Abstract
Human α-defensins are evolutionarily conserved effectors of the innate immune response with broadly acting antibacterial activity. Their role in antiviral immunity is less well understood. We previously showed that these antimicrobial peptides are potent inhibitors of human adenovirus infection. Based on biochemical studies and indirect evidence from confocal microscopy, we proposed that defensins bind to and stabilize the virus capsid and neutralize infection by preventing the release of the endosomalytic protein VI. To determine whether defensin action also restricts exposure of the viral genome, we developed a system to evaluate adenovirus uncoating during cell entry by monitoring the exposure of BrdU-labeled viral genomes. This assay allowed us to determine the kinetics of uncoating of virus particles in single cells. Using this assay, we now provide direct evidence that human α-defensins block adenovirus infection by preventing uncoating during cell entry.
The protein coat of the adenovirus (AdV) particle protects the virus genome from the extracellular environment; however, this protective coat must be shed during AdV entry to initiate infection in a process termed “uncoating.” The molecular basis for AdV uncoating is poorly understood. Current models of AdV entry, based primarily on studies of species B and C human AdVs (HAdVs), postulate that the virus capsid uncoats in discrete steps (10). After binding to high-affinity attachment receptors (e.g., CAR and CD46) and to integrin coreceptors, the virus particle is internalized by clathrin-mediated endocytosis (1, 15). As a consequence of receptor engagement and possibly of other cellular triggers, the pentons at the 12 icosahedral vertices, each comprised of a noncovalently coupled complex of penton base and fiber, are shed at or near the virus surface (10, 19). This process may involve either the sequential dissociation of fiber and then penton base or the dissociation of the penton complex as a unit. Subsequent uncoating, including the release of the internal capsid proteins IIIa, VI, and VIII and some of the major capsid protein (hexon), occurs in the endosome and has been proposed to be triggered in part by acidification during endosomal maturation (10). The released viral proteins, likely through the endosomalytic activity of protein VI (32), mediate endosome disruption and allow the partially uncoated capsid to enter the cytoplasm. The released particle traffics in the cytoplasm along microtubules to the microtubule-organizing center (MTOC) and then to the nuclear pore complex, where additional uncoating events occur to allow translocation of the viral DNA into the nucleus (15).
The term uncoating, defined conceptually here as the removal of the protein capsid shell from the viral genome to the extent that is required to permit viral gene expression and replication, has also been used extensively in the literature as both a functional and a phenotypic descriptor. Uncoating has been measured as an increase in the accessibility of the viral DNA to DNase or DNA-sensitive dyes, either upon recovery from infected cells or in vitro (14, 16, 21, 22, 29, 30, 32). Alternatively, changes in the density of viral protein/DNA complexes or the association of capsid proteins with, or dissociation from, the nucleic acid-containing core have been determined (14, 16, 22, 26, 27, 29, 32). In some cases, these biochemical measurements have been correlated, directly or indirectly, with morphological changes observed by electron microscopy or immunofluorescence (6, 22, 24, 29); however, the structural basis for the various assays and phenotypes of uncoating are frequently unknown. In particular, existing assays that measure DNA accessibility cannot be used to distinguish complete or partial DNA dissociation from the capsid from alterations in the integrity of the capsid that nonetheless maintain the genome within a protein shell. Based on electron microscopy studies and the efficiency of gene delivery by AdV, it has been postulated that infectious particles retain sufficient integrity to sequester the viral genome prior to its docking at the nuclear pore complex; however, the contribution of DNA-capsid dissociation to nonproductive infectious pathways has not been assessed.
Components of the adaptive immune system (e.g., neutralizing antibodies) have been shown to block HAdV infection by impacting one or more steps in this entry pathway (26, 33). We have recently shown that effectors of the innate immune response, members of a family of human antimicrobial peptides known as α-defensins, inhibit HAdV infection at low micromolar concentrations (27). We provided evidence that these naturally occurring antiviral compounds block infection by binding to HAdV outside the cell and preventing escape of the internalized virus/defensin complex from the endosome. This conclusion was based on the failure of HAdV to mediate the translocation of macromolecules, such as the ribotoxin α-sarcin, into the cytoplasm in the presence of inhibitory concentrations of defensin. Consistent with this finding, defensins restricted release of the putative endosomalytic protein VI. Moreover, at late time points postinfection virus particles were strongly colocalized with lysosomes rather than the nucleus. Based on in vitro biochemical studies, we postulated that altered virus trafficking is due to a failure of the virus to uncoat upon defensin binding to the capsid. To test this hypothesis, we developed a system to detect HAdV uncoating in single cells by immunofluorescence based on the prior incorporation of a unique epitope (the thymidine analog 5-bromo-2-deoxyuridine [BrdU]) into the viral genome. This approach allows us to monitor the kinetics of uncoating of HAdV particles at low multiplicities of infection (MOIs) without a requirement for radiolabeling of the viral proteins or nucleic acids. This study provides strong evidence to support a model in which α-defensins prevent HAdV infection by blocking virus uncoating and genome exposure during cell entry.
MATERIALS AND METHODS
Cells and viruses.
Tissue culture reagents were obtained from Invitrogen (Carlsbad, CA). Human A549 cells were from ATCC and were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Omega Scientific, Tarzana, CA), 10 mM HEPES, 4 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1 mM nonessential amino acids (complete DMEM). Stable 293 cells overexpressing the human β5 integrin subunit (293β5) were created by transfecting 293 cells (ATCC) with a cytomegalovirus (CMV) promoter-driven expression plasmid containing the human β5 gene and a neomycin resistance gene (pCDNA3/β5; a gift from David Cheresh, University of California, San Diego, CA). Transfected cells were selected for G418 resistance (700 μg/ml) and sorted for high levels of integrin expression, using an antibody against human β5 integrin (Millipore, Billerica, MA).
