Abstract
Cytomegalovirus (CMV) infection in patients receiving hematopoietic stem cell transplants (HSCT) is associated with morbidity and mortality. Adoptive T cell immunotherapy has been used to treat viral reactivation but is hardly feasible in high-risk constellations of CMV-positive HSCT patients and CMV-negative stem cell donors. We endowed human effector T cells with a chimeric immunoreceptor (cIR) directed against CMV glycoprotein B. These cIR-engineered primary T cells mediated antiviral effector functions such as cytokine production and cytolysis. This first description of cIR-redirected CMV-specific T cells opens up a new perspective for HLA-independent immunotherapy of CMV infection in high-risk patients.
Primary infection by human cytomegalovirus (CMV) and reactivation of latent virus are major problems after hematopoietic stem cell transplantation (HSCT), resulting in inflammation of a wide range of organs, systemic disease, and an increased rate of graft-versus-host disease (GvHD) (3, 5, 21). Antiviral chemotherapy with nucleoside analogs is used prophylactically and preemptively in the early phase after transplantation, but long-term treatment is often associated with toxicity, selection of resistant virus variants, and the inability to prevent all CMV-associated complications (4, 7, 28). Sustained control of latent CMV infection depends on the restoration of a functional antiviral immune response (15, 25).
Adoptive T cell transfer has been used successfully to bridge the critical phase of delayed or insufficient antiviral response in patients with immune suppression. In CMV and Epstein-Barr virus (EBV) infection, the adoptive transfer of ex vivo-expanded, donor-derived, virus-specific T cells reduced virus titers in the recipient to levels similar to those in immunocompetent, healthy, seropositive controls (10, 23, 29, 31). Ex vivo expansion of these cells can be carried out by different procedures (9). In naïve seronegative persons, however, virus-specific T cells occur at very low frequencies, generally insufficient for expansion.
As an alternative, T cells can be grafted with defined specificities using recombinant immunoreceptors (11). The receptor specificity is determined by extracellular single-chain fragments of the variable region (scFv) that recognize predefined antigens and can easily be altered by selecting an appropriate scFv (16). Recombinant immunoreceptors have been successfully developed against a number of tumor antigens (16) but against only a few viral proteins from HIV and hepatitis B virus (6, 20, 22) and not against CMV.
In human CMV infection, the analysis of the physiological cytotoxic T lymphocyte (CTL) response has been focused on a limited set of proteins, namely, the proteins pp65, IE1, and IE2; recent and more extensive studies have shown that glycoprotein B (gB) as well as other CMV glycoproteins is also able to evoke adaptive T cell responses (32, 34). Notably, the CMV gB is expressed at the cell surface during the early or delayed early phase of CMV replication, even in the presence of clinically used inhibitors of viral DNA replication (33).
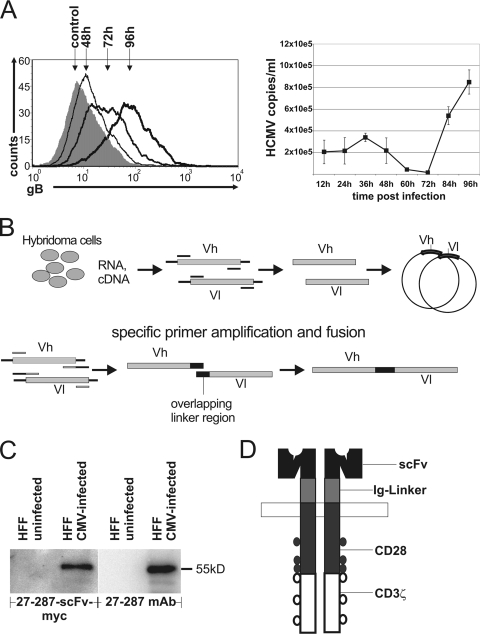
We hypothesize that engineered T cells with specificity for CMV gB have the potential to control CMV infection by specific elimination of infected cells. The time course of gB expression at the surfaces of infected HFF cells was investigated by flow cytometry with gB-specific antibody 27-287 and an anti-mouse immunoglobulin (Ig) Cy5 secondary antibody. Whereas surface expression of CMV gB could be confirmed 48 h after infection (Fig. 1A), one single replication cycle of CMV in cell culture required about 4 days, as detected by quantitative PCR. Supernatants from CMV strain Ad169-infected HFF cells were harvested, debris was removed (720 × g, 10 min), and the virus was pelleted (18,600 × g, 2 h, 4°C). Pellets were subjected to proteinase K digestion (100 μg/ml, 50 min at 56°C and 10 min at 95°C) and analyzed by quantitative real-time PCR (CMV5′, AAGCGGCCTCTGATAACCAAG; CMV3′, GAGCAGACTCTCAGAGGATCGG; 6-carboxyfluorescein [FAM]/6-carboxytetramethylrhodamine [TAMRA]-labeled CMV probe, CATGCAGATCTCCTCAATGCGCGC). CMV gB is thus accessible on the surfaces of infected cells before the viral replication cycle is completed and virus release and spreading occurs.
