Abstract
Foamy virus evolution closely parallels that of the host species, indicating virus-host coadaptation. We studied simian foamy viruses (SFVs) from common marmosets, spider monkeys, and squirrel monkeys, New World monkey (NWM) species that share geographic ranges. The TRIM5α protein from each of these NWM species inhibited the replication of at least one of the SFVs associated with the other two species but did not affect the replication of its own SFV. Thus, TRIM5α has potentially shaped the evolution of SFVs in NWM hosts. Conversely, SFVs may have influenced the evolution of TRIM5 variants in New World primates.
The phylogeny of foamy viruses (FVs), or spumaretroviruses, generally mirrors that of the host species, suggesting coadaptation of virus and host (18). Simian foamy viruses (SFVs) have been isolated from different nonhuman primates (NHPs) (8).
Mammals have evolved proteins, such as TRIM5α, that mediate innate immunity against retroviruses (16). TRIM5α proteins vary in a species-specific manner that can influence the potency of inhibition of infection by particular retroviruses (10, 15, 22, 23). Lineage-specific positive selection has operated on TRIM5 in Old World and New World primates (12, 14). The existence of extant and endogenous retroviruses in Old World primates implicates past retroviral infections in the selective process (14). However, the infectious agents responsible for the selection of TRIM5 variants in New World monkeys (NWMs) have not been identified. Here we examine the relationship between the SFVs and the TRIM5α proteins of NWMs.
While culturing peripheral blood mononuclear cells (PBMCs) from common marmosets (Callithrix jacchus), we detected reverse transcriptase (RT) activity in the supernatant, indicating the presence of a retrovirus in these cell cultures. This retrovirus replicated at a low level and was transmissible to other common marmoset PBMCs and to canine Cf2Th cells. Infection of Cf2Th cells resulted in extensive cytopathic effects (syncytia) and rapid demise of the cultures. In contrast, this retrovirus did not induce many syncytia in marmoset PBMCs, and virus production was sustained for several weeks. The RT of the marmoset retrovirus exhibited higher activity in the presence of Mn2+ than in that of Mg2+ (data not shown).
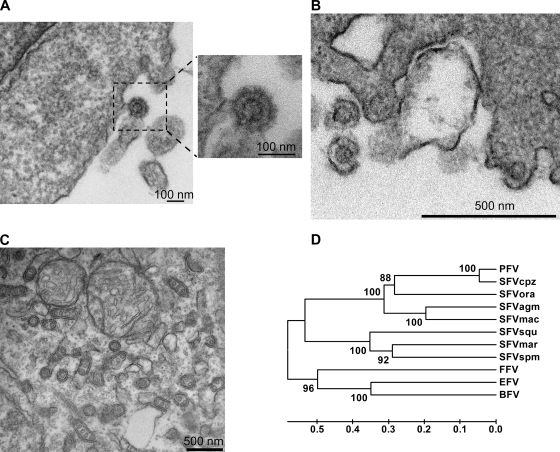
Electron microscopy of ultrathin sections of Cf2Th cells infected with the common marmoset retrovirus revealed the presence of 100- to 110-nm viral particles with 10- to 15-nm envelope spikes and a 55- to 60-nm capsid (Fig. 1A and B). In addition to budding and extracellular particles, we observed naked capsid-like structures in intracellular compartments (Fig. 1C); these were not seen in uninfected control cells. The observed virus ultrastructure was typical of that of FVs (3), suggesting that this marmoset retrovirus could be a foamy virus.
FIG. 1.
Morphological and phylogenetic characterization of the common marmoset retrovirus. (A and B) Extracellular particles and particles budding from the plasma membrane of Cf2Th cells infected with the common marmoset retrovirus are shown. (C) Naked capsids in an intracellular compartment of the Cf2Th cells infected with the common marmoset retrovirus are shown. For panels A to C, thin-sectioned samples were examined with a Tecnai G2 Spirit BioTWIN transmission electron microscope. (D) Phylogenetic analysis of the Gag amino acid sequences from the indicated FVs was conducted with MEGA version 4 software (19). The phylogenetic tree was built using the unweighted pair group method (UPGMA) with 1,000 bootstrap repetitions. The percentage of the replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to each branch. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances are represented in units of amino acid substitutions per site. PFV, prototype primate foamy virus; SFVcpz, simian foamy virus of chimpanzees; SFVora, simian foamy virus of orangutans; SFVmac, simian foamy virus of rhesus macaques.