HAdV-2p was obtained from ATCC. The temperature-sensitive mutant of HAdV type 2 (HAdV-2), HAdV-2ts1, was obtained from Joseph Weber (University of Sherbrooke, Quebec, Canada). The replication-defective HAdV-5 vector used in these studies has an E1/E3 deletion and contains a CMV promoter-driven enhanced green fluorescent protein (eGFP) reporter gene cassette.
HAdV-5P137L was created by replacing the codon CCC (proline) with CTC (leucine) by recombineering (31) in a bacterial artificial chromosome (BAC) construct (pAd5-GFPn1; a gift from Zsolt Ruzsics, Max von Pettenkofer Institute, Munich, Germany) (H. Wodrich, D. Henaff, B. Jammart, C. Segura-Morales, S. Seelmeier, O. Coux, Z. Ruzsics, C. Wiethoff and E. J. Kremer, submitted for publication) containing the entire genome of a HAdV-5 vector with an E1/E3 deletion (derived from AdEasy; Qbiogene, Carlsbad, CA) expressing eGFP-n1 (derived from pEGFP-N1; Clontech, Mountain View, CA). The fidelity of the mutated construct (pAd5GFPn1-P137L) was verified by sequencing the recombineered region and by restriction digestion. 293β5 cells were transfected with the large PacI restriction fragment of pAd5GFPn1-P137L, using Lipofectamine 2000 (Invitrogen). Transfected cells were maintained at 33°C until visible, eGFP-positive plaques formed. The lysate from these transfected cells was serially passaged on 293β5 cells maintained at 33°C until enough virus was produced for purification by CsCl density gradient ultracentrifugation as described below. The identity of the final virus stock was confirmed by restriction digestion, and a PCR product from this stock was sequenced to verify the presence of the engineered mutation.
Virus production and BrdU labeling.
HAdV-2 and the wild-type HAdV-5 vector were propagated by infecting 293β5 cells with 300 particles/cell. When a complete cytopathic effect (CPE) was observed (approximately 48 to 72 h postinfection), cells were harvested, concentrated by low-speed centrifugation, disrupted by three cycles of freezing and thawing, and centrifuged for 10 min at 3,200 × g to remove cell debris. The cleared lysate was layered onto a continuous 15% to 40% CsCl gradient and centrifuged for 2 to 3 h at 111,000 × g (average), using an SW 41 Ti rotor (Beckman Coulter, Inc., Fullerton, CA). The mature virus band was collected and purified in a second CsCl density gradient. The mature virus band was collected, dialyzed against three changes of A195 buffer (8), flash frozen in liquid nitrogen, and stored at −80°C. The processes used for the propagation and purification of HAdV-2ts1 and HAdV-5P137L were identical except that after 2 h of incubation to allow virus internalization, infected cells were shifted to the nonpermissive temperature of 39.5°C and incubated until a complete CPE was observed. To BrdU label the viral genome, the inoculum was removed 2 h postinfection and replaced with complete DMEM containing 4 μg/ml BrdU (Sigma-Aldrich, St. Louis, MO).
Dot blot for BrdU incorporation.
To assay virus for BrdU incorporation, approximately 5 × 1010 particles of HAdV-2 or HAdV-2ts1 were incubated in phosphate-buffered saline (PBS) at the indicated temperatures (Fig. 1) for 10 min or boiled for 5 min, cooled to room temperature (RT), and transferred to a nitrocellulose membrane, using a Bio-Dot apparatus (Bio-Rad, Hercules, CA). Each sample well was treated for 10 min at RT with 500 units/ml DNase in 150 mM NaCl, 5 mM MgCl2, and 10 μM HCl (DNase buffer) and then washed with PBS. The blot was then removed from the Bio-Dot apparatus, blocked with 5% milk in PBS-0.1% Tween-20, and probed for BrdU with a monoclonal antibody (MAb) (BD Biosciences, San Jose, CA). The membrane was then stripped and reprobed with anti-HAdV-2 serum (ATCC).
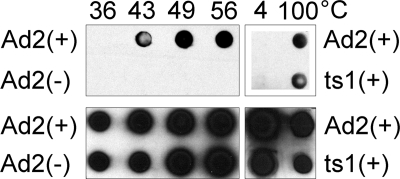
FIG. 1.
BrdU-labeled genomes are exposed upon capsid disassembly. HAdV-2 (Ad2) or HAdV-2ts1 (ts1), labeled (+) or unlabeled (−) with BrdU, was incubated at the indicated temperatures before being analyzed by immunoblotting for BrdU (upper panels) or HAdV-2 proteins (lower panels).
Confocal microscopy.
BrdU-labeled or unlabeled HAdV-5 or HAdV-5P137L particles (3 × 109 particles/sample) in ice-cold complete DMEM (50 μl) were added to glass coverslips sparsely plated with A549 cells. In some experiments, cells were washed twice with serum-free DMEM (SFM) prior to being incubated with virus, and virus was added in SFM. Equivalent results were obtained in complete medium and SFM. Samples were incubated for 45 min at 4°C and then shifted to 37°C and incubated for the periods of time for each figure. In some experiments, cells were washed twice in ice-cold medium prior to being incubated at 37°C. Samples were then fixed in 4% paraformaldehyde (PFA) in PBS for 15 min at RT, quenched and permeabilized with 20 mM glycine-0.5% Triton X-100 for 15 min at RT, treated with 500 units/ml DNase in DNase buffer for 10 min at RT, and probed sequentially with an anti-BrdU MAb (BD Biosciences, San Jose, CA), an Alexa Fluor 647-conjugated anti-mouse secondary antibody (Invitrogen), an anti-lamin B1 rabbit polyclonal antibody (PAb) (Abcam, Cambridge, MA), an Alexa Fluor 488-conjugated anti-rabbit secondary antibody (Invitrogen), and a Cy3-conjugated anti-HAdV-5 hexon MAb (9C12; hybridoma available at the Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City) (26). Samples were mounted for microscopy, using ProLong gold antifade reagent (Invitrogen), and z-series images were acquired with a Zeiss LSM 710 laser scanning confocal microscope (Carl Zeiss, Inc., Thornwood, NY). To maintain consistency, all samples from a single experiment were stained in parallel, and images were acquired with the same microscope using the same settings.
