongly for a direct interaction between the distal 5′ splice site and U1 snRNP. It is possible that the results described here are also transferable to situations where competition exists between functional 3′ splice sites. In other words, the recruitment of U2 snRNP in the context of competing 3′ splice sites may have enhancing functions similar to those observed here for competing 5′ splice sites. In such a scenario, one would predict that a downstream (distal) 3′ splice site could enhance splice site selection at the proximal 3′ splice site.
The systematic analysis carried out here illustrates the importance of evaluating elements of splicing regulation in combination. These elements include splice site strength and competition, the proximity effect between competing splice sites, intronic size, and SR protein mediation by directionally enhancing or inhibiting splice site recognition to regulate the efficiency of splicing. Ultimately, the integration of a combinatorial and context-dependent approach to study pre-mRNA splicing may lead to a clearer understanding of this complex process and suggest additional criteria for a more accurate and predictive splicing code.
Acknowledgments
We are grateful to Matthew Kotlajich for technical assistance and to members of the Hertel laboratory for helpful comments on the manuscript. HeLa cells were obtained from the National Cell Culture Center (Minneapolis, MN).
This work was supported by NIH grants GM 62287 (K.J.H.) and T15LM007443 (P.J.S.).
Footnotes
Published ahead of print on 1 February 2010.
REFERENCES
- 1.Black, D. L. 1995. Finding splice sites within a wilderness of RNA. RNA 1:763-771. [PMC free article] [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 3.Boukis, L. A., N. Liu, S. Furuyama, and J. P. Bruzik. 2004. Ser/Arg-rich protein-mediated communication between U1 and U2 small nuclear ribonucleoprotein particles. J. Biol. Chem. 279:29647-29653. [DOI] [PubMed] [Google Scholar]
- 4.Buratti, E., C. Stuani, G. De Prato, and F. E. Baralle. 2007. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 35:4359-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandler, D. S., J. Qi, and W. Mattox. 2003. Direct repression of splicing by transformer-2. Mol. Cell. Biol. 23:5174-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiara, M. D., and R. Reed. 1995. A two-step mechanism for 5′ and 3′ splice site pairing. Nature 375:510-513. [DOI] [PubMed] [Google Scholar]
- 7.Du, H., and M. Rosbash. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419:86-90. [DOI] [PubMed] [Google Scholar]
- 8.Fox-Walsh, K. L., Y. Dou, B. J. Lam, S. P. Hung, P. F. Baldi, and K. J. Hertel. 2005. The architecture of pre-mRNAs affects mechanisms of splice site pairing. Proc. Natl. Acad. Sci. U. S. A. 102:16176-16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox-Walsh, K. L., and K. J. Hertel. 2009. Splice site pairing is an intrinsically high fidelity process. Proc. Natl. Acad. Sci. U. S. A. 106:1766-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goren, A., O. Ram, M. Amit, H. Keren, G. Lev-Maor, I. Vig, T. Pupko, and G. Ast. 2006. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol. Cell 22:769-781. [DOI] [PubMed] [Google Scholar]
- 11.Hertel, K. J. 2008. Combinatorial control of exon recognition. J. Biol. Chem. 283:1211-1215. [DOI] [PubMed] [Google Scholar]
- 12.Hicks, M. J., B. J. Lam, and K. J. Hertel. 2005. Analyzing mechanisms of alternative pre-mRNA splicing using in vitro splicing assays. Methods 37:306-313. [DOI] [PubMed] [Google Scholar]
- 13.Hiller, M., Z. Zhang, R. Backofen, and S. Stamm. 2007. Pre-mRNA secondary structures influence exon recognition. PLoS Genet 3:e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holste, D., G. Huo, V. Tung, and C. B. Burge. 2006. HOLLYWOOD: a comparative relational database of alternative splicing. Nucleic Acids Res. 34:D56-D62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim, E. C., T. D. Schaal, K. J. Hertel, R. Reed, and T. Maniatis. 2005. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. U. S. A. 102:5002-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931-945. [DOI] [PubMed] [Google Scholar]
- 17.Kanopka, A., O. Muhlemann, and G. Akusjarvi. 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381:535-538. [DOI] [PubMed] [Google Scholar]
- 18.Kim, N., A. V. Alekseyenko, M. Roy, and C. Lee. 2007. The ASAP II database: analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 35:D93-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornblihtt, A. R. 2005. Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 17:262-268. [DOI] [PubMed] [Google Scholar]
- 20.Kuo, H. C., F. H. Nasim, and P. J. Grabowski. 1991. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science 251:1045-1050. [DOI] [PubMed] [Google Scholar]
- 21.Lim, S. R., and K. J. Hertel. 2001. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem. 276:45476-45483. [DOI] [PubMed] [Google Scholar]
- 22.Reed, R., and T. Maniatis. 1986. A role for exon sequences and splice site proximity in splice site selection. Cell 46:681-690. [DOI] [PubMed] [Google Scholar]
- 23.Roca, X., A. J. Olson, A. R. Rao, E. Enerly, V. N. Kristensen, A. L. Borresen-Dale, B. S. Andresen, A. R. Krainer, and R. Sachidanandam. 2008. Features of 5′-splice site efficiency derived from disease-causing mutations and comparative genomics. Genome Res. 18:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roca, X., R. Sachidanandam, and A. R. Krainer. 2005. Determinants of the inherent strength of human 5′ splice sites. RNA 11:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shapiro, M. B., and P. Senapathy. 1987. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 15:7155-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepard, P. J., and K. J. Hertel. 2008. Conserved RNA secondary structures promote alternative splicing. RNA 14:1463-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, Y., and J. L. Manley. 2007. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol. Cell 28:79-90. [DOI] [PubMed] [Google Scholar]
- 28.Thanaraj, T. A., S. Stamm, F. Clark, J. J. Riethoven, V. Le Texier, and J. Muilu. 2004. ASD: the Alternative Splicing Database. Nucleic Acids Res. 32(Database issue):D64-D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, E. T., R. Sandberg, S. Luo, I. Khrebtukova, L. Zhang, C. Mayr, S. F. Kingsmore, G. P. Schroth, and C. B. Burge. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Z., and C. B. Burge. 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14:802-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo, G., and C. B. Burge. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11:377-394. [DOI] [PubMed] [Google Scholar]
- 32.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]