Abstract
The varicella-zoster virus (VZV) IE62 protein is the major transcriptional activator. IE62 is capable of associating with DNA both nonspecifically and in a sequence-specific manner via a consensus binding site (5′-ATCGT-3′). However, the function of the consensus site is poorly understood, since IE62 efficiently transactivates promoter elements lacking this sequence. In the work presented here, sequence analysis of the VZV genome revealed the presence of 245 IE62 consensus sites throughout the genome. Some 54 sites were found to be present within putative VZV promoters. Electrophoretic mobility shift assay (EMSA) experiments using an IE62 fragment containing the IE62 DNA-binding domain and duplex oligonucleotides that did or did not contain the IE62 consensus binding sequence yielded KD (equilibrium dissociation constant) values in the nanomolar range. Further, the IE62 DNA binding domain was shown to have a 5-fold-increased affinity for its consensus site compared to nonconsensus sequences. The effect of consensus site presence and position on IE62-mediated activation of native VZV and model promoters was examined using site-specific mutagenesis and transfection and superinfection reporter assays. In all promoters examined, the consensus sequence functioned as a distance-dependent repressive element. Protein recruitment assays utilizing the VZV gI promoter indicated that the presence of the consensus site increased the recruitment of IE62 but not Sp1. These data suggest a model where the IE62 consensus site functions to down-modulate IE62 activation, and interaction of IE62 with this sequence may result in loss or decrease of the ability of IE62 to recruit cellular factors needed for full promoter activation.
Varicella-zoster virus (VZV) is a human alphaherpesvirus that causes varicella (chicken pox) and establishes latency in cells within dorsal root ganglia during primary infection. Upon reactivation, VZV causes shingles, or zoster, which usually involves productive infection along a single dermatome and infection of the skin innervated by that dermatome (1). Productive VZV infection during either primary infection or reactivation results in the expression of the entire coding capacity of the 125-kb VZV genome, leading to the synthesis of some 70 proteins. The immediate-early 62 (IE62) protein, the major viral transcriptional activator, is capable of transactivating the majority of if not all VZV promoters during lytic infection. It is also capable of transactivating heterologous promoters and model promoters containing a minimal number of cis-acting elements. IE62 is required for viral growth in tissue culture and increases the infectivity of VZV DNA (32, 42). The protein is 1,310 amino acids in length and contains a potent N-terminal acidic activation domain and a centrally located DNA binding domain. It is believed to be a native homodimer, based on comparison with its putative homolog HSV-1 ICP4 and the experimentally established dimerization of a bacterially expressed fragment containing the DNA-binding domain and sequences corresponding to the ICP4 dimerization domain (50).
IE62 is divided into five regions (I to V) based on sequence alignments of it and its alphaherpesvirus orthologues. In the case of IE62, region I (amino acids [aa] 1 to 467) contains an acidic activation domain recently shown to interact with the human Mediator complex (7, 38, 53, 55) and other sequences necessary for direct interaction with the cellular transcription factors Sp1 and USF (35, 56). Region III (aa 641 to 734) contains a nuclear localization signal and sites for phosphorylation by viral kinases (20, 21). The function of region IV (aa 11735 to 1147) is unknown, but mutations in this region result in alteration of IE62 activity (3). Region V (aa 1148 to 1310) is the site of numerous mutations in VZV vaccine strains, although the significance of this remains unknown (14). The DNA binding domain (DBD) of IE62 falls within region II of the protein, which is the most highly conserved region among the alphaherpesvirus major transactivators, and overlaps the region responsible for dimerization (6). DNA binding appears to be important for IE62 function, as mutations which diminished DNA binding caused a corresponding loss in transactivation in experiments utilizing the HSV-1 gD promoter (48).
Previous studies have shown that partially purified bacterially expressed fragments corresponding to amino acids 417 to 646, which contains the complete DBD, were capable of binding to DNA in a sequence-specific manner. Using DNase I footprinting, Wu and Wilcox identified a consensus sequence (ATCGT) to which IE62 bound within its own promoter (52). This sequence corresponds to the first 5 bases of the bipartite consensus sequence of the HSV-1 homolog ICP4 (ATCGTCNNNNYCGRC), where N is any nucleotide, Y is a pyrimidine, and R is a purine (8, 10, 11, 28, 30). The IE62 DBD was also shown to interact with this sequence within the ICP4-binding sites present in the HSV ICP4 and gD promoters. Site-specific mutation of each nucleotide within the ICP4 consensus site showed that only mutations within the 5′ ATCGT sequence affected IE62 DBD binding, whereas mutations in both the 5′ and 3′ sequences affected ICP4 binding (4, 48, 49, 50). Tyler and Everett (49) showed that the DBD fragment interacted with several sequences within the ORF62 promoter, including three ATCGT consensus sites. The role of these sites in expression of ORF62 was not established, although it was concluded that IE62 likely transactivated via a site-specific mechanism.
The function of the IE62 consensus site in transcription has been the subject of only one previous study, and that study did not directly test the effect of the IE62 consensus site (4). The heterologous HSV-1 gD promoter, which contains one copy of the HSV ICP4 consensus site, was used, and the results showed that deletion of that consensus site (containing the ATCGT sequence) resulted in a decrease of IE62-mediated transactivation, whereas addition of tandem ICP4 consensus sites increased IE62-mediated transactivation of the gD promoter. This is in contrast to data concerning the function of the HSV-1 ICP4 consensus binding sequence in the context of ICP4-mediated transcriptional control. In vitro and in vivo studies collectively suggest that the binding of ICP4 to its consensus binding site is not required for activation (15, 45). Rather, specifically positioned ICP4 consensus sites near the transcription initiation sites of the HSV-1 ICP4 transcript, latency-associated transcript (LAT), and L/ST transcript are involved in ICP4-mediated repression of transcription (2, 5, 17, 23, 29, 41, 57). Finally, work from several laboratories has shown that the IE62 consensus binding site is not required for IE62-mediated transactivation, since IE62 is capable of efficiently activating many promoters which do not contain this sequence (18, 35, 56). Thus, the role of the IE62-consensus site in IE62 function and viral replication is not well characterized or understood.
In the work presented here, we examined the role of the IE62 consensus site with respect to the interaction of the IE62 DNA-binding domain (DBD) with DNA and IE62-mediated transactivation of model and native VZV promoters. Using computer analysis, we identified a total of 245 consensus IE62 binding sites distributed throughout the VZV genome. We expressed an IE62 fragment containing the DBD (aa 417 to 647) used in previous studies and employed a dual chromatographic purification scheme to produce a highly purified protein preparation. This recombinant polypeptide was used in electrophoretic mobility shift assay (EMSA) experiments to analyze its association with oligonucleotides containing consensus and nonconsensus sequences. Our results show that the DBD interacts both specifically and nonspecifically with DNA and that the difference in affinity between these interactions is ∼5-fold under our experimental conditions.
Insertion of the ATCGT consensus sequence resulted in a decrease of expression from both native and model promoters, and this suppression is distance dependent relative to the position of TATA elements. Mutation of native consensus sites within the VZV ORF62 and gC promoters resulted in an increase of expression from these promoters in the context of both ectopic IE62 expression and VZV superinfection, yielding further evidence that the role of the IE62 consensus site is to down modulate viral gene expression. In the case of the ORF62 promoter, while all three consensus sites play a role in down modulation of the promoter, the most influential site in this regard abuts the enhancer core, which is the binding site for the tripartite Oct-1/HCF-1/VZV ORF10 complex (31, 33, 34). These results have led to a model in which the IE62 consensus binding site is a cis-acting repressor that increases occupancy of the DNA by IE62 and may interfere with the ability of IE62 to interact with cellular factors.
MATERIALS AND METHODS
Cells and viruses.
MeWo cells, a human melanoma cell line that supports the replication of VZV, were grown in Eagle's minimal essential medium supplemented with 10% fetal bovine serum as previously described (46). VZV strain MSP was propagated in MeWo cell monolayers as described by Lynch et al. (26) and Peng et al. (35). Neurons derived from fetal rat dorsal root ganglia were isolated and cultured as described by Kleitman et al. (22).
Plasmids.