To measure the effect of the CRM1 inhibitor leptomycin B (LMB; Sigma-Aldrich), cells were incubated with 20 nM LMB for 30 min at 37°C prior to the addition of virus in medium containing 20 nM LMB. To measure the effect of HD5, A549 cells on coverslips were washed twice with SFM, and BrdU-labeled HAdV-5 in SFM was added. Samples were incubated for 45 min at 4°C to allow virus binding, washed twice with ice-cold SFM, and then incubated in SFM with or without 10 μM HD5 (Peptides International, Inc., Louisville, KY) for 45 min at 4°C. Samples were then immediately fixed in 4% PFA or incubated for 45 min at 37°C prior to being fixed in 4% PFA.
Image analysis.
A z-profile from the confocal images was created with Zen software (Carl Zeiss, Inc.). The signal threshold of the z-profile for each channel was determined by comparison with that of stained, uninfected control cells, using ImageJ software (W. S. Rasband, National Institutes of Health, Bethesda, MD [http://rsb.info.nih.gov/ij/]). The same threshold values were used for all images in a given experiment. One exception was the experiments used to test the effect of HD5. HD5 binding results in a lower-intensity signal for the anti-hexon antibody, perhaps due to competition for similar binding sites on the virus capsid (27); therefore, the hexon thresholds for the HD5-negative and -positive samples were set independently such that the mean particle size in the hexon channel at time zero, when virus is mainly monodispersed on the cell surface, was equivalent for the two conditions. Cell borders were delineated from bright-field images. For each cell, the total pixel area for each channel and the total area of colocalized pixels between each pair of channels (e.g., red/blue) above the threshold values were determined by using ImageJ software. The percent colocalized particles was determined by dividing the area of the colocalized pixels between two channels by the total pixel area for either of those channels. This value is not weighted for signal intensity. The percent uncoated particles was calculated by dividing the BrdU (blue) pixel area by the sum of the hexon (red) and BrdU pixel areas minus the area of red/blue, colocalized pixels to avoid counting the area of red/blue, colocalized pixels twice. To normalize the uncoating values between time course experiments, the average percent uncoating at the zero time point was taken as the minimum value, and the average percent uncoating at 60 min was taken as the maximum value. For the figures, all pixels above the threshold values for the red and blue channels were set to white, and all pixels below the threshold values were set to black.
RESULTS
Genomic labeling of AdV particles.
We sought to incorporate a unique epitope into the AdV genome as a means to monitor uncoating of AdV particles during entry. The thymidine analog 5-bromo-2-deoxyuridine (BrdU) seemed a likely candidate to measure exposure of the viral genome, based on the availability of specific antibodies to detect BrdU and on reports from early literature that AdVs grown in the presence of BrdU retain the ability to package their genomes (13). Based on preliminary studies, we chose 4 μg/ml BrdU as the labeling condition for all future experiments. This concentration provided a balance between adequate particle production and signal intensity.
To confirm that the virus genome was labeled, we assayed virus particles by immunoblotting for BrdU. Previous studies showed that HAdV-5 is stable to thermal denaturation up to 43°C (26, 27). We exposed BrdU-labeled and unlabeled viruses to increasing temperatures, immobilized the viruses on nitrocellulose using a dot blot apparatus, and probed the membrane for BrdU or for viral capsid proteins (Fig. 1). No signal was observed for the unlabeled virus when probed with the BrdU antibody or with BrdU-labeled virus incubated at 36°C. In contrast, a positive signal was observed for the BrdU-labeled virus exposed to temperatures at or above 43°C. Capsid proteins were detected in both samples at all temperatures, confirming the presence of viruses. These results demonstrate that virus particles produced in cells exposed to BrdU package a genome incorporating the label. Moreover, they confirm that the BrdU epitope is masked in intact virions and can be exposed only upon capsid disruption.
Detection of virus uncoating in single cells.
Having established conditions to produce labeled virus particles, we next sought to ensure that the immunofluorescence signal from BrdU was specific to uncoated virus in infected cells and that the labeling procedure did not alter the entry phenotype of the virus. One tool to aid these studies was a well-characterized HAdV-2 temperature sensitive mutant (HAdV-2ts1) that encodes a mutation in the viral protease gene (P137L) that greatly reduces the infectivity of virus grown at the nonpermissive temperature due to a failure to uncoat and escape the endosome (12, 21, 25). HAdV-2ts1 was produced under the same labeling conditions (4 μg/ml BrdU) as wild-type HAdV-2 and was also found to contain labeled genomic DNA. Consistent with findings from previous studies (27), the HAdV-2ts1 particles were very stable to thermal denaturation; however, the BrdU label could be detected upon boiling (Fig. 1). In order to use this mutant virus in immunofluorescence studies, we required a virus with the same phenotype as HAdV-2ts1 but with a HAdV-5 capsid to enable detection by a HAdV-5-specific antihexon MAb. To this end, we engineered the protease mutation P137L, which alone is responsible for the HAdV-2ts1 phenotype (12), into a HAdV-5-based vector expressing GFP. The high level of sequence conservation between HAdV-2 and HAdV-5 protease (99.5% identical amino acids) suggested that the phenotype of HAdV-5 bearing this mutation would be identical to that of HAdV-2ts1. Indeed, the HAdV-5 vector bearing this mutation (HAdV5-P137L), when propagated at the nonpermissive temperature, contained the precursor forms of proteins VI, VII, and VIII, a hallmark of the HAdV-2ts1 phenotype (data not shown).