FIG. 1.
CMV gB expression on surfaces of infected cells and construction of chimeric immunoreceptors. (A, left) Time course of gB surface expression on CMV-infected HFF cells as determined by fluorescence-activated cell sorting (FACS) analysis using the 27-287 antibody and a secondary Cy5-labeled anti-mouse antibody compared to that for an isotype control. (Right) Release of viral particles from infected cells into the supernatant determined by quantitative real-time PCR at the indicated time points. Small amounts of human CMV (HCMV) genomes detected within the first 48 h postinfection are due to residual input virus. (B) Construction of an scFv. RNA was isolated from the hybridoma cell line 27-287, which produces an antibody against CMV gB. cDNA was generated, and the antibodies' variable regions were amplified by PCR, cloned, and sequenced. This was followed by fusion with specific primers in an overlap extension PCR. PCR products were cloned into the vector pBullet #607 (14), which already contained the cIR signaling domains, and sequenced. Vh, variable region of immunoglobulin heavy chain; Vl, variable region of immunoglobulin light chain. (C) Specific binding of 27-287-scFv-myc (gB-scFv-myc) protein to CMV gB was monitored by Western blotting. Protein lysates of uninfected and CMV-infected HFF cells were prepared in radioimmunoprecipitation assay (RIPA) buffer. Twenty micrograms of protein was used for SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, blocked, and incubated (1 h) with the 27-287 hybridoma supernatant or the pCDNA4-27-287-scFv-MH-transfected 293T cell supernatant. gB-scFv-myc protein was detected with anti-myc antibody and anti-mouse horseradish peroxidase (HRP) antibody; 27-287 monoclonal antibody (MAb) was detected with anti-mouse HRP antibody by enhanced chemiluminescence and documented with a Fuji LAS-1000 system. (D) Schematic representation of a chimeric immunoreceptor. The extracellular gB scFv was fused to an Ig hinge region and intracellular signaling domains of the CD28 and CD3ζ cDNAs.