To clone the proviral DNA of the marmoset retrovirus and confirm its identity as a FV, we designed primers that target genomic regions that are well conserved in different SFVs (see Fig. S1A in the supplemental material). Using these primers, we amplified by PCR, cloned, and sequenced a few fragments from the DNA of the infected common marmoset PBMCs. Sequence analysis indicated that the proviral DNA belonged to a new SFV and exhibited 60 to 70% identity with that of spider monkey FV (SFVspm), the only sequenced NWM FV (20). We used these partial sequences to design new primers to amplify the full provirus of this common marmoset FV (SFVmar). Three PCR fragments containing the long terminal repeats (LTRs) and the 5′ and 3′ halves of SFVmar were cloned and sequenced.
To study the evolutionary relationship of SFVmar to other foamy viruses, we constructed a phylogenetic tree, using the Gag, Pol, or Env amino acid sequences of different FVs. To add to this phylogenetic tree, the provirus of SFVsqu (a FV of the squirrel monkey, a NWM) (4) was amplified from infected Cf2Th cells and sequenced. The phylogenetic tree for Gag is shown in Fig. 1D. SFVmar was, as expected, most closely related to other NWM FVs (SFVspm and SFVsqu). Progressively more distant relatedness to the FVs of Old World primates and to the nonprimate FVs (feline foamy virus [FFV], bovine foamy virus [BFV], and equine foamy virus [EFV]) was observed. Thus, as expected (13, 18), FV phylogeny parallels that of the host species. Similar results were obtained when the Pol and Env sequences were analyzed (data not shown). Comparisons of nucleotide and amino acid sequence similarities among FVs (see Table S1 in the supplemental material) indicated that SFVmar is a new FV.
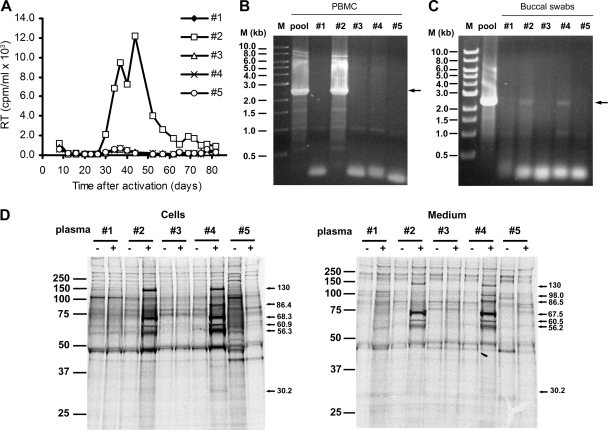
Our initial common marmoset PBMC culture was a pool of PBMCs from different common marmosets. To investigate the frequency of SFVmar infection, the PBMCs of five common marmosets were individually cultured. We detected RT activity only in the supernatant of PBMCs cultured from marmoset number 2 (Fig. 2A). Genomic DNA from the cultured PBMCs of marmoset number 2 was analyzed by PCR, using primers that specifically target the SFVmar pol gene (see Fig. S1C in the supplemental material). We detected a specific band of about 2.3 kb (the expected size of the PCR product) in the DNA derived from the PBMCs of monkey number 2 as well as from our initial pool of PBMCs (Fig. 2B). A specific 2.3-kb band was also amplified from the buccal swab DNAs of marmoset number 2 and marmoset number 4 (Fig. 2C). Apparently, marmoset number 2, and perhaps marmoset number 4, are infected with SFVmar.
FIG. 2.