The cloning of the pCMV62 plasmid expressing IE62 under the control of the cytomegalovirus (CMV) immediate-early (IE) promoter was described previously by Perera et al. (37, 39). The following plasmids were constructed in this study: pTA-Luc/5DS, p10US-Luc, p20US-Luc, p50US-Luc, p100US-Luc, p2xUS-Luc, p3xUS-Luc, pgC-Luc, pgC-Luc/mcon, p62prom/m1, p62prom/m2, p62prom/m1-2, p62prom/m3, p62prom/m1-2-3, pgI-Luc/ATCGT, and p62DBD/WT.
The construction of the minimal model promoter plasmid pTA-Luc, which contains a consensus TATA box with no other known binding sites, was previously described by Yang et al. (56). A set of plasmids containing a single IE62 consensus sites at positions +5, −10, −20, −50, and −100 (p5DS-Luc, p10US-Luc, p20US-Luc, p50US-Luc, and p100US-Luc, respectively) relative to the TATA box was generated from pTA-Luc using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Plasmids p2xUS-Luc, containing two tandem IE62 consensus sites, and p3xUS-Luc, containing three tandem consensus sites at the −10 position, were constructed by insertion of additional sites into p10US-Luc.
A set of luciferase reporter plasmids was constructed by using the pGL-2 basic vector (Promega, Madison, WI). A 222-bp region upstream of the ORF14 translational start site was amplified and inserted between the XhoI and HindIII sites of the pGL-2 basic vector to produce the gC-Luc vector. The single consensus occurring in this sequence was mutated to an ATCAT to form the gC-Luc/mcon vector.
A luciferase reporter plasmid containing the promoter region of ORF62 was described previously (34). This plasmid (pIE62PR) contains two of the three consensus sites within the promoter region (referred to as p62prom/m1 in this study); the third site was added through site-directed mutagenesis to form p62prom/WT. The remaining two consensus sites were mutated in p62prom/m1 to generate plasmids p62prom/m1-2 and p62prom/m1-2-3. These two consensus sites were also mutated in the p62prom/WT plasmid to form p62prom/m2 and p62prom/m3. In each case, the consensus ATCGT was mutated to ATCAT. The plasmid pgI-Luc was constructed by inserting the glycoprotein I (VZV ORF67) promoter sequence obtained from the pgI3.4CAT plasmid previously described by Peng et al. (35) into the pGL-2 basic luciferase reporter vector. This promoter contains a partial consensus sequence which was mutated into a full consensus site to generate pgI-Luc/ATCGT.
IE62 DNA binding domain expression and purification.
The DNA sequence corresponding to the DNA-binding domain (aa 417 to 647) of IE62 was amplified and inserted in-frame between the HindIII and BglII sites of the pRSET bacterial expression plasmid (Invitrogen, Carlsbad, CA) to produce the p62DBD/WT plasmid. This expression plasmid adds an N-terminal polyhistidine tag containing the Xpress epitope to the DBD fragment.
The p62DBD/WT expression plasmid was transformed into E. coli host strain BL21 and maintained as glycerol stocks. Bacterial colonies were inoculated into 1 liter of LB broth with 100 μg/ml ampicillin and grown with shaking at 37°C until the optical density at 600 nm (OD600) was 0.4. IPTG (isopropyl-β-d-thiogalactopyranoside) was then added to a final concentration of 0.1 mM, and induction followed at 37°C for a further 4 h. Bacteria were harvested by centrifugation and resuspended in 50 ml of nickel-Sepharose column start buffer 10 mM sodium phosphate ([pH 7.4], 500 mM NaCl, and 40 mM imidazole). The bacteria were disrupted by eight rounds of sonication for 15 s each time at a duty cycle of 10% and output of 4. After sonication, lysates were clarified by filtration through a 0.20-μm syringe filter and applied to a 1-ml nickel-Sepharose column. Following a 10-ml wash with start buffer, protein was eluted with 300 mM imidazole and dialyzed into heparin-Sepharose column start buffer (20 mM sodium phosphate [pH 7.4] and 200 mM NaCl), and the partially purified DBD was applied to a 1-ml heparin-Sepharose column. Following a 10-ml wash with start buffer, elution was performed with start buffer containing 1 M NaCl and yielded a highly purified preparation of the DBD at average concentrations of 4 mg/ml (8 mg total). Purity of the DBD was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie staining or Western blotting, and protein concentrations were determined through Bradford assays. Peak fractions were pooled and dialyzed against buffer containing 50% glycerol and stored at −20°C.
EMSAs.
Various amounts of the purified DBD were incubated at room temperature with 1 ng of end-labeled oligonucleotide probes in buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 100 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol [DTT], and 50 μg/ml bovine serum albumin [BSA]). After incubation for 15 min, 0.2 volume of 10% Ficoll was added to the incubation mix. Samples were applied to a nondenaturing 5% polyacrylamide gel with 0.25× Tris-borate-EDTA running buffer and run at 150 V at 4°C for 3 h. Complexes and unbound probe were detected and quantified by autoradiography using a personal FX phosphorimager (Bio-Rad, Hercules, CA).
Transient transfections and reporter gene assays.
MeWo cells were transfected with specific plasmids using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Six microliters of Lipofectamine reagent was used per microgram of transfected DNA in each transfection. In transfections performed in 12-well plates, 6.9 × 105 MeWo cells per well were seeded in 1 ml of complete growth medium. The cells were 80% confluent at the time of transfection. One microgram of a given firefly luciferase or dual-luciferase reporter vector was cotransfected with various amounts of the pCMV62 plasmid and complementary amounts of the pCDNA3.1 plasmid to equalize the total amount of DNA and CMV IE promoter sequences in each transfection. In superinfection experiments, melanoma cells in 12-well plates were transfected with reporter plasmids, and 24 h later the monolayer was overlaid with 1.5 × 105 VZV-infected cells. Cells were collected 48 h after transfection and were lysed by alternating freeze-thaw cycles.
Extracts of cells transfected with luciferase reporter vectors were prepared in 200 μl of lysis buffer (50 mM HEPES [pH 7.4], 250 mM NaCl, 1% NP-40, 1 mM EDTA) followed by dual-luciferase assays. Dual-luciferase assays were performed using the dual-luciferase reporter assay system (Promega, Madison, WI) with 20 μl of cell extracts per the manufacturer's instructions. All transfections were performed in triplicate, and superinfections were performed with nine samples. All results were confirmed by independent experiments, and firefly luciferase activities were normalized to Renilla luciferase activity by using a constant amount of pFRL plasmid in the transfections. Firefly luciferase activities were represented as fold induction of the measured activity over basal activity, with the wild-type reporter vector in the absence of effector vector.
Magnetic bead recruitment assays.
Magnetic bead recruitment assays were performed as described by Peng et al. (35) using 128-bp biotinylated DNA fragments containing either the wild-type gI promoter or the mutant promoter containing the IE62 consensus sequence. The presence of IE62 and Sp1 in the eluates from the beads was determined by immunoblotting.
RESULTS
IE62 consensus sites are distributed throughout the VZV genome.
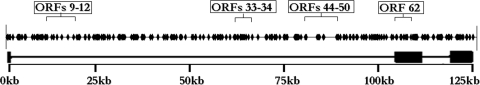
Previous work suggested that the IE62 consensus sequence may act as a positive cis element in the mechanism of IE62 transactivation. However, several well-characterized VZV promoters lack IE62 consensus sites yet are still efficiently activated by IE62 (18, 37). Further, IE62 has been shown to activate model promoters containing only the adenovirus late promoter TATA element (36, 56). As a first step in assessing the role of the consensus site in IE62-mediated gene regulation, we wished to determine the number of consensus sites occurring within the entire VZV genome. The VZV genome (human herpesvirus 3 [strain Dumas]; GenBank no. X04370.1) was searched for the IE62 consensus site (ATCGT) using the NCBI Blastn program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The total number of sites identified was 245, and these sites were distributed throughout the VZV genome in both noncoding and coding regions (Fig. 1). This number is in close agreement with the 260 sites predicted using the method developed by von Hippel (51) for estimation of the number of sites which will occur at random within a genome based on the length and sequence of the site and the size and G+C content of the genome being analyzed. Thus, there appears to be no obvious enrichment or dearth of IE62 binding sites within the VZV genome based on this analysis.