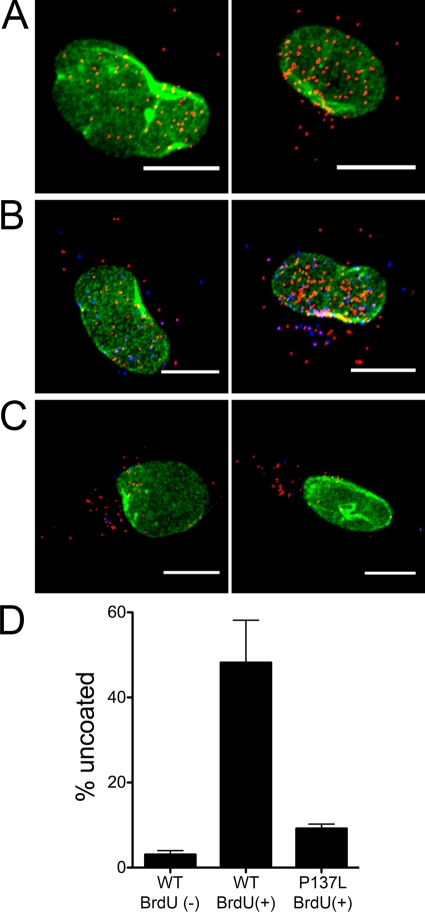
To demonstrate that exposure of the BrdU epitope in infected cells is dependent upon virus uncoating, we compared the BrdU signals from cells infected with BrdU-labeled and unlabeled wild-type HAdV-5 vectors and BrdU-labeled HAdV5-P137L at 45 min postinfection (Fig. 2). These HAdV-5-based vectors had similar growth characteristics and were labeled under the same conditions as HAdV-2 and HAdV-2ts1. As expected, no BrdU signal above background was detected in cells infected with unlabeled virions. In contrast, a large number of discrete BrdU-positive puncta (Fig. 2, blue areas) were observed in cells infected with BrdU-labeled HAdV-5. The numbers of these puncta were dramatically reduced in samples infected with BrdU-labeled HAdV5-P137L. We observed equivalent levels of bright puncta upon staining for hexon (Fig. 2, red areas) in all samples. The hexon signal for HAdV-5 was strongly colocalized with the nucleus (Fig. 2, green areas), while most of the hexon signal for HAdV-P137L remained cytoplasmic, confirming the defective entry phenotype of this virus. These results are consistent with a specific signal from BrdU-labeled viruses that uncoat during cell entry and provide an assay to monitor the effect of antiviral agents on this process.
FIG. 2.
Detection of BrdU in labeled, uncoated virus particles in infected cells. Representative images, which are stained for hexon (red), BrdU (blue), and lamin (green), are shown for unlabeled HAdV-5 wild type (WT) (A), labeled HAdV-5 WT (B), and labeled HAdV-5P137L (C) viruses. Scale bars are 10 μm. (D) Percent uncoating was determined for HAdV-5 (WT) or HAdV-5P137L (5P137L) vectors, labeled (+) or unlabeled (−) with BrdU, at 45 min postinfection in A549 cells. Data are the means and standard errors of the means of the results from three independent experiments in which ∼30 cells were evaluated for each condition. Note that the low value for the unlabeled WT virus in this assay does not indicate that the virus failed to uncoat but reflects the absence of BrdU in its genome.
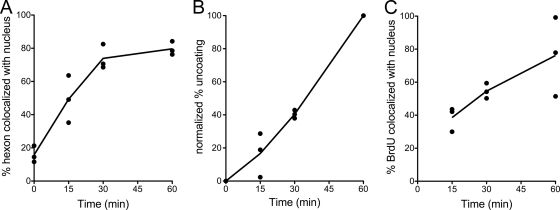
To confirm that encapsidation of the labeled genome did not affect virus entry, we measured the kinetics of hexon colocalization with the nucleus. In confocal images of cells infected with the BrdU-labeled HAdV-5 vector, hexon accumulated progressively at the nucleus (stained with lamin) for the first 30 min postinfection, was half of maximal at ∼15 min, and reached a maximal plateau of ∼80% colocalization at 30 min, which was maintained at later time points (Fig. 3A). In a single experiment that was assessed at 2 and 4 h postinfection, we observed that no additional virus accumulated beyond the maximum observed at 60 min (data not shown). The ∼20% nuclear colocalization measured at the initial time points of these experiments is due to coincident colocalization of the virus with the lamin signal in the z-projections of the confocal images used for this analysis. The observed kinetics of hexon colocalization with the nucleus are comparable to those measured previously with unlabeled virus (Fig. 4B) (27), confirming that the BrdU labeling did not grossly affect virus binding, internalization, and cytoplasmic translocation.
FIG. 3.
Kinetics of hexon trafficking and genome exposure and translocation in infected cells. A549 cells were infected with BrdU-labeled HAdV-5 and analyzed for hexon colocalization with the nucleus (A), normalized percent uncoating (B), and BrdU colocalization with the nucleus (C) at the indicated times postinfection. Points represent the mean values from each of three independent experiments in which ∼30 cells were evaluated at each time point. Lines connect the averages of these mean values. In panel C, no value is given for the zero time point, as no uncoating occurred.
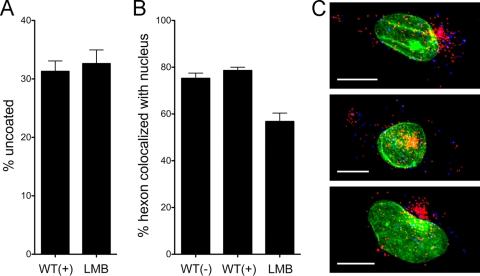
FIG. 4.
BrdU labeling does not alter virus trafficking to the MTOC. A549 cells infected with HAdV-5 (WT), labeled (+) or unlabeled (−) with BrdU, or with BrdU-labeled HAdV-5 in the presence of 20 nM LMB (LMB) were evaluated at 45 min postinfection for percent uncoating (A) and hexon colocalization with the nucleus (B). Data are the mean values and standard errors for ∼30 cells for each condition. Note that the WT(−) and WT(+) controls from this experiment were included as one of the three experiments averaged for Fig. 2. (C) Three cells representative of the LMB phenotype, colored as in Fig. 2, are shown. Scale bar is 10 μm.