Detection of SFVmar in different common marmosets. (A) RT activity in the culture supernatant is shown as a function of time after isolation and activation (in 1 μg/ml phytohemagglutinin [PHA-P] for 3 days) of the PBMCs from five common marmosets (number 1 to number 5). (B) The SFVmar Pol region was PCR amplified from DNA isolated from PBMCs prepared from the original pool of blood from eight common marmosets (pool) and from the blood of five individual common marmosets in the original pool (numbers 1 to 5). The PCR primers used for the amplification are SFVmar3440-F and SFVmar5754-R (see Fig. S1C in the supplemental material). The arrow indicates the expected authentic PCR product. (C) Buccal swabs from the five individual marmosets (number 1 to number 5) were used to prepare DNA, which was PCR amplified with the same primers as described for panel B. As a control, DNA isolated from the pooled PBMCs from eight common marmosets (pool) was used. M, 1-kb DNA ladder (New England Biolabs). (D) Cf2Th cells infected with SFVmar were radiolabeled with [35S]methionine-[35S]cysteine for 16 h. Radiolabeled SFVmar proteins from cell lysates and medium were immunoprecipitated with plasma from marmosets number 1 to number 5. The molecular weight markers are shown on the left of each gel, and the calculated molecular mass of each specific band (in kilodaltons) is shown on the right of the gels. −, uninfected cells; +, infected cells.
To analyze the immunoreactivity of the plasma from these common marmosets with SFVmar proteins, we metabolically radiolabeled virus-infected cells with [35S]methionine and [35S]cysteine. Cell lysates and the detergent-lysed virion proteins were precipitated with plasma from each of the five marmosets studied. Plasma from monkeys number 2 and number 4 precipitated several proteins with molecular weights expected for SFVmar products; these proteins were not precipitated from uninfected cells (Fig. 2D). Thus, at least two of the five monkeys tested were seropositive for antibodies directed against SFVmar proteins.
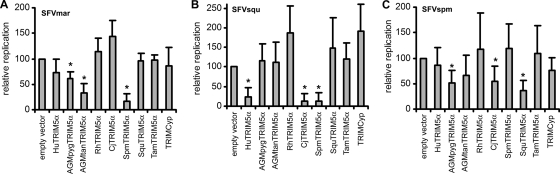
The TRIM5α proteins of two NWMs (tamarins and marmosets) can inhibit infection by FVs of Old World primates (21). We asked whether TRIM5α proteins from different species, especially from NWMs, were able to restrict SFVs from NWMs. Equivalent amounts of SFVmar, SFVsqu, and SFVspm were incubated with Cf2Th cells expressing TRIM5α proteins from different species. Figure 3 shows the amount of virus produced from these cells 3 or 4 days after incubation with the virus, relative to that produced from control cells containing the empty control vector. We observed that in cells expressing the TRIM5α protein (SpmTRIM5α) from the spider monkey, a NWM, the level of SFVmar replication was only 16% of that in the control cells (Fig. 3A). SFVmar infection was not apparently inhibited by TRIM5α from common marmosets, tamarins, and squirrel monkeys, other NWMs. African green monkey (AGMpyg and AGMtan) TRIM5α proteins, but not that of the rhesus monkey, another Old World monkey (OWM), modestly inhibited SFVmar replication.
FIG. 3.
Restriction of NWM SFV infection by TRIM5α and TRIMCyp proteins. Cf2Th cells stably expressing TRIMCyp from owl monkeys or comparable levels of TRIM5α proteins from different primate species or transduced with the empty control vector were infected with SFVmar (A), SFVsqu (B), or SFVspm (C). Cell supernatants were collected 3 or 4 days later and analyzed for RT activity. Values represent the RT level of each virus at 3 to 4 days postinfection relative to that seen in the cells containing the empty vector. Values are the means derived from 5 to 11 different experiments, each performed in duplicate. Error bars represent the standard deviation. Significant reductions in virus replication, determined by using the Bonferroni t test (2) and indicated by a P value of 0.0056, are marked by asterisks. CjTRIM5α (Callithrix jacchus TRIM5α) cDNA was prepared using the following primers: CjTrim-F, 5′-GGGAATTCATGGCTTCCAGAATCCTGGT-3′, and CjTrim-R, 5′-GGAGCGGCCGCATCAAGAGCTTGGTGA-3′. In the process of producing the cDNA, an N-terminal hemagglutinin (HA) epitope tag was added to the encoded CjTRIM5α protein to facilitate detection. Hu, human; Rh, rhesus monkey; AGMpyg, African green monkey (Chlorocebus pygerythrus); AGMtan, African green monkey (Chlorocebus tantalus); Spm, spider monkey; Cj, Callithrix jacchus (common marmoset); Squ, squirrel monkey; Tam, tamarin.