T cells were redirected against an immunodominant gB epitope which is conserved between various CMV strains and clinical isolates (17). A functional single-chain antibody molecule (gB scFv) (Fig. 1B and C) was derived from the hybridoma cell line 27-287 by PCR with specific oligonucleotides (Vh27-287f, 5′-GCCACCATGGAATGCAGCTGGGTCTT-3′; Vh27-287rLINKER, 5′-ACCCGACCCGCCACCGCCCGATCCACCACCTCCTGAGGAGACGGTGACTGAGG-3′; Vl27-287fLINKER, 5′-TCGGGCGGTGGCGGGTCGGGTGGCGGCGGATCTGACATTGTGCTGACACAGTCTCCT-3′; and Vl_rBamHI, 5′-GGATCCCCATCAGCCCGTTTTATTTCC-3′) and cloned into pSTBlue-1. The scFv was excised with PmlI and BamHI and cloned into the pBullet #607 vector (14). The resulting chimeric immunoreceptor (cIR) consists of variable regions of the gB scFv molecule, an Ig hinge region, CD28 transmembrane and costimulatory domains, and signal-transducing elements of the CD3ζ chain (Fig. 1D). For lentiviral expression, the gB cIR was excised from the pBullet vector by HincII and EcoRV and cloned into the PmeI site of the pWPI vector. Lentiviral vector particles were generated by transfection of 293T cells with a three-plasmid HIV-derived lentiviral system using Lipofectamine 2000 (Invitrogen) with plasmids pWPI, psPAX2, and pMD2.G (D. Trono, Geneva, Switzerland) in a molar ratio of 4:3:1. Supernatants were concentrated by ultracentrifugation for 2 h at 4°C and 20,000 rpm in a Beckmann SW28 rotor and used for infection of lymphocytes (3 × 106 cells with 3 μg p24). This transduction of Jurkat and primary human T cells resulted in high cIR surface expression (Fig. 2). Engineered T cells were specifically activated by the gB cIR since lentivirally transduced T cells cocultured with CMV-infected HFF cells produced large amounts of gamma interferon (IFN-γ), whereas untransduced T cells showed only background amounts of cytokine production (data not shown).
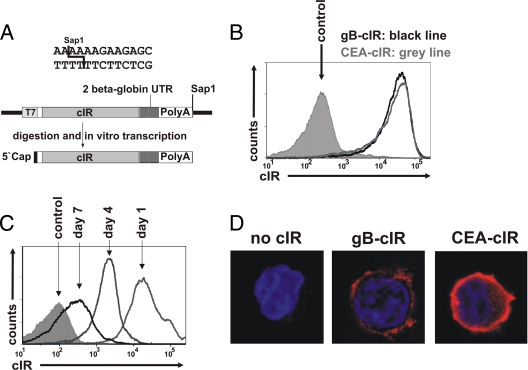
FIG. 2.
Expression of cIR in CD8+ T cells after RNA transfer. (A) Schematic diagram of the vector pST1 (kindly provided by U. Sahin) coding for the anti-gB cIR. T7 promoter-mediated in vitro transcription results in 5′-capped RNA with two copies of the 3′ β-globin UTR and a 120-nt poly(A) tail. (B) Expression of gB cIR and a carcinoembryonic-antigen-specific control receptor (CEA cIR) (14) on the surfaces of CD8+ T cells 1 day postelectroporation, detected with an anti-human Ig Cy5 antibody. Cells electroporated without RNA served as a control. (C) Time course of surface cIR expression after electroporation of in vitro-transcribed RNA into CD8+ T cells detected with an anti-human Ig Cy5 antibody. Cells electroporated with green fluorescent protein (GFP) RNA served as a control. (D) Surface expression of cIR on lentivirally transduced Jurkat T cells. Jurkat T cells transduced with the empty vector or with a vector expressing the gB cIR or the CEA cIR were centrifuged onto glass slides, fixed with 2% paraformaldehyde, and stained with an anti-human Ig Cy5 antibody. Cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole; 1 μg/ml). The samples were analyzed with a Leica TCS SP5 microscope.
As an alternative strategy, we expressed the anti-gB cIR by RNA transfer (1). Electroporation of GMP-grade cIR RNA has several advantages over lentiviral vectors with regard to its use in clinical trials (35). Persistence of transferred RNA and therefore of cIR expression is limited to a period of several days. It does not raise the issue of gene vector safety, nor does it require additional safety measures, such as the inclusion of a suicide gene. On the other hand, the shorter expression period following RNA transfer might necessitate repeated transfer of RNA-transfected effector T cells. The cIR constructs were excised from the pBullet vector and cloned into the vector pST1 (13). RNA ivT was obtained with the mMessage mMachine T7 Ultra mRNA in vitro transcription kit (Ambion) using a linear DNA template derived from the vector pST1. T7 RNA polymerase-mediated in vitro transcription yielded respective mRNAs with two RNA-stabilizing 3′ β-globin untranscribed regions (UTRs) and a 120-nucleotide (nt) poly(A) tail (Fig. 2A). Electroporation of 8 μg of mRNA (500 V, 5-ms square-wave pulse, 4-mm cuvette) resulted in high-level cIR expression in >97% of cells as detected with a goat anti-human Ig Cy5 antibody (Fig. 2B). cIR surface expression lasted for 7 days, although with decreasing expression densities over time (Fig. 2C).
