Abstract
Herpes simplex virus (HSV) entry into cells requires four membrane glycoproteins: gD is the receptor binding protein, and gB and gH/gL constitute the core fusion machinery. Crystal structures of gD and its receptors have provided a basis for understanding the initial triggering steps, but how the core fusion proteins function remains unknown. The gB crystal structure shows that it is a class III fusion protein, yet unlike other class members, gB itself does not cause fusion. Bimolecular complementation (BiMC) studies have shown that gD-receptor binding triggers an interaction between gB and gH/gL and concurrently triggers fusion. Left unanswered was whether BiMC led to fusion or was a by-product of it. We used gB monoclonal antibodies (MAbs) to block different aspects of these events. Non-virus-neutralizing MAbs to gB failed to block BiMC or fusion. In contrast, gB MAbs that neutralize virus blocked fusion. These MAbs map to three functional regions (FR) of gB. MAbs to FR1, which contains the fusion loops, and FR2 blocked both BiMC and fusion. In contrast, MAbs to FR3, a region involved in receptor binding, blocked fusion but not BiMC. Thus, FR3 MAbs separate the BiMC interaction from fusion, suggesting that BiMC occurs prior to fusion. When substituted for wild-type (wt) gB, fusion loop mutants blocked fusion and BiMC, suggesting that loop insertion precedes BiMC. Thus, we postulate that each of the gB FRs are involved in different aspects of the path leading to fusion. Upon triggering by gD, gB fusion loops are inserted into target lipid membranes. gB then interacts with gH/gL, and this interaction is eventually followed by fusion.
Entry of herpes simplex virus (HSV) into cells requires four viral glycoproteins, gB, gD, gH, and gL, plus one of several cell receptors, either herpesvirus entry mediator (HVEM), nectin-1, or 3-OST (45). Crystal structures and other studies have documented that receptor binding triggers conformational changes to gD that trigger the downstream events leading to fusion (10, 11, 18, 26, 28, 52). Moreover, when HSV receptor-bearing cells are transfected with expression plasmids for glycoproteins gB, gD, gH, and gL, the cells fuse to form multinucleated giant cells or syncytia (39, 48). However, the precise series of events that take place after receptor binding have not yet been fully elucidated. What we do know is that both gB and a heterodimer of gH/gL constitute the core fusion machinery that is conserved and required for the fusion step of entry of all herpesviruses (18, 26, 30, 46, 49).
Thus far, we know the crystal structure of one form of the gB ectodomain of HSV type 1 (HSV-1) (19). This protein has the characteristics of a fusion protein and is a charter member of the class III group of viral fusion proteins (4). Others in this class include Epstein-Barr virus gB, vesicular stomatitis virus (VSV) G, and baculovirus gp64 (5, 22, 41). Like VSV G and gp64, gB has two putative fusion loops at the base of each protomer of the crystallized trimer. Single-amino-acid mutations in many of the hydrophobic residues of the putative fusion loops of gB ablate its ability to function in cell-cell fusion assays (16, 17). Moreover, these mutants are unable to complement the entry of a gB-null virus (16). Finally, the ectodomains of these mutants, unlike wild-type protein, failed to coassociate with liposomes, indicating that the putative fusion loops do insert into membranes (16, 17). Recently, it was shown that several of these mutants are also defective for fusion events involved in virus egress (51). Together, these studies provide compelling evidence that HSV gB functions as a fusion protein and that the fusion loops are critical for this function. However, unlike VSV G and baculovirus gp64, gB does not function on its own in entry but, rather, requires the participation of gH/gL. In the absence of crystallographic data for gH/gL, it is not yet clear what role it plays in herpesvirus fusion. In a previous study, we used bimolecular complementation (BiMC) to examine protein-protein interactions that occur among the viral glycoproteins during fusion (1). A similar study was carried out by Avitabile et al. (2). The BiMC assay is based on the observation that N- and C-terminal fragments of green fluorescent protein (GFP) (and derivatives such as enhanced yellow fluorescent protein [EYFP]) do not spontaneously reconstitute a functional fluorophore (20, 29, 40). However, the codons for each half can be appended to the genes for two interacting proteins (23, 24). When these are cotransfected, an interaction between the two proteins of interest brings the two halves of the fluorophore in close enough contact to restore fluorescence.