FIG. 1.
Schematic of the VZV genome indicating the positions of all IE62 consensus sites. Boxes represent regions with sparse or dense consensus sites. Sites were determined to be potentially within promoter regions when positioned within 200 to 300 bp upstream of the translational start site of an open reading frame.
The focus of the work reported here was to determine the effect of the presence of the IE62 consensus sequence within promoter regions on IE62-mediated transactivation. A relatively small number of VZV promoter regions have been extensively characterized, with the emphasis being on identification of the TATA elements and the roles of binding sites for ubiquitous cellular factors such as Sp1, USF, and AP1 (35, 40, 54, 56). Therefore, in order to estimate the number of consensus sites occurring in VZV promoters, we used a conservative putative promoter size of either 200 or 300 bp upstream from the translational start sites of the VZV ORFs. Using these parameters, between 26 (200 bp) and 39 (300 bp) VZV promoters contain 28 to 54 consensus sites, respectively. Therefore, it is possible that over one-half of the 71 VZV promoter regions contain at least one IE62 consensus binding site.
Purification of the IE62 DBD fragment.
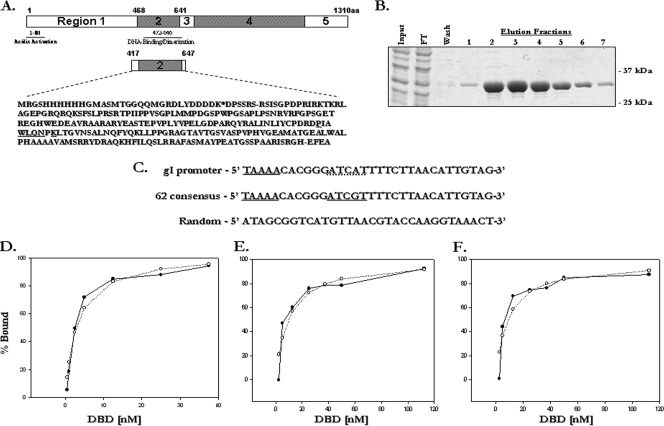
All previous studies examining IE62 DNA-binding were performed using partially purified IE62 fragments containing the IE62 DBD (48, 49, 50, 52). Attempts during the course of this work to purify full-length baculovirus-expressed IE62 free of nucleases and minor contaminants resulted in low yields and preparations containing numerous breakdown products. Therefore, to study the binding of IE62 to duplex DNA oligonucleotides containing or lacking the ATCGT consensus site, an N-terminal His-Xpress-tagged construct expressing an IE62 fragment containing the IE62 DBD was generated (Fig. 2). This fragment encompasses amino acids 417 to 647 and was previously used in partially purified form by Tyler and Everett (49) to identify the IE62 consensus binding site.
FIG. 2.
Expression of the IE62 DBD and characterization of its DNA-binding Activity. (A) Schematic showing the five regions spanning the length of the IE62 protein and the amino acid sequence of the tagged recombinant IE62-DBD. The region of homology with the helix-turn-helix motif of the Drosophila AntP homeodomain is underlined. (B) Coomassie-stained SDS-PAGE gels of fractions from a typical elution of a nickel-Sepharose column with 300 mM imidazole. (C) Sequence of the 62 consensus, gI promoter, and random oligonucleotide probes used in gel shift assays. Underlining indicates the partial IE62 consensus site within the gI promoter region and the full consensus site within the 62 consensus probe. (D, E, and F) Typical results from EMSAs for the 62 consensus, gI promoter, and random oligonucleotide probes, respectively. The data (•) were fit to a single-site-binding model (○) using the SigmaPlot (Systat) program. DNA binding is shown as the fraction of total probe bound. KDs for each probe were calculated using the SigmaPlot program from data obtained from three independent experiments.
A dual chromatographic purification procedure utilizing nickel-Sepharose and heparin-Sepharose chromatography was used to obtain the purest possible product. Peak fractions from the heparin-Sepharose column were combined, dialyzed, and stored prior to use as described in Materials and Methods. The two columns were chosen not only for their specific properties but also for their opposing overall charges in order to decrease nonspecific protein contamination in the final preparation. This procedure, yielding an average of 8 mg of DBD per liter of bacterial culture, resulted in a highly pure product that exhibited no contaminating bands upon SDS-PAGE analysis and displayed no evidence of nuclease activity in EMSAs.
Interaction of the IE62 DBD with consensus and nonconsensus DNA sequences.
To determine the effect of the presence of the consensus site on the affinity of the IE62 DBD for DNA, three 32-nucleotide duplex probes were generated for use as binding substrates (Fig. 2C). Two contained sequences derived from the VZV gI promoter. This promoter is responsive to IE62 and contains the sequence 5′-ATCAT-3′, which differs from the IE62 consensus site by the substitution of A for G in the fourth position. The first probe contained the native VZV gI promoter sequence (gIprom). The second probe consisted of the same sequence with a single nucleotide substitution of a G for an A, thus forming a full consensus sequence of ATCGT (62con). A third probe containing random sequence that lacked any full or partial IE62 consensus sequences was generated using the Random DNA Oligo Generator program (http://www-personal.umich.edu/∼kning/random.html). These probes were then employed in EMSA experiments using the purified IE62 DBD fragments.
In these experiments, a fixed amount of the individual 32P-labeled probes was incubated with increasing amounts of the IE62 DBD. The percentage of DBD-DNA complex formation at each DBD concentration was quantified using an FX phosphorimager (Bio-Rad, Hercules, CA) and was fitted to a single-site ligand-binding equation using the SigmaPlot program (Systat Software, Chicago, IL) (Fig. 2D to F). The calculated KDs (equilibrium dissociation constants) using data from triplicate EMSAs show that the DBD has a 4- to 5-fold-greater affinity for the 62con probe (KD = 2.6 ± 0.4 nM) than for the gIprom probe (KD = 10.3 ± 0.8 nM). The affinity for the random probe (KD = 10.2 ± 1.9 nM) was virtually identical to that for the gIprom probe, suggesting that a single base substitution within the consensus sequence results in binding that is indistinguishable from that to random DNA. These results represent important baseline parameters with respect to the DBD and IE62 binding but do not discount the possibility that greater differences may occur with full-length IE62, where the presence of other domains may influence DNA-binding.
The IE62 consensus site represses IE62-mediated activation when placed within native and model promoters.
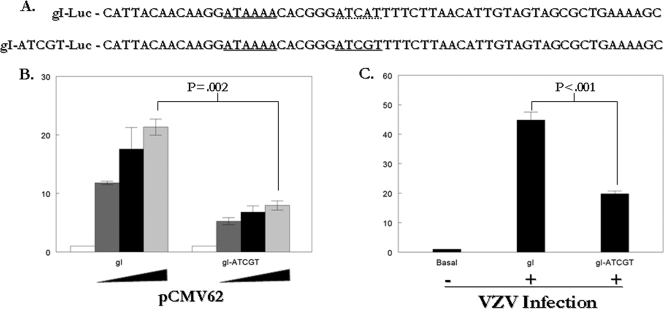
The next series of experiments examined the effect of the IE62 consensus site when it was placed within native and model promoter systems. The gI-Luc reporter contains the VZV gI promoter, which has been extensively characterized and has been previously shown to be transactivated by IE62 (18, 19, 35). In order to address the question of the effect of the presence of the true 5-bp IE62 binding site on expression from a native promoter element, a reporter plasmid, pgI-Luc/ATCGT, was generated by mutation of the ATCAT sequence within the gI-Luc reporter plasmid to the ATCGT IE62 consensus sequence (Fig. 3A). No transcription factor binding sites were created or lost upon mutation of the sequence based on an analysis with the AliBaba2 program (http://www.gene-regulation.com) using 80% matrix conservation. These reporter plasmids were transfected into MeWo cells with increasing amounts of the pCMV62 expression plasmid and assayed for luciferase activity. The presence of the IE62 consensus site resulted in an approximately 50% decrease in IE62-mediated transactivation relative to the wild-type promoter at all levels of the pCMV62 plasmid added (Fig. 3B). Since the only VZV protein present in these experiments was IE62, a second series of experiments was performed in which transfection of the reporter plasmid was followed by superinfection with VZV-infected cell stocks. Once again, the level of luciferase activity observed with the pgI-Luc/ATCGT was approximately 50% of that observed with the wild-type promoter (Fig. 3C).