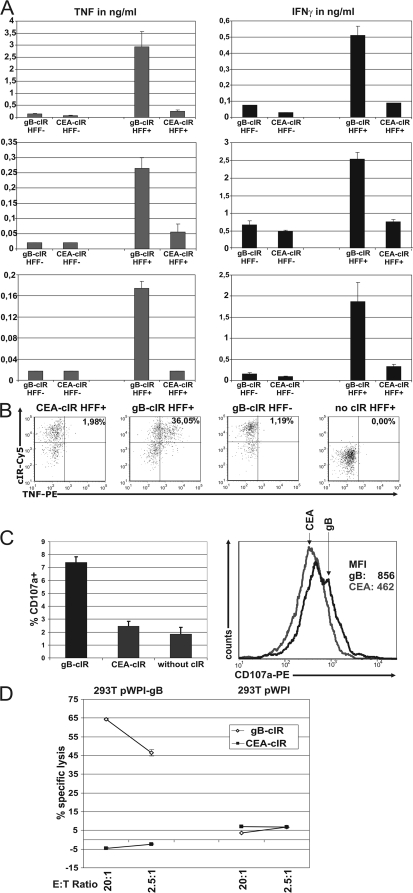
Coincubation of gB-cIR-expressing T cells with infected HFF cells resulted in specific production of tumor necrosis factor (TNF) and IFN-γ in T cells of CMV-seronegative donors (Fig. 3A). Intracellular staining with an anti-human TNF antibody confirmed that only gB-cIR-expressing T cells, and not T cells with irrelevant cIR, responded to stimulation with CMV-infected HFF cells with production of TNF (Fig. 3B). Moreover, coincubation of gB-cIR-engineered T cells with infected HFF cells led to specific degranulation, as estimated by CD107a surface staining (Fig. 3C). gB-cIR-expressing T cells are able to kill gB-positive (gB+) target cells efficiently at a low effector-to-target (E/T) ratio of 2.5:1. Carcinoembryonic antigen (CEA)-cIR-carrying T cells showed no cytotoxicity toward gB+ cells (Fig. 3D), illustrating again the specificity of target recognition.
FIG. 3.