When HSV receptor-bearing cells, such as B78H1 cells that are engineered to express nectin-1, are transfected with plasmids that express gB, gD, gH, and gL, they undergo cell-cell fusion (13, 15, 27, 31, 48). When gD is omitted, no fusion occurs. We found that fusion of these transfected cells could be triggered by addition of a soluble form of gD (the gD ectodomain). We then used this approach to examine interactions between gB and gH/gL during cell fusion (1). Therefore, we tagged gB with the C-terminal half of EYFP and gH with the N-terminal half. When plasmids bearing these forms were cotransfected into C10 cells along with a plasmid for untagged gL, no fusion occurred, but importantly, no BiMC occurred. However, when we added gD306, cells began to fuse within 10 min, and all of the syncytia that formed exhibited bright EYFP fluorescence indicative of BiMC. We concluded that gD triggers both fusion and a physical interaction between gB and gH/gL. However, these experiments did not separate these two events, so we were unable to determine if the interaction preceded fusion or merely was a by-product of it.
The purpose of this study was to determine if the gB-gH/gL interaction is essential for fusion and if it occurs prior to fusion. We focused on gB because its structure is known and we have a panel of well-characterized monoclonal antibodies (MAbs) to gB. Our approach was to determine which of these MAbs, if any, could block fusion and also block the interaction with gH/gL. We also examined the effect of mutations to the fusion loops of gB on its interaction with gH/gL. We previously mapped these MAbs to four functional regions (FR) of gB, three of which were resolved in the crystal structure (6, 19). Of these, FR1 contains the fusion loops, FR2 is in the center of the gB structure with no known function, and FR3 is at in the crown of the protein and may be involved in binding to cells (7). Our rationale was that if the interaction between gB and gH/gL is important for fusion, then it should not be blocked by nonneutralizing anti-gB MAbs. At the same time, we thought that some neutralizing MAbs might not only block fusion but also block BiMC. We found that neutralizing MAbs to FR1 and FR2 inhibited both BiMC and fusion. In contrast, we found that neutralizing MAbs that map to FR3 blocked fusion but failed to block the interaction between gB and gH/gL, thereby dissociating the two events. Finally, we found that gB mutants with changes in the fusion loops that were fusion negative were also unable to bind to gH/gL. The latter results suggest that insertion of gB into the target membrane precedes its interaction with gH/gL.
MATERIALS AND METHODS
Cells and plasmids.
Mouse melanoma cells (B78H1) expressing nectin-1 (designated here as C10 cells) were grown in 5% fetal bovine serum-Dulbecco's modified Eagle's medium (FBS-DMEM) containing 250 μg/ml G418 (31). Chinese hamster ovary cells (CHO-K1) were grown in 10% fetal calf serum (FCS)-Ham's F12 medium. African green monkey kidney cells (Vero) were grown in DMEM with 5% FBS. CHO-K1 and Vero cells were obtained from ATCC.
The construction of HSV-1 EYFP-tagged gB (Bc) and gH (Hn) was described previously (1). Briefly, the glycoproteins were PCR amplified such that the natural stop codons were excluded to allow in-frame ligation with N- or C-terminal fragments of EYFP. The N-terminal EYFP (EYFPN; 1 to 173) or C-terminal EYFP (EYFPC; 173 to 239) halves were PCR amplified using pCS2 as a template (33). A translation stop codon was incorporated after codon 173. To construct C-EYFP-tagged gB fusion loop mutants, plasmids pBH732 (A261D), pBH739 (W174R), and pBH777 (Y179S) were digested with EcoRI/PmlI restriction enzymes. The 1,715-bp fragments replaced the similar region in pCW803 (1). Plasmids pT7EMCLuc (luciferase), pCAGT7 (T7 promoter), pBG38 (nectin-1), pPEP98 (gB), pPEP99 (gD), pPEP100 (gH), and pPEP101 (gL) were gifts of P. G. Spear (15, 34, 39).