The replication of SFVsqu was greatly diminished by spider monkey TRIM5α (12% of that of the control) (Fig. 3B). SFVsqu replication was also inhibited by marmoset TRIM5α (12% of that of the control) and human TRIM5α (23% of that of the control).
The inhibition of SFVspm replication by TRIM5α proteins was not as strong as that seen for SFVmar and SFVsqu; however, we observed a reduction in SFVspm replication in cells expressing squirrel monkey TRIM5α (36% of that of the control) and modest reductions in cells expressing AGM and marmoset TRIM5α proteins (Fig. 3C). Our results indicate that the replication of particular SFVs indigenous to the New World is inhibited by specific TRIM5α proteins from NWMs.
Until now the only foamy virus cloned from a NWM was SFVspm (20); however, the presence of FVs in other NWMs, such as common marmosets, capuchins, and squirrel monkeys, has been described (3, 4, 7). Here we describe the cloning and characterization of a FV from common marmosets (SFVmar). This new FV is, as expected, phylogenetically closer to the SFVs of spider and squirrel monkeys (two NWM species) than to other characterized FVs from Old World primates. Although our sample is small, the observed incidence of infection/seropositivity suggests that exposure to SFV is frequent in common marmosets, at least in captivity.
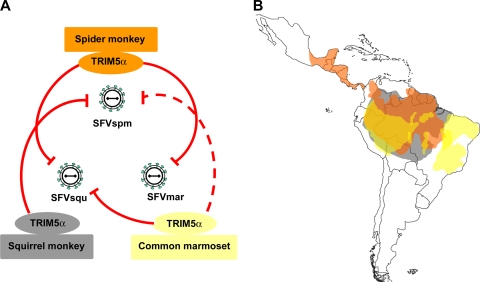
Here we show that the replication of SFVs indigenous to the New World is inhibited by NWM TRIM5α variants. The observed patterns of New World SFV sensitivity and resistance to NWM TRIM5α proteins provide insights into the coevolution of FVs and their primate hosts. Although cross-species transmission of FVs does occur, phylogenetic studies have revealed ancient cospeciation of simian FVs and their hosts (13, 18). Thus, in the majority of cases, FVs adapt to one or a small number of host species. Although spider monkeys, squirrel monkeys, and common marmosets retain TRIM5α proteins that are able to restrict some New World SFVs, these TRIM5α proteins exhibited no antiviral activity against the SFVs indigenous to the species (Fig. 4A). In successfully adapting to a new host species, each SFV has apparently evolved to minimize the detrimental impact of the particular TRIM5α protein encountered. Such escape from TRIM5α restriction may involve changes in the capsid, the binding target for TRIM5α (5, 6, 9, 11, 17).
FIG. 4.
TRIM5α restriction patterns and habitats of New World monkeys. (A) The ability of the TRIM5α proteins from three NWMs to inhibit infection with SFVs from these monkey species is depicted by red lines; a weaker level of restriction is depicted by a broken line. The figure is based upon the data shown in Fig. 3. Note that none of the TRIM5α proteins from these monkeys inhibits the replication of the SFV indigenous to the species. (B) The ranges of the three NWMs shown in panel A are shaded as follows: orange, spider monkeys; gray, squirrel monkeys; and yellow, marmosets. Note the large areas of overlap of these ranges.
Throughout primate evolution, the capsid-interacting B30.2(SPRY) domain of TRIM5α has been subjected to strong positive selection (12, 14). Even after the separation of NWMs from Old World primates, NWM TRIM5α proteins continued to diversify. A striking example is the length expansion of the B30.2(SPRY) V3 variable loop in the NWM TRIM5α proteins (14). Of note, a sequence triplication that occurred in the spider monkey lineage created a TRIM5α protein with the longest known V3 loop (96 residues); this feature may contribute to the strong level of restriction of SFVmar and SFVsqu by spider monkey TRIM5α. If past or current retroviral infections drove TRIM5 evolution in NWMs, the possible identity of the retroviruses has been unclear. Although the TRIM5α proteins of some NWM species can restrict infections with different retroviruses, like simian immunodeficiency virus (SIV), N-tropic murine leukemia virus (N-MLV), Mason-Pfizer monkey virus (MPMV), and Old World primate FVs (1, 15, 21), NWMs are not known to be exposed to these viruses. Our results hint that FVs may have influenced TRIM5α diversification in NWM species. Marmosets, squirrel monkeys, and spider monkeys occupy overlapping geographic ranges in South America (Fig. 4B), making likely the possibility that exposure to the SFVs of these other species occurred at some point in time. Any detrimental effects of heterologous SFV infection could have selected for more potently restricting TRIM5α variants.