FIG. 3.
Insertion of an IE62 consensus site lowers transactivation of the gI promoter in transfection and superinfection experiments. (A) Sequences of the native (top) and mutated (bottom) gI promoter regions. Underlining indicates the positions of the wild-type partial consensus site, the mutated full consensus site, and the atypical gI promoter TATA sequence ATAAA. (B) Results of reporter assays in MeWo cells transfected with 1 μg of the pgI-Luc and pgI-Luc/ATCGT reporter plasmids and 0.01 μg, 0.02 μg, or 0.04 μg of pCMV62. (C) Results of reporter assays in MeWo cells transfected with 1 μg of the pgI-Luc and pgI-Luc/ATCGT reporter plasmids followed by superinfection with VZV-infected cells 24 h following the initial transfection. The luciferase activity from the reporter in the absence of infection or cotransfection was normalized to 1, and experimental activities are reported as n-fold activation over this level.
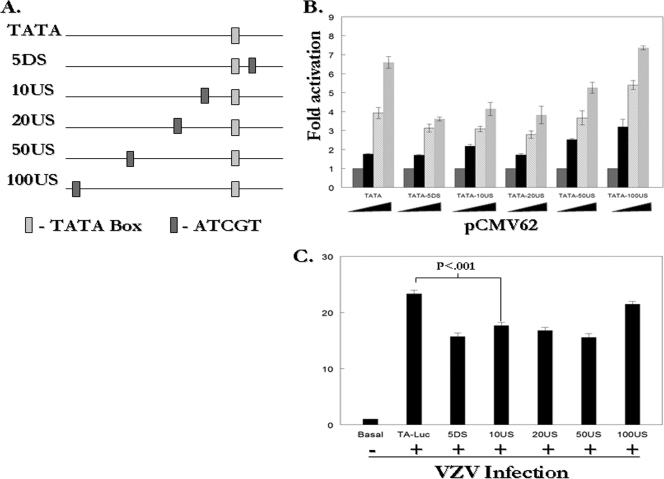
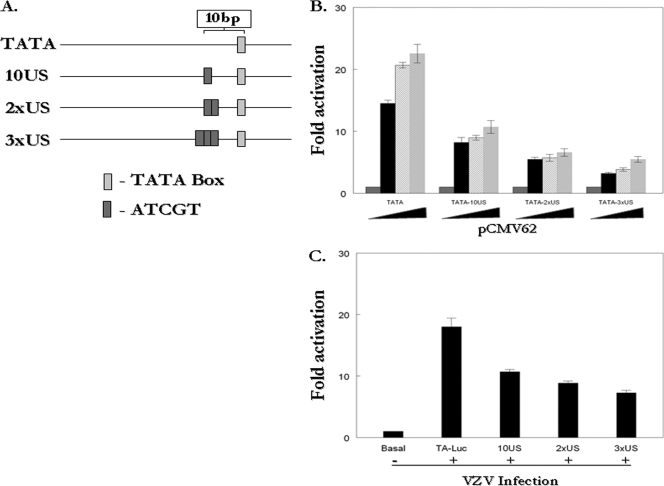
These results suggested that the IE62 consensus site may have a repressive function. However, the consensus site generated in the gI-Luc/ATCGT plasmid lies 6 bp downstream of the gI promoter TATA element and 10 bp upstream of the transcription start site (25). Thus, the consensus site-specific repression observed in the context of the gI promoter could be due to interference with binding of the basal transcriptional machinery based on both the position of the site and increased IE62 affinity. Therefore, the effect of consensus site position within a promoter was examined using reporter constructs based on the minimal model promoter pTA-Luc, which contains only a TATA box and is responsive to IE62 (56). The plasmids p5DS-Luc, p10US-Luc, p20US-Luc, p50US-Luc, and p100US-Luc were generated with consensus sites placed within the pTA-Luc plasmid at positions 5 bp downstream or 10, 20, 50, or 100 bp upstream of the TATA box, respectively (Fig. 4A). The IE62-mediated activation of each reporter was then assessed relative to the pTA-Luc promoter. When the site was placed 5 bp downstream, a position similar to that in the gI promoter, we again observed a decrease in activation in the presence of increasing amounts of pCMV62, with approximately 50% reduction in luciferase activity compared to that seen with pTA-Luc at the highest levels of the IE62 expression plasmid used (Fig. 4B). Similar levels of repression were observed with consensus sites at the 10US and 20US positions. This effect was reduced at the 50US position and lost at the 100US position. In order to be certain that the observed repression was not an artifact of overexpression of IE62 from the pCMV62 plasmid, the experiments with the model promoters were repeated in the context of VZV superinfection. Repression was observed with all consensus site-containing reporters except for p100US-Luc (Fig. 4C). The levels of repression were less than those obtained with IE62 alone, suggesting that the suppression seen with a single consensus site may be blunted by competition for IE62 by other viral promoters and viral proteins and/or cellular factors modified by infection. However, the effects were statistically significant, again indicating that the IE62 consensus site acted as a repressive or down-modulatory cis element.
FIG. 4.
The IE62 consensus site has a position-dependent inhibitory effect on the activity of a minimal model promoter containing only an IE62-responsive TATA box. (A) Consensus site positions in model promoter constructs relative to the TATA box. (B) Results from reporter assays in MeWo cells transfected with 1 μg of the pTA-Luc, p5DS-Luc, p10US-Luc, p20US-Luc, p50US-Luc and p100US-Luc reporter plasmids and 0.01 μg, 0.02 μg or 0.04 μg of pCMV62. (C) Results of reporter assays in MeWo cells transfected with 1 μg of the reporter plasmids followed by superinfection with VZV infected cells 24 h following the initial transfection. The luciferase activity from the reporter in the absence of infection or cotransfection was normalized to 1 and experimental activities are reported as n-fold activation over this level.
In the final series of experiments with model promoters, the effects of the presence of multiple tandem copies of the consensus on IE62-mediated activation were determined. In these experiments, reporter plasmids were generated with one, two, or three consensus sites beginning 10 bp upstream of the TATA box in the pTA-Luc reporter plasmid (Fig. 5A). The results of expression in the presence of increasing amounts of the pCMV62 plasmid are presented in Fig. 5B and show increasing repression with increasing numbers of consensus sites at all levels of pCMV62 used. Very similar results were obtained in superinfection experiments with multiple consensus sites (Fig. 5C).
FIG. 5.
Tandem consensus sites exacerbate the inhibitory effect seen on IE62 transactivation of a model TATA promoter in transfection experiments. (A) Schematic showing position of the IE62 consensus site relative to the TATA box in the model promoter constructs. (B) Results of reporter assays. One microgram of pTA-Luc, p10US-Luc, p2xUS-Luc, or p3xUS-Luc reporter plasmid was cotransfected with 0.01 μg, 0.02 μg, or 0.04 μg of pCMV62 into MeWo cells. The luciferase activity from the reporters in the absence of infection or cotransfection was normalized to 1, and experimental activities are reported as n-fold activation over this level. (C) Results of reporter assays in MeWo cells transfected with 1 μg of the reporter plasmids followed by superinfection with VZV infected cells 24 h following the initial transfection. The luciferase activity from the reporter in the absence of infection or cotransfection was normalized to 1, and experimental activities are reported as n-fold activation over this level.
The three consensus sites within the ORF62 promoter have repressive effects.