We also assessed the kinetics of uncoating. For these experiments, the percent uncoating was defined as the number of BrdU-positive pixels in each cell divided by the total number of hexon-positive and BrdU-positive pixels. This formula counts all single-positive (either only hexon-positive or only BrdU-positive) pixels as contributing to the total amount of virus in the cell, and pixels that were positive for both hexon and BrdU were counted only once. Using these criteria, we observed that the rate of uncoating was comparable between experiments (Fig. 3B), increasing linearly up to 60 min postinfection with a half-life (t1/2) between 30 and 45 min postinfection. However, because of variations in the microscope settings and staining intensities between experiments, the raw values calculated using this formula varied considerably between experiments (data not shown). Unlike that of hexon, BrdU colocalization with the nucleus was less rapid and more variable between experiments (Fig. 3C). Maximal colocalization ranged from 50 to 100% at 60 min postinfection, and the t1/2 was approximately 30 min. These data demonstrate that genome exposure begins almost immediately postinfection, likely coincident with exposure to cellular triggers (e.g., acidification) and endosome escape, and that exposed genomes traffic to the nucleus.
The fidelity of cytoplasmic trafficking of BrdU-positive virions was further assessed in experiments using the CRM1 inhibitor leptomycin B (LMB). In enucleated cells or in cells exposed to CRM1 inhibitors, HAdV has been shown to accumulate at the microtubule-organizing center (MTOC) (2, 28). Based on these observations, it has been proposed that HAdV is actively rescued from the MTOC in a CRM1-dependent fashion prior to docking at the nuclear pore complex, although the molecular basis for this process is unclear. At 45 min postinfection, inhibitory concentrations of LMB reduced the nuclear localization of hexon without affecting the extent of uncoating (Fig. 4A and B). The modest quantitative effect of LMB on nuclear colocalization is primarily due to the fact that in the z-projections of confocal images used for these analyses, the virus that has accumulated at the MTOC often either partially or completely overlaps the nucleus (Fig. 4C, yellow areas); nonetheless, the dramatic effect of LMB on the subcellular distribution of HAdV is readily apparent (compare Fig. 4C with Fig. 2). Thus, intracellular trafficking of HAdV to the MTOC is not disrupted by BrdU labeling.
The human α-defensin HD5 prevents HAdV uncoating during cell entry.
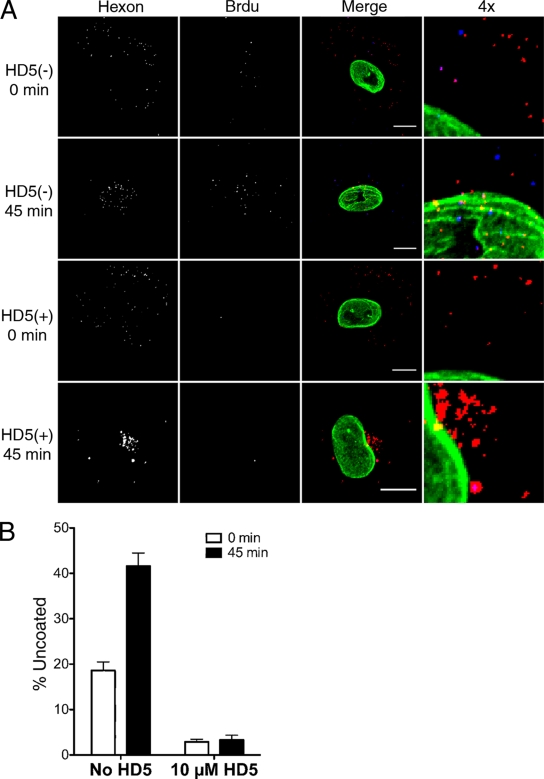
Having verified that our assay can be used to monitor virus uncoating in single cells, we next tested the effects of the human α-defensin HD5 on this process during HAdV-5 entry. For these experiments, virus was bound to A549 cells at 4°C to prevent internalization and then incubated for an additional 45 min at 4°C in the presence or absence of an inhibitory concentration of HD5 (10 μM). Cells were then fixed and assessed for uncoated virus immediately or after being warmed to 37°C for 45 min to allow internalization and infection (Fig. 5). At the zero time point, hexon-positive pixels were randomly dispersed in both samples, consistent with virus binding to the plasma membrane. A low number of BrdU-positive pixels was observed in the sample incubated without defensin, which was virtually absent in the samples incubated with HD5. At 45 min postinfection, the number of BrdU-positive signals increased substantially in the defensin-minus sample, and both hexon- and BrdU-positive pixels were largely colocalized with the nucleus. In contrast, no increase in the numbers of BrdU signals were observed in the samples incubated with HD5, and the hexon in these samples accumulated in a perinuclear compartment. These results revealed for the first time that the α-defensin HD5 blocks HAdV uncoating and genome exposure, in particular, during cell entry and are consistent with our previous studies indicating that the virus accumulates in lysosomes as a consequence of a failure to uncoat.
FIG. 5.
The α-defensin HD5 blocks genome exposure during cell entry. A549 cells were infected with BrdU-labeled HAdV-5 in the presence (+) or absence (−) of 10 μM HD5. (A) Representative cells, separated by color channel as indicated or merged and colored as in Fig. 2, are shown for the indicated time points. A section of the merged image for each condition was enlarged fourfold (4x). Scale bar, 10 μm. (B) Percent uncoating was evaluated at 0 and 45 min postinfection. Data are the mean values and standard errors for ∼30 cells for each condition from one experiment representative of three independent experiments.