Nucleotide sequence accession numbers.
The complete sequences of SFVmar and SFVsqu have been deposited in GenBank under accession numbers GU356395 and GU356394, respectively.
Supplementary Material
Acknowledgments
We thank Yvette McLaughlin and Elizabeth Carpelan for manuscript preparation. We acknowledge Angela Carville and the New England Regional Primate Center for providing the common marmoset blood and buccal swabs. We acknowledge ATCC for the SFVsqu and SFVspm viruses.
This study was supported by the National Institutes of Health (grants AI 063987 and AI 076094 and Center for AIDS Research Award AI 06354), the International AIDS Vaccine Initiative, and the late William F. McCarty-Cooper.
Footnotes
Published ahead of print on 3 February 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Diehl, W. E., E. Stansell, S. M. Kaiser, M. Emerman, and E. Hunter. 2008. Identification of postentry restrictions to Mason-Pfizer monkey virus infection in New World monkey cells. J. Virol. 82:11140-11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glantz, S. A. 1981. The special case of two groups: the t test, p. 63-93. In Primer of biostatistics. McGraw-Hill Book Company, New York, NY.
- 3.Hooks, J. J., and C. J. Gibbs, Jr. 1975. The foamy viruses. Bacteriol. Rev. 39:169-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston, P. B. 1971. Taxonomic features of seven serotypes of simian and ape foamy viruses. Infect. Immun. 3:793-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kar, A. K., F. Diaz-Griffero, Y. Li, X. Li, and J. Sodroski. 2008. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J. Virol. 82:11669-11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langelier, C. R., V. Sandrin, D. M. Eckert, D. E. Christensen, V. Chandrasekaran, S. L. Alam, C. Aiken, J. C. Olsen, A. K. Kar, J. G. Sodroski, and W. I. Sundquist. 2008. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J. Virol. 82:11682-11694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marczynska, B., C. J. Jones, and L. G. Wolfe. 1981. Syncytium-forming virus of common marmosets (Callithrix jacchus jacchus). Infect. Immun. 31:1261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meiering, C. D., and M. L. Linial. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U. S. A. 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U. S. A. 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweizer, M., and D. Neumann-Haefelin. 1995. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 207:577-582. [DOI] [PubMed] [Google Scholar]
- 14.Song, B., B. Gold, C. O'Huigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J. Virol. 79:6111-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song, B., H. Javanbakht, M. Perron, D. H. Park, M. Stremlau, and J. Sodroski. 2005. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J. Virol. 79:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 17.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U. S. A. 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Switzer, W. M., M. Salemi, V. Shanmugam, F. Gao, M. E. Cong, C. Kuiken, V. Bhullar, B. E. Beer, D. Vallet, A. Gautier-Hion, Z. Tooze, F. Villinger, E. C. Holmes, and W. Heneine. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376-380. [DOI] [PubMed] [Google Scholar]
- 19.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 20.Thümer, L., A. Rethwilm, E. C. Holmes, and J. Bodem. 2007. The complete nucleotide sequence of a New World simian foamy virus. Virology 369:191-197. [DOI] [PubMed] [Google Scholar]
- 21.Yap, M. W., D. Lindemann, N. Stanke, J. Reh, D. Westphal, H. Hanenberg, S. Ohkura, and J. P. Stoye. 2008. Restriction of foamy viruses by primate Trim5alpha. J. Virol. 82:5429-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 23.Ylinen, L. M., Z. Keckesova, S. J. Wilson, S. Ranasinghe, and G. J. Towers. 2005. Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5alpha alleles. J. Virol. 79:11580-11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.