All of the experiments described above involved insertion of IE62 consensus sites into IE62-responsive promoters. The next series of experiments examined the effect of mutation of existing consensus sites. The promoter chosen was that controlling expression of ORF62. IE62 both positively and negatively regulates its own expression, and the ORF62 promoter contains three consensus binding sites (9, 39). These three binding sites were first identified by Tyler and Everett (49) and were confirmed to be present in the ORF62 promoter construct generated as described in Materials and Methods. The ORF62 promoter region also contains binding sites for numerous cellular transcription factors (GA-binding protein, Sp1, CCAAT-binding proteins, and ATF/CRE factors) and the ORF10/Oct-1/HCF-1 (enhancer core) complex (27, 33) (Fig. 6). The three consensus sites within the ORF62 promoter are positioned as follows: two occur 267 (site 1) and 252 (site 2) bp upstream of the previously identified TATA box. The third site (site 3) occurs 72 bp upstream relative to the TATA box and is adjacent to the enhancer core site. Sites 1 and 3 are oriented in the same direction relative to the TATA box, whereas site 2 is oriented in the opposite direction.
FIG. 6.
The ORF 62 promoter sequence. Binding sites for the following viral and cellular factors are underlined: GA-binding protein, CCAAT-binding proteins, ATF/CRE factors, Sp1, and the VZV IE enhancer core (EC) complex composed of Oct-1/HCF-1/ORF10. The three consensus sites are located 267 (1), 252 (2), and 72 (3) base pairs upstream of the reported TATA box. Site 2 is in the opposite orientation relative to sites 1 and 3.
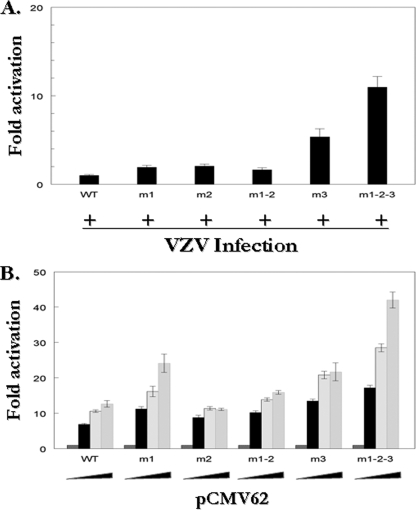
Site-directed mutagenesis was used to functionally remove the sites, both individually and collectively, by altering the ATCGT sequence to ATCAT, creating the reporters m1, m2, m1-2, m3, and m1-2-3. The effects of these mutations were initially evaluated in transfection/superinfection experiments, since the VZV ORF10 protein would be present allowing formation of the enhancer core complex. As shown in Fig. 7A, when distal sites 1 and 2 were mutated individually, a 2-fold increase in promoter activity was observed in each case. In the case of both sites 1 and 2 being mutated in unison (m1-2), the same 2-fold increase in activity was seen. This suggested that these sites had a repressive effect on the ORF62 promoter and that both sites are required to exert that effect. When the more proximal site 3 was mutated, a 5-fold increase in luciferase activity was observed, again indicating a repressive function of this consensus site within the ORF62 promoter and suggesting that it is the major repressive site. When all three sites were mutated in concert, a combined effect was seen, yielding a 10-fold increase in promoter activity.
FIG. 7.
Consensus sites suppress transactivation of the ORF62 promoter. The native ORF62 promoter consensus binding sequences (1, 2, and 3) were mutated alone or in unison to generate the m1, m2, m1-2, m3, and m1-2-3 mutant promoters. One microgram of wild-type or mutant reporter plasmid was transfected into MeWo cells with 0.01 μg, 0.02 μg, or 0.04 μg of the pCMV62 expression plasmid (B) or followed by superinfection 24 h posttransfection (A). The luciferase activity from the reporters in the absence of cotransfection was normalized to 1. In superinfection experiments, activity from the wild-type promoter in the presence of infection was set to 1, and experimental activities are reported as n-fold activation over these levels.
A second series of experiments examined the function of the three IE62 consensus sites within the ORF62 promoter, using pCMV62 transfection experiments in order to assess the effect of IE62 alone. In the absence of viral infection and therefore in the lack of the enhancer core complex, sites 1 and 3 showed the repressive effect observed in the superinfection experiments (Fig. 7B). Mutation of site 2 no longer showed an increase in expression in these assays. Also, the m1-2 double mutation consistently showed lower activity than the m1 promoter mutation. This would suggest that the relief of repression seen when mutating site 1 is somehow reduced by mutating site 2 along with it. Finally, the increase in expression seen with mutation of site 3 in these assays was 2-fold compared to 5-fold in superinfection assays, and the level of increased expression from the m1-2-3 promoter element was less than that observed with superinfection. These results suggest that other viral proteins may be involved in the function of the IE62 consensus site.
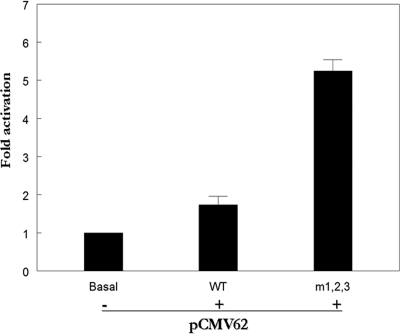
The IE62 consensus site represses the ORF62 promoter in primary fetal rat DRG neurons.
Currently very little is known regarding the details of VZV gene expression in neurons of the dorsal root ganglia (DRG), the primary target of VZV latency. The IE62 consensus sites present in the ORF62 promoter could play a role in establishment of VZV latency through their repressive effect on the activity of the promoter during primary infection of DRG neurons. To gain insight into the role these sites might play, primary fetal rat DRG neuronal cultures were transfected with the p62prom/WT and p62prom/m1-2-3 reporter plasmids and pCMV62. The mutant promoter exhibited a 3-fold increase in reporter activity compared to the wild type (Fig. 8), results very similar to those obtained in MeWo cells (Fig. 7B). Thus, no significant increase or decrease in repression associated with the presence of these sites was observed. However, the wild-type promoter was less responsive to IE62 in the DRG neurons. In MeWo cells, IE62 was capable of transactivating to 10-fold over basal levels at the highest (0.04/1) effector-to-reporter plasmid ratio used (Fig. 7B), whereas a ratio of effector to reporter plasmid that was 2-fold higher (0.08/1) resulted in at best a 2-fold increase over basal levels in the neurons. Thus, IE62-mediated activation of its own promoter may be decreased in neurons compared to other cell types.
FIG. 8.
Consensus sites suppress IE62-mediated transactivation of the ORF62 promoter in primary rat dorsal root ganglion neurons. Two micrograms of either the wild-type or m1-2-3 reporter plasmid was cotransfected with 0.16 μg of pCMV62 expression plasmids into primary DRG neurons. The luciferase activity from the reporters in the absence of cotransfection was normalized to 1. Experimental activities are reported as n-fold activation over this level.
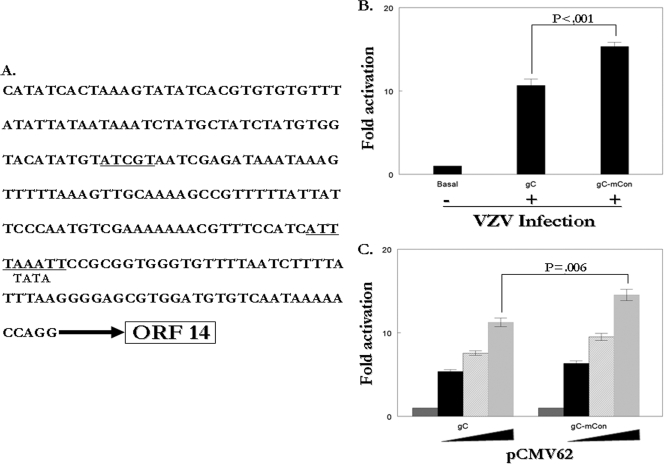
The single consensus site within the gC promoter has a repressive effect.
Due to the complexity of the ORF62 promoter, a much less complex native VZV promoter containing an IE62 consensus site was also chosen for analysis. The expression of VZV gC is currently obscure. Even late in infection, few cells express gC, and this expression occurs at various levels. The VZV ORF14 promoter, which drives the expression of VZV glycoprotein C, is responsive to IE62-mediated activation and has previously been reported to contain an atypical TATA box near the transcriptional start site and sites for inducible cellular Hox family factors far upstream (47). Our computer analysis identified an IE62 consensus site 76 bp upstream from the putative TATA box within the gC promoter (Fig. 9A). Mutation of this site to ATCAT resulted in a 20 to 40% increase in activation of the promoter by either VZV superinfection or transfection with pCMV62 (Fig. 9B and C). Therefore, it appears this site within the gC promoter has a repressive function similar to all other sites in both model and native VZV promoters tested in this study. While the repressive effects reported here are modest, this work is the first identification of an IE62 site possibly influencing gC expression. Since IE62 exits the nucleus late in infection (20, 21, 26), it is possible that the decrease in nuclear concentration of IE62 could relieve the repression identified here and be one of the factors involved in expression of VZV gC.