DISCUSSION
We developed an assay to measure the uncoating of HAdV particles during cell entry by monitoring the exposure of the virus genome labeled with BrdU by immunofluorescence. We demonstrated that the BrdU-labeled genome is encapsidated and that BrdU epitopes are completely masked in intact, mature virus particles. The presence of the labeled genome did not impact the kinetics of virus entry and the accumulation of hexon at the nucleus. The BrdU signal was specific to labeled, uncoated virus that could be observed in single cells. Labeled virus appeared to traffic first to the MTOC and then to the nucleus, consistent with current models of HAdV trafficking during cell entry. Finally, using this assay we confirmed our model of defensin-mediated neutralization, which was based on in vitro biochemical studies, by demonstrating directly that these antiviral molecules prevent HAdV uncoating and genome exposure during cell entry.
BrdU labeling was previously proposed in studies of human CMV (HCMV) as a general method to monitor early events in DNA virus entry (23). We have now shown that this approach, with some modifications, is applicable to HAdV and may provide an avenue to investigate several aspects of HAdV entry. In the course of these studies, we considered a number of technical aspects of the labeling and detection protocols that have bearing on the interpretation of the results.
First, one caveat of this genome-labeling technique is that double-stranded DNA (dsDNA) must be denatured to expose BrdU epitopes for immunodetection. In preliminary in vitro studies, we tested several denaturation methods, including exposure to acid or base. Because the stability of the virus capsid can be affected by extremes of pH, we chose to denature the DNA by partial nuclease digestion. This approach prevents potential artifacts from harsh chemical treatment and is analogous to the classical biochemical method to measure virus uncoating by determining the sensitivity of radiolabeled viral genomic DNA to nuclease digestion. Therefore, we anticipated that our results obtained using this technique to monitor uncoating of particles at a single-cell level would be comparable to those of studies of virus uncoating that measured the behavior of the entire virus inoculum of a bulk population of cells.
Second, we chose our labeling condition based on a moderate reduction in particle yield and a detectable signal by immunofluorescence but did not optimize it further. Increased BrdU concentration should lead to a greater degree of labeling, both of individual particles and of the total virus population, and to brighter particles. Cells may remain viable, but essentially growth arrested, at higher concentrations of BrdU; however, the length of time that cells can be maintained under high concentrations of BrdU without substantial reductions in particle production has not been determined. Kjellen et al. recovered HAdV-5 labeled with 30 μg/ml BrdU in HeLa cells after 70 h with only a sevenfold reduction in particle yield, although the entry phenotype of this virus was not tested (13). This time frame is suitable for HAdV-5, but other serotypes and/or mutants with delayed growth kinetics may require longer incubation times; therefore, the BrdU concentration and the frequency of replenishment of BrdU in the media would need to be determined to make this technique more broadly applicable. Other than a reduction in particle number due to cytotoxicity, high BrdU concentrations may have additional effects on core protein binding and DNA packaging. Therefore, the fidelity of the entry kinetics of labeled virus should be tested as a control for any labeling procedure. Moreover, BrdU-labeled DNA is sensitive to UV cross-linking, which may affect DNA dynamics during entry.
Third, we did not quantify the amount of label incorporated into the virus particles used for these studies. The degree of labeling could be determined by measuring the density of labeled virus compared to that of an unlabeled control, as has been done for adeno-associated virus (AAV) (3, 4). That the bulk of the particles were labeled was indicated by our ability to isolate the labeled virus in a single band in CsCl density gradients. The HAdV-5 genome is 22.4% thymidine, and the DNA genome is 16% of the mass of the particle (14). Given the density of HAdV-5 of 1.335 g/ml (14) and a difference in mass between BrdU and thymidine of 65 Da, a particle with complete substitution of BrdU by thymidine should have a mass of 1.355 g/ml. This difference is readily detectable in the CsCl gradients used for purification. A similar rationale was used to demonstrate the existence of two types of AAV particles, encapsidating either strand of single-stranded DNA (ssDNA), by BrdU labeling (3).
Fourth, although it was not our intent to optimize this assay beyond the level needed for these studies, direct conjugation of fluorescent dyes to the virus may broaden the utility of this technique and could be used to address the origin of the BrdU-positive but hexon-negative particles that were commonly observed in our studies. BrdU-positive pixels that are not colocalized with hexon represent either uncoated virions, which for technical reasons do not stain for hexon, or are genomes that have dissociated from the capsid. A failure to stain for hexon may be due to a steric block imposed by DNA extruding from the capsid, as has been observed with viruses disrupted by heat or other treatments in vitro. Such an artifact could be avoided using a hexon-conjugated fluorescent dye rather than an antibody to label the capsid. If these events are not due to an artifact, a decrease in BrdU colocalization with hexon may reflect separation of the viral genome from the capsid. Although variable from experiment to experiment, there was a general trend toward increased colocalization of hexon and BrdU at early time points, which diminished over time postinfection (data not shown). This separation could occur at the nuclear membrane, consistent with the prevailing model of AdV entry, or it may be due to dissociation of some genomes from the capsid in the cytoplasm. Based on our studies, the latter seems to be more predominant than the former. This aspect of AdV entry has not been previously examined at the single-particle level other than by electron microscopy, although it has been suggested by biochemical analyses (16, 17). Recognition of viral DNA by endosomal (e.g., TLR9) and cytoplasmic innate immune sensors supports the idea that at least some viral DNA is either entirely dissociated from the capsid or sufficiently exposed during infection (5, 9, 11, 18, 20, 34). Early studies showed that the viral genome remains associated with viral proteins, but these radiolabeling experiments could not always distinguish core proteins from the capsid (16, 29), and although electron microscopy studies have demonstrated largely intact capsids with dense centers in the cytoplasm, it is unclear whether they have undergone uncoating (6, 29). Dissociation from the capsid may result in failure or a delay in the trafficking of the genome, which would likely retain association with core proteins, through the cytoplasm to the nucleus. Indeed, we observed that colocalization of BrdU with the nucleus and with the MTOC was delayed relative to that of hexon and was more variable between experiments (Fig. 3C and 4). Therefore, genome dissociation from the capsid may result in nonproductive infection and contribute to an increase in the apparent particle-to-PFU ratio during virus infection.