Antibodies used.
Polyclonal antibody R137 was prepared against purified gH1t/gL1 (35, 36). gB and gH/gL monoclonal antibodies used were all characterized previously (6, 8, 9, 43). Neutralizing antibodies H1781 and H1838 (25, 38) were purchased from Virusys Corporation. IgGs were prepared from all antibodies for use in all the studies.
Production and purification of gD protein.
Soluble gD306t from HSV-1 was purified from baculovirus-infected insect cells (Sf9) as previously described (44, 50).
Quantitative assay of cell-cell fusion.
We used a modified version of a previously described luciferase reporter gene activation assay (12, 13, 34, 39). Briefly, effector cells (CHO-K1 cells) were transfected with plasmids encoding T7 RNA polymerase, gBc, gD, gHn, and gL. To prepare receptor-bearing target cells, CHO-K1 cells were transfected with a plasmid expressing nectin-1 (pBG38) and a plasmid encoding the firefly luciferase gene under the control of the T7 promoter (pT7EMCLuc). At 5 hours posttransfection, target cells were trypsinized, 105 cells were added to each well of effector cells, and the plates were incubated at 37°C for 18 h. Cells were lysed with lysis buffer (luciferase assay system; Promega). Luciferase production was measured with luciferase substrate (Promega) using a BioTek plate reader. For quantification of blocking by MAbs, we added 100 μg/ml of the appropriate antibody IgG when target and effector cells were mixed.
Synchronization and blocking of fusion.
We used a previously described assay to synchronize fusion (1). Briefly, C10 cells were seeded on glass coverslips and transfected with gL, EYFP-tagged gB (Bc), and gH (Hn) using GenePorter (Gene Therapy Systems) according to the manufacturer's instructions. At 20 hours posttransfection, cells were triggered for fusion with 250 μg/ml soluble gD (gD306t). For blocking of BiMC, cells were exposed to 100 μg/ml gB monoclonal antibodies for 1 h at 37°C before the addition of soluble gD. Cells were then incubated with soluble gD for 1 h at 37°C and processed for immunofluorescence. This incubation time with gD306t was chosen because longer exposure to gD306 resulted in the formation of too many syncytia to count.
Immunofluorescence.
We carried out a previously described assay to examine the transfected cells by immunofluorescence (IF). Briefly, C10 cells were fixed with 3% paraformaldehyde at room temperature (RT) and quenched with 50 mM NH4Cl. Cells were washed with phosphate-buffered saline (PBS) and incubated with 10% normal goat serum and then labeled with anti-gB monoclonal antibody (MAb) SS55 (6) or anti-gH/gL Pab R137. Coverslips were washed with PBS and then incubated with Alexa Fluor 594-conjugated goat anti-IgG (Invitrogen) secondary antibody diluted in 10% goat serum-PBS. The coverslips were rinsed three times with PBS and once with H2O and mounted in ProLong Gold antifade reagent (Invitrogen). Samples were examined by confocal microscopy with a Nikon TE-300 inverted microscope coupled to a Perkin Elmer imaging system. A two-line argon krypton laser (488/514 and 568/647 nm) was used to excite the fluorescence of Alexa Fluor 594 (590/617 nm) and EYFP (515/528 nm).
RESULTS
Is the gB-gH/gL interaction an important step toward fusion?