FIG. 9.
Mutation of a native consensus site increases IE62-mediated transactivation of the gC promoter. (A) Position of the IE62 consensus site relative to the gC TATA box. One microgram of wild-type or mutant reporter plasmid was transfected into MeWo cells with 0.01 μg, 0.02 μg, or 0.04 μg of the pCMV62 expression plasmid (C) or followed by superinfection 24 h posttransfection (B). The luciferase activity from the reporters in the absence of infection or cotransfection was normalized to 1, and experimental activities are reported as n-fold activation over this level.
The presence of the IE62 consensus site increases binding of IE62 but not Sp1 to the gI promoter.
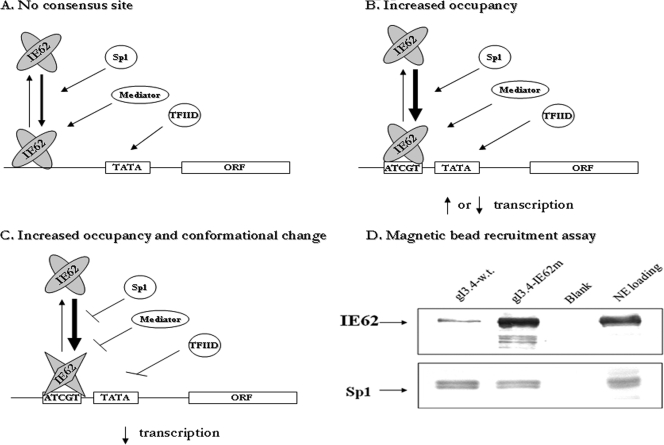
The increased binding of the IE62 DBD to the IE62 consensus sequence suggests a model in which the full-length IE62 protein binds more tightly to promoters containing such sites and increases its occupancy (Fig. 10). Since IE62 is known to recruit cellular factors such as Sp1 (34, 35), increased IE62 binding could increase binding of such cellular factors. This increase in binding of cellular factors and IE62 could result in either an increase or decrease in activation depending upon whether they enhance or occlude binding of the general transcription apparatus (Fig. 10B). Alternatively, the increased affinity of IE62 for its consensus site could result in a situation where IE62 undergoes a conformational change that decreases or eliminates its ability to recruit cellular factors (Fig. 10C). In this case, the presence of the consensus site would be expected to have a repressive effect. In order to test these possibilities, the binding of IE62 and Sp1 to the native gI-Luc promoter and the gI-Luc/ATCGT promoter was examined in magnetic bead recruitment assays. As shown in Fig. 10D, the significant increase in IE62 binding to the gI-Luc/ATCGT sequence (8-fold) did not result in an observable increase in Sp1 recruitment, suggesting that the latter possibility may be correct.
FIG. 10.
(A) Model of IE62-mediated transactivation in the absence of a consensus site. (B) Model of simple increased occupancy of the promoter in the presence of a consensus site, resulting in either increased or decreased transcription depending on whether the site properly positions IE62 to make its protein-protein contacts. (C) Model of increased occupancy coupled with a conformational change which interrupts IE62 protein-protein interactions upon binding to the consensus site, resulting in the reduced activity observed in almost all tested positions within native and model promoters. (D) Results from magnetic bead recruitment assays examining IE62 and Sp1 binding to the wild-type gI and gI62con promoter elements. The presence of Sp1 and IE62 in eluants of bound proteins was detected by immunoblotting with rabbit polyclonal antibodies specific for the two proteins.
DISCUSSION
The IE62 protein is the major viral transactivator encoded by VZV (18, 39, 47, 54). Despite the importance of IE62 in VZV replication and the intense and extensive study of which it has been the subject, the fundamental question of the function of IE62 DNA binding and role played by the IE62 consensus binding site (5′-ATCGT-3′) has remained largely unexplored. The results presented here indicate that the site is present throughout the VZV genome and is found in both coding and noncoding regions. In contrast to the findings of Betz and Wydoski (4), predictions concerning the role of the IE62 consensus site by Tyler and Everett (49) and data obtained with ICP4 (15, 45), the presence of the true IE62 consensus site invariably had a repressive effect on IE62-mediated activation. This was true whether the site was incorporated into model and native promoters or a native consensus site was removed through mutation.
Our analysis of the total number of IE62 consensus sites identified 254 such sequences within the VZV genome. There does not appear to be a significant enrichment of sites within the 200- and 300-bp regions used to designate potential promoters. The 200-bp limit for 71 open reading frames amounts to 11.36% of the genome, and 28 (11.4%) of the consensus sites are present in these sequences. Similarly the 300-bp limit comprises 17% of the genome, and 54 (22%) of the consensus sites fall within these limits. Individual promoters, however, may contain clusters of sites, as evidenced by the ORF62 promoter, which contains three, or 5.5%, of the sites found using the 300-bp limit. Further analysis suggested a 2-fold enrichment in the frequency of sites containing either the sequence ATCGTCT or ATCGTTA. However, mutation of these sequences in the sixth and seventh positions had no effect on DNA binding (data not shown), in agreement with the findings of Tyler and Everett (49).
The EMSA experiments represent the first quantification of the binding affinity of the IE62 DBD. The value of 2.6 nM for the dissociation constant of the DBD with an oligomer derived from the VZV gI promoter and mutated by a single base substitution to contain the consensus site falls within the range of KDs (1 to 35 nM) found by Kuddus and DeLuca (24) for interaction of HSV-1 ICP4 with oligonucleotides of various lengths containing the ICP4 consensus site. Essentially identical KDs of approximately 10 nM were obtained for the oligonucleotide derived from the wild-type gI promoter sequence containing a site differing from the consensus by a single base and an oligonucleotide containing random sequence. Thus, it appears that a single base substitution in the IE62 consensus sequence renders its recognition indistinguishable from random sequence by the DBD.
A second important observation is that the binding isotherms were best fitted with a single-binding-site model. Since two DNA-binding sequences would be present per DBD homodimer, this suggests that a single binding site could be created by the interaction of the monomeric DBD polypeptides. Alternatively, there could be two sites present per homodimer but binding of DNA to one site precludes binding to the second site. The latter possibility may be supported by the results of Shepard and DeLuca (43, 44), who showed that heterodimers of HSV-1 ICP4 comprised of one monomer which retained DNA-binding activity and a mutant which did not resulted in complementation and restoration of function, although the DNA-binding activity of the heterodimer was weaker than that of the wild type.
The placement of the consensus site within the VZV gI promoter resulted in suppression of IE62-mediated transactivation. The fact that similar data were obtained with the mutant versus wild-type promoters in reporter assays involving superinfection indicates that the suppression also occurs upon expression of the remainder of the VZV genome and suggests that IE62 is likely responsible for this suppression via interaction with the consensus site. Since the consensus site generated within the gI promoter falls just downstream of the TATA box and just upstream of the start site of transcription, it was possible that this specific position was a factor in determining suppression. The results from the placement of the consensus site at various distances relative to the TATA box within the minimal model pTA-Luc reporter show this was not the case. A positional effect was seen only in the sense that suppression decreased with distance. Insertion of tandem copies of the consensus sequence resulted in increased suppression. This increased suppression could be due to multiple molecules of IE62 bound, more than one of which can interfere with transcription, or an increase in the probability that a single IE62 molecule is bound. There is also no evidence for the influence of rotational position of the site. The sites at +10 and −5 relative to the TATA box are predicted to be on opposite sides of the DNA helix, yet both show similar levels of repression in both the transfection and superinfection experiments. Finally, no binding sites for other activators, such as Sp1 and USF, were present in the model promoters used. Thus, in contrast to ICP4 (16, 17), IE62 is capable of repression of basal as well as activated transcription in the context of the consensus site.