In these studies, the overall extent of uncoating was low, both as measured by the amount of hexon colocalized with BrdU (data not shown) and by using the uncoating formula defined in Results. This low percentage may be real and due to variations in the capacity of individual virions to uncoat, perhaps from differential exposure to cellular triggers, or may be due to a technical artifact. Every uncoated particle may not be detected in this assay due to the degree of BrdU labeling, the distribution and continued association of core proteins that mask BrdU epitopes, and the accessibility of DNA to DNase and antibody. Early studies measuring DNase sensitivity of radiolabeled viral genomes from cytoplasmic extracts of infected cells in some cases reported a greater extent of uncoating (up to 45 to 85% of input virus at 1 h postinfection) than was measured in our studies (14, 29, 30), although the amount of viral DNA associated with the nucleus in cell fractionation experiments was similar to that observed in our studies (6, 16). It is not clear how much capsid perturbation is required to generate nuclease sensitivity and to be measured as uncoated for either assay. A hole created by removal of just the fiber is unlikely to be sufficient to allow DNA out of or enzymes and antibodies into the interior of the capsid. Therefore, it is likely that at least one or more of the vertices (penton base and fiber) is removed. Indeed, genome sensitivity to nuclease has been shown to correlate with vertex removal (22, 24, 29). BrdU may become detectable in this assay, therefore, only upon extensive capsid disruption or when the viral genome has separated or become partially externalized from the capsid. Less-drastic uncoating events, although sufficient to release the viral membrane lytic proteins and mediate endosomalysis, may not be detected.
We also observed that uncoating continues to increase once maximal hexon colocalization with the nucleus has been reached. Uncoating may be asynchronous, and the ∼20% of virus that does not reach the nucleus may contribute to this effect. Alternatively, this effect may be due to additional uncoating events that occur at the nuclear membrane. Although beyond the scope of our current studies, this technique may allow for a more-careful analysis of the subcellular localization of uncoating as well as the fate of the viral DNA in the nucleus at late times postinfection.
We developed this uncoating assay in order to more fully explore the mechanism of defensin-mediated neutralization of AdV infection. Our previous confocal studies provided indirect evidence that AdV fails to uncoat in the presence of inhibitory concentrations of defensin, because we observed that most of the virus became trapped in the lysosome rather than reaching the nucleus (27). This effect was also measured as a reduction in the capacity of AdV to mediate sarcin translocation into the cytoplasm in the presence of defensin. From these studies, we proposed that a failure to mediate endosomalysis was due to a failure to uncoat. Biochemical studies showing that defensin binding increased the thermostability of AdV also supported this model. Nonetheless, these studies provided no direct evidence that AdV fails to uncoat during entry. An alternative model is that defensin could block the membrane lytic activity of proteins (e.g., VI) released from the uncoated virus rather than uncoating per se. The marked reduction in BrdU signals in cells infected with the virus in the presence of defensin, however, provides strong evidence that these molecules directly block uncoating during cell entry. Although we cannot formally exclude the possibility that the reduction in BrdU signals is due to HD5 binding to and shielding the DNA directly rather than preventing uncoating, we feel that this is unlikely, given the number of defensin molecules that would be needed to uniformly bind the nucleic acid for this to occur and our previously published studies demonstrating that endosome lysis is also impaired. In our current studies, virus was prebound to cells and monodispersed prior to exposure to defensin. These conditions mimic those used in our previous studies to measure the time course of defensin-mediated inhibition of infection and provide further evidence that AdV is neutralized by defensin due to a discrete block of uncoating rather than a more-general effect such as virus aggregation, which has been proposed as the mechanism of defensin-mediated neutralization of polyomavirus (7). In addition to a blockage of BrdU exposure at the 45-min time point postinfection, we also observed a reduction in the number of BrdU signals at the zero time point. Up to 20% uncoating at time zero was also observed in early studies, as measured by DNase sensitivity (14). In both treated and untreated samples, virus was first incubated with cells for 45 min at 4°C to allow binding and to synchronize the infection. Therefore, a certain amount of uncoating must occur during the subsequent 45-min incubation at 4°C, and defensin stabilization of the capsid also prevents this from occurring. Taken together, these observations provide evidence to support the concept that defensins neutralize AdV infection by stabilizing the capsid and preventing uncoating. Although not amenable to live-cell imaging, the assay described here may be used to evaluate the effects of other molecules, including antibodies, clotting factors, complement components, and small-molecule inhibitors, or mutations in viral proteins, such as the membrane lytic protein VI, on AdV uncoating. In addition, this assay may provide insight into the subcellular localization of uncoating in relation to innate immune sensors of viral DNA during infection.
Acknowledgments
We thank Milena Iacobelli-Martinez for the creation of the 293β5 cells and Crystal Moyer for helpful discussions.
This work was supported by NIH grants HL054352 and EY11431 to G.R.N. E.K.N. was supported by the Immunology and Microbial Sciences Summer Program at TSRI. The authors declare no competing financial interests.
This is Scripps Research Institute manuscript number 20478.
Footnotes
Published ahead of print on 3 February 2010.