gB-specific T cells engineered by RNA transfer are functionally redirected. (A) RNA-electroporated T cells with gB and CEA cIR (1 × 105 T cells) were coincubated with, respectively, CMV-infected (HFF+) and uninfected (HFF−) HFF cells (5,000 HFF cells/well) in medium containing 5% (vol/vol) human serum of CMV-negative donors for 18 to 20 h. IFN-γ and TNF in supernatants were quantified by sandwich enzyme-linked immunosorbent assay (ELISA) (human IFN-γ and TNF OptEIA ELISA set; BD Biosciences). Data represent the means of results from triplicates plus standard deviations (SD). Three different T cell-HFF combinations are shown. First row, T cells derived from cord blood (50% CD8+ and 50% CD4+); second and third rows, magnetic-bead-purified CD8+ T cells (Miltenyi) from CMV-negative donors. (B) TNF is produced by gB-cIR-engineered T cells after coincubation with CMV-infected HFF cells, as determined by intracellular flow cytometry with a phycoerythrin (PE)-labeled anti-human TNF antibody (clone mab11). Staining was performed after incubation with 10 μg/ml Brefeldin A for 5 h at 37°C. (C) Specific degranulation of cIR-engineered T cells was determined by FACS analysis with a PE-labeled anti-CD107a antibody (clone H4A3). cIR-expressing T cells were coincubated with CMV-infected HFF cells at an E/T ratio of 5:1 for 6 h in the presence of 5 μM monensin and with 5% serum from CMV-negative individuals. T cells with gB cIR show degranulation compared to T cells with control CEA cIR and without cIR. (Left) Fraction of CD107a-positive cells (above cutoff value, which is set to 98% of measured events of the negative control). The mean values from three independent experiments are shown. (Right) Histogram data of a representative experiment. MFI, mean fluorescence intensity. (D) Antigen-specific lysis of CMV gB-expressing cells by cIR RNA-electroporated T cells. Europium-based cytotoxicity assay showing specific lysis after coincubation of gB cIR or control anti-CEA-cIR-engineered T cells with 293T cells with or without gB expression. Cytotoxicity assays were performed as described previously (19). Specific lysis was calculated by the formula percent lysis = (experimental counts − spontaneous release) × 100/(maximum release − spontaneous release). Data represent the means of results of triplicates plus SD.
We assume that targeting gB by cIR-redirected T cells, engineered by lentiviral or RNA-mediated gene transfer, has the potential to prevent CMV replication and limit the spread of CMV in vivo. Murine CMV (MCMV) is the established and convenient model for acute and persistent herpesviral infection (27). However, preclinical animal studies of the MCMV model would be complicated by the fact that the MCMV gB homolog shares only poor sequence similarity to human CMV gB. This precludes cross-reactivity of reagents specific for human CMV gB. Moreover, the MCMV gB gene is a true late and lytic gene (26), thereby differing significantly from the human CMV gB gene. Although beyond the scope of this paper, it is conceivable that one could develop a model based on a recombinant MCMV expressing the human CMV gB. Such an approach would still be complicated by the fact that MCMV gB cannot be simply replaced by human CMV gB in functional terms and would further require appropriate transcriptional regulation of gB expression. Rhesus CMV (RhCMV), another model of acute and persistent cytomegalovirus infection, is more closely related to human CMV (30). RhCMV gB is similarly processed and shares higher homology to human CMV gB (24). RhCMV gB transcription and expression patterns are not known in detail, and further studies on this promising macaque model are required to determine its suitability in this setting. Nevertheless, the potential use of our cIR in an RhCMV infection model warrants further studies, as the antibody 27-287 has shown cross-reactivity with RhCMV gB (18).
The strategy of eliminating CMV-infected cells by cIR-redirected T cells has the advantage of HLA-independent recognition of gB. This is of particular relevance since CMV has evolved several efficient mechanisms to downregulate the major histocompatibility complex (MHC) (8) and to prevent peptide presentation (12). The anti-gB cIR, however, enables the cytotoxic T cell to recognize gB on the cell surface, independently of processed antigen presentation by the MHC. This principle is not used by naturally occurring adaptive T-cell immunity; CMV has thus not been under selective pressure to evolve a counterstrategy.
A further concern may be the blockade of the cIR by soluble antigen, e.g., viral particles containing gB. However, the load of human CMV particles in infected individuals is low, virus is mostly cell associated (2), and soluble forms of gB or shedding from the surfaces of cells or particles have not been reported despite extensive research. It was shown that cIR function of T cells redirected against tumor antigen is unimpaired by soluble protein present in serum (14, 36).
Taken together we show that CMV gB, which is expressed by infected cells early in the replication cycle, is an attractive target for redirected, engineered T cells endowed with a specific cIR. cIR-redirected target recognition would allow adoptive transfer with T cells from CMV-negative donors and, more remarkably, independently of HLA restriction. This approach has the potential to pave the way for a new therapeutic option in the treatment of one of the most dreaded complications after stem cell transplantation.