ICP4 significantly downregulates its own expression through interaction with cellular factors and a consensus site located downstream of the ICP4 promoter TATA box (16). IE62, in contrast, activates expression of its own promoter (3, 39) via a mechanism involving the cellular factors Sp1 and HCF-1 (33). However, in reporter gene assays, the use of increasing amounts of effector plasmids expressing IE62 resulted in response curves with high levels of expression using small amounts of the plasmid and lower levels of expression at the highest levels of plasmid used. This effect was attributed to sequestration of cellular factors by excess IE62 by analogy with other transcriptional activators (12, 13, 39). The results obtained in this study, however, indicate that this dampening of IE62 response is also in part due to the presence of three consensus binding sites present in the IE62 promoter. Mutation of the three sites individually showed minimal to moderate effects on IE62-mediated activation. Simultaneous mutation of all three sites in the context of superinfection, however, resulted in 10-fold increased expression, suggesting that the three sites act in concert to downregulate expression.
Mutation of site 3 within the ORF62 promoter had a much greater effect than mutation of the two distal sites when superinfection rather than ectopic expression was used as the source of IE62. Site 3 is adjacent to the ORF10/Oct-1/HCF-1 complex binding site, which has been shown to be required for one of the two mechanisms for ORF62 expression (31, 33). Thus, binding of IE62 to this site could interfere with the interaction of the heterotrimeric complex with the enhancer core sequence and decrease the level of IE62 expression during infection. Presumably this would occur at high concentrations of IE62 within the cell. Such a mechanism could be involved in modulation of IE62 levels in infected cells, allowing maximal replication while decreasing potential problems with overexpression of viral and possibly cellular genes by IE62. This mechanism could also play a role in limiting IE62 expression and therefore transactivation of other viral promoters during establishment of latency, based on the data from the transfection experiments using fetal rat DRG neurons.
The IE62 consensus site may contribute to downregulation or down modulation of promoters by at least two mechanisms. One is being positioned near critical binding sites for other factors needed for expression, such as site 3 within the ORF62 promoter, and occluding that site. The second mechanism would be much more general, as evidenced by the results from the model promoters and the VZV gI and gC promoters. This mechanism could involve a conformational change within the IE62 molecule upon binding to its consensus site, which makes it less able to interact with or recruit elements of the cellular transcription machinery. Such a mechanism is supported by the fact that the difference in KD between oligonucleotides containing the consensus site and those containing other sequences is only 5-fold. This hypothesis is further strengthened by the recruitment assays using the gI promoter fragment, where introduction of the full consensus sequence resulted in an increase of IE62 binding without a concomitant increase in Sp1 recruitment.
The repressive role of IE62 consensus sites could be useful for examination of the function of VZV gene products. Single or multiple tandem consensus sites could be inserted into the promoters of individual genes, resulting in a graded downregulation of expression but not necessarily complete shutdown. Thus, the effects of lower levels of specific viral gene products could be assessed in the context of viral pathogenesis in specific tissue types by modulated repression of skin-, T-cell-, and neuron-specific viral factors. Such a strategy could be used to attenuate the virus in a highly specific and controlled manner. Thus, judicious placement of the consensus site could be used to reduce viral spread and the incidence of latency in future vaccine strains.
Acknowledgments
This work was supported by grant AI18449 from the National Institutes of Health and grants from the John R. Oishei Foundation and the National Shingles Foundation. K.W. was supported by NIH predoctoral Training Grant T32AI07614.
Footnotes
Published ahead of print on 3 February 2010.
REFERENCES
- 1.Arvin, A. M. 2001. Varicella-zoster virus, p. 2731-2767. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Lippincott-Raven, Philadelphia, Pa.
- 2.Batchelor, A. H., and P. O'Hare. 1990. Regulation and cell-type-specific activity of a promoter located upstream of the latency-associated transcript of herpes simplex virus type 1. J. Virol. 64:3269-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudoux, L., P. Defechereux, S. Schoonbroodt, M. P. Merville, B. Rentier, and J. Piette. 1995. Mutational analysis of varicella-zoster virus major immediate-early protein IE62. Nucleic Acids Res. 23:1341-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betz, J. L., and S. G. Wydoski. 1993. Functional interaction of varicella zoster virus gene 62 protein with the DNA sequence bound by herpes simplex virus ICP4 protein. Virology 195:793-797. [DOI] [PubMed] [Google Scholar]
- 5.Bohenzky, R. A., A. G. Papavassiliou, I. H. Gelman, and S. Silverstein. 1993. Identification of a promoter mapping within the reiterated sequences that flank the herpes simplex virus type 1 UL region. J. Virol. 67:632-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. K. 1989. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 17:4637-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. I., D. Heffel, and K. Seidel. 1993. The transcriptional activation domain of varicella-zoster virus open reading frame 62 protein is not conserved with its herpes simplex virus homolog. J. Virol. 67:4246-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiDonato, J. A., and M. T. Muller. 1989. DNA binding and gene regulation by the herpes simplex virus type 1 protein ICP4 and involvement of the TATA element. J. Virol. 63:3737-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Disney, G. H., T. A. McKee, C. M. Preston, and R. D. Everett. 1990. The product of varicella-zoster virus gene 62 autoregulates its own promoter. J. Gen. Virol. 71:2999-3003. [DOI] [PubMed] [Google Scholar]
- 10.Everett, R. D., M. Elliott, G. Hope, and A. Orr. 1991. Purification of the DNA binding domain of herpes simplex virus type 1 immediate-early protein Vmw175 as a homodimer and extensive mutagenesis of its DNA recognition site. Nucleic Acids Res. 19:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faber, S. W., and K. W. Wilcox. 1986. Association of the herpes simplex virus regulatory protein ICP4 with specific nucleotide sequences in DNA. Nucleic Acids Res. 14:6067-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelman, I. H., and S. Silverstein. 1986. Co-ordinate regulation of herpes simplex virus gene expression is mediated by the functional interaction of two immediate early gene products. J. Mol. Biol. 191:395-409. [DOI] [PubMed] [Google Scholar]
- 13.Gill, G., and M. Ptashne. 1988. Negative effect of the transcriptional activator GAL4. Nature 334:721-724. [DOI] [PubMed] [Google Scholar]
- 14.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu, B., and N. DeLuca. 1994. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. J. Virol. 68:7953-7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu, B., R. Kuddus, and N. A. DeLuca. 1995. Repression of activator-mediated transcription by herpes simplex virus ICP4 via a mechanism involving interactions with the basal transcription factors TATA-binding protein and TFIIB. Mol. Cell. Biol. 15:3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu, B., R. Rivera-Gonzalez, C. A. Smith, and N. A. DeLuca. 1993. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proc. Natl. Acad. Sci. U. S. A. 90:9528-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, H., D. Boucaud, J. Hay, and W. T. Ruyechan. 2001. Cis and trans elements regulating expression of the varicella zoster virus gI gene. Arch. Virol. Suppl. 2001(17):57-70. [DOI] [PubMed] [Google Scholar]
- 19.Ito, H., M. H. Sommer, L. Zerboni, H. He, D. Boucaud, J. Hay, W. Ruyechan, and A. M. Arvin. 2003. Promoter sequences of varicella-zoster virus glycoprotein I targeted by cellular transactivating factors Sp1 and USF determine virulence in skin and T cells in SCIDhu mice in vivo. J. Virol. 77:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinchington, P. R., K. Fite, and S. E. Turse. 2000. Nuclear accumulation of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, is inhibited by phosphorylation mediated by the VZV open reading frame 66 protein kinase. J. Virol. 74:2265-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinchington, P. R., and S. E. Turse. 1998. Regulated nuclear localization of the varicella-zoster virus major regulatory protein, IE62. J. Infect. Dis. 178(Suppl. 1):S16-S21. [DOI] [PubMed] [Google Scholar]
- 22.Kleitman, N., P. M. Wood, and R. P. Bunge. 1998. Tissue culture methods for the study of myelination, p. 545-594, In G. Banker and K. Goslin (ed.), Culturing nerve cells. MIT Press, Cambridge, MA.