REFERENCES
- 1.Arnberg, N. 2009. Adenovirus receptors: implications for tropism, treatment and targeting. Rev. Med. Virol. 19:165-178. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, C. J., R. G. Crystal, and P. L. Leopold. 2003. Association of adenovirus with the microtubule organizing center. J. Virol. 77:13275-13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns, K. I., and S. Adler. 1972. Separation of two types of adeno-associated virus particles containing complementary polynucleotide chains. J. Virol. 9:394-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns, K. I., and J. A. Rose. 1970. Evidence for a single-stranded adenovirus-associated virus genome: isolation and separation of complementary single strands. J. Virol. 5:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerullo, V., M. P. Seiler, V. Mane, N. Brunetti-Pierri, C. Clarke, T. K. Bertin, J. R. Rodgers, and B. Lee. 2007. Toll-like receptor 9 triggers an innate immune response to helper-dependent adenoviral vectors. Mol. Ther. 15:378-385. [DOI] [PubMed] [Google Scholar]
- 6.Chardonnet, Y., and S. Dales. 1970. Early events in the interaction of adenoviruses with HeLa cells. I. Penetration of type 5 and intracellular release of the DNA genome. Virology 40:462-477. [DOI] [PubMed] [Google Scholar]
- 7.Dugan, A. S., M. S. Maginnis, J. A. Jordan, M. L. Gasparovic, K. Manley, R. Page, G. Williams, E. Porter, B. A. O'Hara, and W. J. Atwood. 2008. Human alpha-defensins inhibit BK virus infection by aggregating virions and blocking binding to host cells. J. Biol. Chem. 283:31125-31132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, R. K., D. K. Nawrocki, L. A. Isopi, D. M. Williams, D. R. Casimiro, S. Chin, M. Chen, D. M. Zhu, J. W. Shiver, and D. B. Volkin. 2004. Development of stable liquid formulations for adenovirus-based vaccines. J. Pharm. Sci. 93:2458-2475. [DOI] [PubMed] [Google Scholar]
- 9.Fejer, G., L. Drechsel, J. Liese, U. Schleicher, Z. Ruzsics, N. Imelli, U. F. Greber, S. Keck, B. Hildenbrand, A. Krug, C. Bogdan, and M. A. Freudenberg. 2008. Key role of splenic myeloid DCs in the IFN-alphabeta response to adenoviruses in vivo. PLoS Pathog. 4:e1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 11.Iacobelli-Martinez, M., and G. R. Nemerow. 2007. Preferential activation of Toll-like receptor nine by CD46-utilizing adenoviruses. J. Virol. 81:1305-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imelli, N., Z. Ruzsics, D. Puntener, M. Gastaldelli, and U. F. Greber. 2009. Genetic reconstitution of the human adenovirus type 2 temperature-sensitive 1 mutant defective in endosomal escape. Virol. J. 6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kjellen, L., H. G. Pereira, R. C. Valentine, and J. A. Armstrong. 1963. An analysis of adenovirus particles and soluble antigens produced in the presence of 5-bromodeoxyuridine. Nature 199:1210-1211. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence, W. C., and H. S. Ginsberg. 1967. Intracellular uncoating of type 5 adenovirus deoxyribonucleic acid. J. Virol. 1:851-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leopold, P. L., and R. G. Crystal. 2007. Intracellular trafficking of adenovirus: many means to many ends. Adv. Drug Deliv. Rev. 59:810-821. [DOI] [PubMed] [Google Scholar]
- 16.Lonberg-Holm, K., and L. Philipson. 1969. Early events of virus-cell interaction in an adenovirus system. J. Virol. 4:323-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirza, M. A., and J. Weber. 1979. Uncoating of adenovirus type 2. J. Virol. 30:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muruve, D. A., V. Petrilli, A. K. Zaiss, L. R. White, S. A. Clark, P. J. Ross, R. J. Parks, and J. Tschopp. 2008. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452:103-107. [DOI] [PubMed] [Google Scholar]
- 19.Nakano, M. Y., K. Boucke, M. Suomalainen, R. P. Stidwill, and U. F. Greber. 2000. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 74:7085-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nociari, M., O. Ocheretina, J. W. Schoggins, and E. Falck-Pedersen. 2007. Sensing infection by adenovirus: Toll-like receptor-independent viral DNA recognition signals activation of the interferon regulatory factor 3 master regulator. J. Virol. 81:4145-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Berná, A. J., R. Marabini, S. H. Scheres, R. Menendez-Conejero, I. P. Dmitriev, D. T. Curiel, W. F. Mangel, S. J. Flint, and C. San Martin. 2009. Structure and uncoating of immature adenovirus. J. Mol. Biol. 392:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prage, L., U. Pettersson, S. Hoglund, K. Lonberg-Holm, and L. Philipson. 1970. Structural proteins of adenoviruses. IV. Sequential degradation of the adenovirus type 2 virion. Virology 42:341-358. [DOI] [PubMed] [Google Scholar]
- 23.Rosenke, K., and E. A. Fortunato. 2004. Bromodeoxyuridine-labeled viral particles as a tool for visualization of the immediate-early events of human cytomegalovirus infection. J. Virol. 78:7818-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell, W. C., R. C. Valentine, and H. G. Pereira. 1967. The effect of heat on the anatomy of the adenovirus. J. Gen. Virol. 1:509-522. [DOI] [PubMed] [Google Scholar]
- 25.Silvestry, M., S. Lindert, J. G. Smith, O. Maier, C. M. Wiethoff, G. R. Nemerow, and P. L. Stewart. 2009. Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect. J. Virol. 83:7375-7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, J. G., A. Cassany, L. Gerace, R. Ralston, and G. R. Nemerow. 2008. Neutralizing antibody blocks adenovirus infection by arresting microtubule-dependent cytoplasmic transport. J. Virol. 82:6492-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, J. G., and G. R. Nemerow. 2008. Mechanism of adenovirus neutralization by human alpha-defensins. Cell Host Microbe 3:11-19. [DOI] [PubMed] [Google Scholar]
- 28.Strunze, S., L. C. Trotman, K. Boucke, and U. F. Greber. 2005. Nuclear targeting of adenovirus type 2 requires CRM1-mediated nuclear export. Mol. Biol. Cell 16:2999-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sussenbach, J. S. 1967. Early events in the infection process of adenovirus type 5 in HeLa cells. Virology 33:567-574. [DOI] [PubMed] [Google Scholar]
- 30.Svensson, U., and R. Persson. 1984. Entry of adenovirus 2 into HeLa cells. J. Virol. 51:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiethoff, C. M., H. Wodrich, L. Gerace, and G. R. Nemerow. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79:1992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu, J., X. Huang, and Y. Yang. 2007. Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and -independent pathways. J. Virol. 81:3170-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]