Acknowledgments
We acknowledge Niels Schaft for helpful discussions; Barbara Alberter for helpful discussions and critical reading of the manuscript; Didier Trono, Geneva, Switzerland, for providing the plasmids of the lentiviral vector system; and Ugur Sahin, Mainz, Germany, for providing the pST1 plasmid.
This work was supported in part by the Wilhelm Sander Foundation (grants 2002.033.1, 2002.034.1, and 2009.002.1), the Interdisciplinary Center for Clinical Research (IZKF; Genesis, Diagnostics and Therapy of Inflammation Processes), the Universitätsbund Erlangen-Nürnberg, and the Deutsche Forschungsgemeinschaft (grants SFB 796 and EN423/2-1).
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Birkholz, K., A. Hombach, C. Krug, S. Reuter, M. Kershaw, E. Kampgen, G. Schuler, H. Abken, N. Schaft, and J. Dorrie. 2009. Transfer of mRNA encoding recombinant immunoreceptors reprograms CD4+ and CD8+ T cells for use in the adoptive immunotherapy of cancer. Gene Ther. 16:596-604. [DOI] [PubMed] [Google Scholar]
- 2.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeckh, M., W. Leisenring, S. R. Riddell, R. A. Bowden, M. L. Huang, D. Myerson, T. Stevens-Ayers, M. E. Flowers, T. Cunningham, and L. Corey. 2003. Late cytomegalovirus disease and mortality in recipients of allogeneic hematopoietic stem cell transplants: importance of viral load and T-cell immunity. Blood 101:407-414. [DOI] [PubMed] [Google Scholar]
- 4.Boeckh, M., and P. Ljungman. 2009. How we treat cytomegalovirus in hematopoietic cell transplant recipients. Blood 113:5711-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeckh, M., W. G. Nichols, G. Papanicolaou, R. Rubin, J. R. Wingard, and J. Zaia. 2003. Cytomegalovirus in hematopoietic stem cell transplant recipients: current status, known challenges, and future strategies. Biol. Blood Marrow Transplant. 9:543-558. [DOI] [PubMed] [Google Scholar]
- 6.Bohne, F., M. Chmielewski, G. Ebert, K. Wiegmann, T. Kurschner, A. Schulze, S. Urban, M. Kronke, H. Abken, and U. Protzer. 2008. T cells redirected against hepatitis B virus surface proteins eliminate infected hepatocytes. Gastroenterology 134:239-247. [DOI] [PubMed] [Google Scholar]
- 7.Boutolleau, D., C. Deback, C. Bressollette-Bodin, S. Varnous, N. Dhedin, B. Barrou, J. P. Vernant, I. Gandjbakhch, B. M. Imbert-Marcille, and H. Agut. 2009. Resistance pattern of cytomegalovirus (CMV) after oral valganciclovir therapy in transplant recipients at high-risk for CMV infection. Antiviral Res. 81:174-179. [DOI] [PubMed] [Google Scholar]
- 8.del Val, M., K. Munch, M. J. Reddehase, and U. H. Koszinowski. 1989. Presentation of CMV immediate-early antigen to cytolytic T lymphocytes is selectively prevented by viral genes expressed in the early phase. Cell 58:305-315. [DOI] [PubMed] [Google Scholar]
- 9.Einsele, H., and H. Hebart. 2004. CMV-specific immunotherapy. Hum. Immunol. 65:558-564. [DOI] [PubMed] [Google Scholar]
- 10.Einsele, H., E. Roosnek, N. Rufer, C. Sinzger, S. Riegler, J. Loffler, U. Grigoleit, A. Moris, H. G. Rammensee, L. Kanz, A. Kleihauer, F. Frank, G. Jahn, and H. Hebart. 2002. Infusion of cytomegalovirus (CMV)-specific T cells for the treatment of CMV infection not responding to antiviral chemotherapy. Blood 99:3916-3922. [DOI] [PubMed] [Google Scholar]