- 23.Kuddus, R., B. Gu, and N. A. DeLuca. 1995. Relationship between TATA-binding protein and herpes simplex virus type 1 ICP4 DNA-binding sites in complex formation and repression of transcription. J. Virol. 69:5568-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuddus, R. H., and N. A. DeLuca. 2007. DNA-dependent oligomerization of herpes simplex virus type 1 regulatory protein ICP4. J. Virol. 81:9230-9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling, P., P. R. Kinchington, M. Sadeghi-Zadeh, W. T. Ruyechan, and J. Hay. 1992. Transcription from varicella-zoster virus gene 67 (glycoprotein IV). J. Virol. 66:3690-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch, J. M., T. K. Kenyon, C. Grose, J. Hay, and W. T. Ruyechan. 2002. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 302:71-82. [DOI] [PubMed] [Google Scholar]
- 27.McKee, T. A., G. H. Disney, R. D. Everett, and C. M. Preston. 1990. Control of expression of the varicella-zoster virus major immediate early gene. J. Gen. Virol. 71:897-906. [DOI] [PubMed] [Google Scholar]
- 28.Michael, N., and B. Roizman. 1989. Binding of the herpes simplex virus major regulatory protein to viral DNA. Proc. Natl. Acad. Sci. U. S. A. 86:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michael, N., and B. Roizman. 1993. Repression of the herpes simplex virus 1 alpha 4 gene by its gene product occurs within the context of the viral genome and is associated with all three identified cognate sites. Proc. Natl. Acad. Sci. U. S. A. 90:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michael, N., D. Spector, P. Mavromara-Nazos, T. M. Kristie, and B. Roizman. 1988. The DNA-binding properties of the major regulatory protein alpha 4 of herpes simplex viruses. Science 239:1531-1534. [DOI] [PubMed] [Google Scholar]
- 31.Moriuchi, H., M. Moriuchi, and J. I. Cohen. 1995. Proteins and cis-acting elements associated with transactivation of the varicella-zoster virus (VZV) immediate-early gene 62 promoter by VZV open reading frame 10 protein. J. Virol. 69:4693-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriuchi, M., H. Moriuchi, S. E. Straus, and J. I. Cohen. 1994. Varicella-zoster virus (VZV) virion-associated transactivator open reading frame 62 protein enhances the infectivity of VZV DNA. Virology 200:297-300. [DOI] [PubMed] [Google Scholar]
- 33.Narayanan, A., M. L. Nogueira, W. T. Ruyechan, and T. M. Kristie. 2005. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J. Biol. Chem. 280:1369-1375. [DOI] [PubMed] [Google Scholar]
- 34.Narayanan, A., W. T. Ruyechan, and T. M. Kristie. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 104:10835-19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng, H., H. He, J. Hay, and W. T. Ruyechan. 2003. Interaction between the varicella zoster virus IE62 major transactivator and cellular transcription factor Sp1. J. Biol. Chem. 278:38068-38075. [DOI] [PubMed] [Google Scholar]
- 36.Perera, L. P. 2000. The TATA motif specifies the differential activation of minimal promoters by varicella zoster virus immediate-early regulatory protein IE62. J. Biol. Chem. 275:487-496. [DOI] [PubMed] [Google Scholar]
- 37.Perera, L. P., J. D. Mosca, W. T. Ruyechan, and J. Hay. 1992. Regulation of varicella-zoster virus gene expression in human T lymphocytes. J. Virol. 66:5298-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perera, L. P., J. D. Mosca, W. T. Ruyechan, G. S. Hayward, S. E. Straus, and J. Hay. 1993. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J. Virol. 67:4474-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perera, L. P., J. D. Mosca, M. Sadeghi-Zadeh, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate early protein, IE62, can positively regulate its cognate promoter. Virology 191:346-354. [DOI] [PubMed] [Google Scholar]
- 40.Rahaus, M., and M. H. Wolff. 2003. Reciprocal effects of varicella-zoster virus (VZV) and AP1: activation of jun, fos and ATF-2 after VZV infection and their importance for the regulation of viral genes. Virus Res. 92:9-21. [DOI] [PubMed] [Google Scholar]
- 41.Roberts, M. S., A. Boundy, P. O'Hare, M. C. Pizzorno, D. M. Ciufo, and G. S. Hayward. 1988. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (alpha 4) promoter and a specific binding site for the IE175 (ICP4) protein. J. Virol. 62:4307-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoonbroodt, S., J. Piette, L. Baudoux, P. Defechereux, B. Rentier, and M. P. Merville. 1996. Enhancement of varicella-zoster virus infection in cell lines expressing ORF4- or ORF62-encoded proteins. J. Med. Virol. 49:264-273. [DOI] [PubMed] [Google Scholar]
- 43.Shepard, A. A., and N. A. DeLuca. 1989. Intragenic complementation among partial peptides of herpes simplex virus regulatory protein ICP4. J. Virol. 63:1203-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shepard, A. A., and N. A. DeLuca. 1991. Activities of heterodimers composed of DNA-binding- and transactivation-deficient subunits of the herpes simplex virus regulatory protein ICP4. J. Virol. 65:299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smiley, J. R., D. C. Johnson, L. I. Pizer, and R. D. Everett. 1992. The ICP4 binding sites in the herpes simplex virus type 1 glycoprotein D (gD) promoter are not essential for efficient gD transcription during virus infection. J. Virol. 66:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spengler, M. L., W. T. Ruyechan, and J. Hay. 2000. Physical interaction between two varicella zoster virus gene regulatory proteins, IE4 and IE62. Virology. 272:375-381. [DOI] [PubMed] [Google Scholar]
- 47.Storlie, J., W. Jackson, J. Hutchinson, and C. Grose. 2006. Delayed biosynthesis of varicella-zoster virus glycoprotein C: upregulation by hexamethylene bisacetamide and retinoic acid treatment of infected cells. J. Virol. 80:9544-9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyler, J. K., K. E. Allen, and R. D. Everett. 1994. Mutation of a single lysine residue severely impairs the DNA recognition and regulatory functions of the VZV gene 62 transactivator protein. Nucleic Acids Res. 22:270-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyler, J. K., and R. D. Everett. 1993. The DNA binding domain of the varicella-zoster virus gene 62 protein interacts with multiple sequences which are similar to the binding site of the related protein of herpes simplex virus type 1. Nucleic Acids Res. 21:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyler, J. K., and R. D. Everett. 1994. The DNA binding domains of the varicella-zoster virus gene 62 and herpes simplex virus type 1 ICP4 transactivator proteins heterodimerize and bind to DNA. Nucleic Acids Res. 22:711-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Hippel, P. H. 1979. On the molecular basis of the specificity of interaction of transcriptional proteins with genome DNA, p. 279-347. In R. F. Goldberger (ed.), Biological regulation and development, vol. 1. Plenum, New York, NY.
- 52.Wu, C. L., and K. W. Wilcox. 1991. The conserved DNA-binding domains encoded by the herpes simplex virus type 1 ICP4, pseudorabies virus IE180, and varicella-zoster virus ORF62 genes recognize similar sites in the corresponding promoters. J. Virol. 65:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto, S., A. Eletsky, T. Szyperski, J. Hay, and W. T. Ruyechan. 2009. Analysis of the varicella-zoster virus IE62 N-terminal acidic transactivating domain and its interaction with the human mediator complex. J. Virol. 83:6300-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, M., J. Hay, and W. T. Ruyechan. 2004. The DNA element controlling expression of the varicella-zoster virus open reading frame 28 and 29 genes consists of two divergent unidirectional promoters which have a common USF site. J. Virol. 78:10939-10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, M., J. Hay, and W. T. Ruyechan. 2008. Varicella-zoster virus IE62 protein utilizes the human mediator complex in promoter activation. J. Virol. 82:12154-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, M., H. Peng, J. Hay, and W. T. Ruyechan. 2006. Promoter activation by the varicella-zoster virus major transactivator IE62 and the cellular transcription factor USF. J. Virol. 80:7339-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeh, L., and P. A. Schaffer. 1993. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J. Virol. 67:7373-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]