- 11.Eshhar, Z., T. Waks, G. Gross, and D. G. Schindler. 1993. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc. Natl. Acad. Sci. U. S. A. 90:720-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hengel, H., and U. H. Koszinowski. 1997. Interference with antigen processing by viruses. Curr. Opin. Immunol. 9:470-476. [DOI] [PubMed] [Google Scholar]
- 13.Holtkamp, S., S. Kreiter, A. Selmi, P. Simon, M. Koslowski, C. Huber, O. Tureci, and U. Sahin. 2006. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108:4009-4017. [DOI] [PubMed] [Google Scholar]
- 14.Hombach, A., D. Koch, R. Sircar, C. Heuser, V. Diehl, W. Kruis, C. Pohl, and H. Abken. 1999. A chimeric receptor that selectively targets membrane-bound carcinoembryonic antigen (mCEA) in the presence of soluble CEA. Gene Ther. 6:300-304. [DOI] [PubMed] [Google Scholar]
- 15.Kapp, M., S. M. Tan, H. Einsele, and G. Grigoleit. 2007. Adoptive immunotherapy of HCMV infection. Cytotherapy 9:699-711. [DOI] [PubMed] [Google Scholar]
- 16.Kershaw, M. H., M. W. Teng, M. J. Smyth, and P. K. Darcy. 2005. Supernatural T cells: genetic modification of T cells for cancer therapy. Nat. Rev. Immunol. 5:928-940. [DOI] [PubMed] [Google Scholar]
- 17.Kniess, N., M. Mach, J. Fay, and W. J. Britt. 1991. Distribution of linear antigenic sites on glycoprotein gp55 of human cytomegalovirus. J. Virol. 65:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kropff, B., and M. Mach. 1997. Identification of the gene coding for rhesus cytomegalovirus glycoprotein B and immunological analysis of the protein. J. Gen. Virol. 78:1999-2007. [DOI] [PubMed] [Google Scholar]
- 19.Lehner, M., C. Grillhoesl, F. Full, B. Vogel, P. Weller, I. Muller-Fleckenstein, M. Schmidt, B. Fleckenstein, W. Holter, and A. Ensser. 2009. Transformation efficiency by herpes-virus saimiri is not a limiting factor in clonal CD8pos T cell outgrowth. Virology 388:15-20. [DOI] [PubMed] [Google Scholar]
- 20.Masiero, S., V. C. Del, R. Gavioli, G. Mattiuzzo, M. G. Cusi, L. Micheli, F. Gennari, A. Siccardi, W. A. Marasco, G. Palu, and C. Parolin. 2005. T-cell engineering by a chimeric T-cell receptor with antibody-type specificity for the HIV-1 gp120. Gene Ther. 12:299-310. [DOI] [PubMed] [Google Scholar]
- 21.Matthes-Martin, S., S. W. Aberle, C. Peters, W. Holter, T. Popow-Kraupp, U. Potschger, G. Fritsch, R. Ladenstein, A. Rosenmayer, K. Dieckmann, and H. Gadner. 1998. CMV-viraemia during allogenic bone marrow transplantation in paediatric patients: association with survival and graft-versus-host disease. Bone Marrow Transplant. 21(Suppl. 2):S53-S56. [PubMed] [Google Scholar]
- 22.Patel, S. D., M. Moskalenko, T. Tian, D. Smith, R. McGuinness, L. Chen, G. A. Winslow, S. Kashmiri, J. Schlom, C. P. Stanners, M. H. Finer, and J. G. McArthur. 2000. T-cell killing of heterogenous tumor or viral targets with bispecific chimeric immune receptors. Cancer Gene Ther. 7:1127-1134. [DOI] [PubMed] [Google Scholar]
- 23.Peggs, K. S., S. Verfuerth, A. Pizzey, N. Khan, M. Guiver, P. A. Moss, and S. Mackinnon. 2003. Adoptive cellular therapy for early cytomegalovirus infection after allogeneic stem-cell transplantation with virus-specific T-cell lines. Lancet 362:1375-1377. [DOI] [PubMed] [Google Scholar]
- 24.Powers, C., and K. Fruh. 2008. Rhesus CMV: an emerging animal model for human CMV. Med. Microbiol. Immunol. 197:109-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinnan, G. V., Jr., N. Kirmani, A. H. Rook, J. F. Manischewitz, L. Jackson, G. Moreschi, G. W. Santos, R. Saral, and W. H. Burns. 1982. Cytotoxic t cells in cytomegalovirus infection: HLA-restricted T-lymphocyte and non-T-lymphocyte cytotoxic responses correlate with recovery from cytomegalovirus infection in bone-marrow-transplant recipients. N. Engl. J. Med. 307:7-13. [DOI] [PubMed] [Google Scholar]
- 26.Rapp, M., M. Messerle, B. Buhler, M. Tannheimer, G. M. Keil, and U. H. Koszinowski. 1992. Identification of the murine cytomegalovirus glycoprotein B gene and its expression by recombinant vaccinia virus. J. Virol. 66:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddehase, M. J., C. O. Simon, C. K. Seckert, N. Lemmermann, and N. K. Grzimek. 2008. Murine model of cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325:315-331. [DOI] [PubMed] [Google Scholar]
- 28.Reusser, P., H. Einsele, J. Lee, L. Volin, M. Rovira, D. Engelhard, J. Finke, C. Cordonnier, H. Link, and P. Ljungman. 2002. Randomized multicenter trial of foscarnet versus ganciclovir for preemptive therapy of cytomegalovirus infection after allogeneic stem cell transplantation. Blood 99:1159-1164. [DOI] [PubMed] [Google Scholar]
- 29.Riddell, S. R., K. S. Watanabe, J. M. Goodrich, C. R. Li, M. E. Agha, and P. D. Greenberg. 1992. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science 257:238-241. [DOI] [PubMed] [Google Scholar]
- 30.Rivailler, P., A. Kaur, R. P. Johnson, and F. Wang. 2006. Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J. Virol. 80:4179-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rooney, C. M., C. A. Smith, C. Y. Ng, S. Loftin, C. Li, R. A. Krance, M. K. Brenner, and H. E. Heslop. 1995. Use of gene-modified virus-specific T lymphocytes to control Epstein-Barr-virus-related lymphoproliferation. Lancet 345:9-13. [DOI] [PubMed] [Google Scholar]
- 32.Scheinberg, P., J. J. Melenhorst, J. M. Brenchley, B. J. Hill, N. F. Hensel, P. K. Chattopadhyay, M. Roederer, L. J. Picker, D. A. Price, A. J. Barrett, and D. C. Douek. 2009. The transfer of adaptive immunity to cytomegalovirus (CMV) during hematopoietic stem cell transplantation is dependent on the specificity and phenotype of CMV-specific T cells in the donor. Blood 114:5071-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smuda, C., E. Bogner, and K. Radsak. 1997. The human cytomegalovirus glycoprotein B gene (ORF UL55) is expressed early in the infectious cycle. J. Gen. Virol. 78:1981-1992. [DOI] [PubMed] [Google Scholar]
- 34.Sylwester, A. W., B. L. Mitchell, J. B. Edgar, C. Taormina, C. Pelte, F. Ruchti, P. R. Sleath, K. H. Grabstein, N. A. Hosken, F. Kern, J. A. Nelson, and L. J. Picker. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202:673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Tendeloo, V., P. Ponsaerts, and Z. N. Berneman. 2007. mRNA-based gene transfer as a tool for gene and cell therapy. Curr. Opin. Mol. Ther. 9:423-431. [PubMed] [Google Scholar]
- 36.Zhang, T., A. Barber, and C. L. Sentman. 2006. Generation of antitumor responses by genetic modification of primary human T cells with a chimeric NKG2D receptor. Cancer Res. 66:5927-5933. [DOI] [PubMed] [Google Scholar]