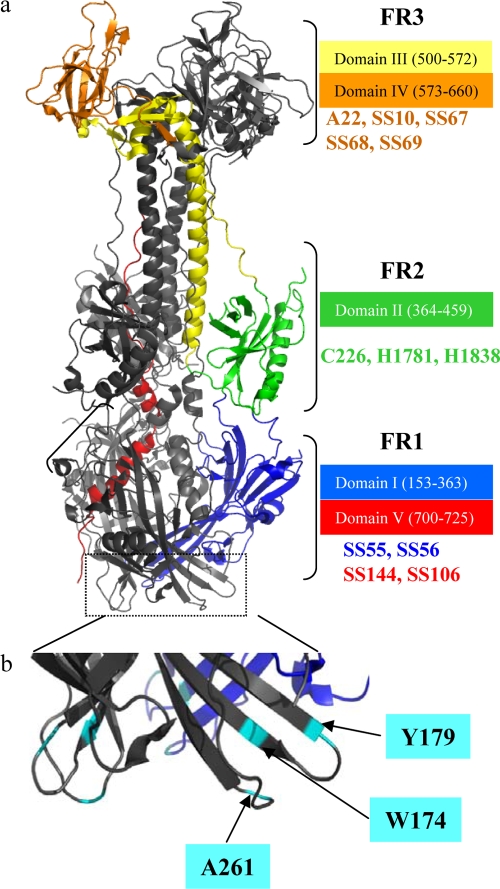
Based on the crystal structure (Fig. 1), the gB trimer can be divided into six distinct structural domains: I, base, residues 153 to 363 (blue); II, middle, residues 364 to 459 (green); III, helical core, residues 500 to 572, (yellow); IV, crown, residues 573 to 660, (orange); and V, arm, residues 700 to 725 (red) (19). Amino acids 31 to 110 were not present in the crystal structure, and those residues were designated structural domain VI. Four functional regions (FR) were also defined, based on mapping of anti-gB neutralizing MAbs to the crystal structure (Fig. 1) (6). The positions of some of these MAbs as well as other nonneutralizing MAbs used in this study are indicated on Fig. 1. FR1 is composed of amino acids in structural domains I and V, FR2 is within domain II, FR3 is within domains III and IV, and FR4 is in the N terminus that is not in the crystal structure. Figure 1 also indicates the positions of three mutations to the fusion loops (A261D, Y179S, and W174R) that ablate gB function (16, 17, 51) and were studied here.
FIG. 1.
Crystal structure of gB. (a) Ribbon representation of a gB trimer. Functional regions were defined based on antibody mapping (6). Functional region 1 (FR1) comprises domains I (blue) and the C terminus of domain V (red). FR2 contains domain II (green). FR3 comprises amino acids located between domain III (yellow) and domain IV (orange). Color code for the structural domains is as originally published (19). Monoclonal antibodies used in this study representative of each FR are indicated. (b) Close view of fusion loops region with fusion loop mutants used (cyan).
Effect of antibodies to FR1 of gB on its interaction with gH/gL and fusion.
We hypothesized that virus-neutralizing antibodies to gB should block either BiMC due to the interaction between gB and gH/gL, the fusion event, or both. In contrast, antibodies that bind gB but have no ability to neutralize virus infectivity should have no effect on either function. To test our hypothesis, we selected neutralizing and nonneutralizing antibodies that map to each FR to determine which ones block the interaction between gB and gH/gL and/or fusion.
Neutralizing MAbs to FR1 block the association of gB with lipid membranes (16). Presumably, this is the mechanism whereby these antibodies neutralize HSV. To test their effects on BiMC and fusion, nectin-1-expressing cells (C10) were transfected with plasmids for gL and YFP-tagged gB (Bc) and gH (Hn) (Fig. 2). Twenty hours later, cells were treated with a test antibody for 1 h at 37°C followed by a further 1 h of incubation with gD306. Cells were then fixed, immunostained for gH/gL with PAb R137, followed by a secondary antibody, and examined by confocal microscopy for both antibody staining (red) and BiMC (green). The number of syncytia on each coverslip was counted by direct examination. Fusion was also quantified using a luciferase-based fusion assay with a separate transfection mixture.
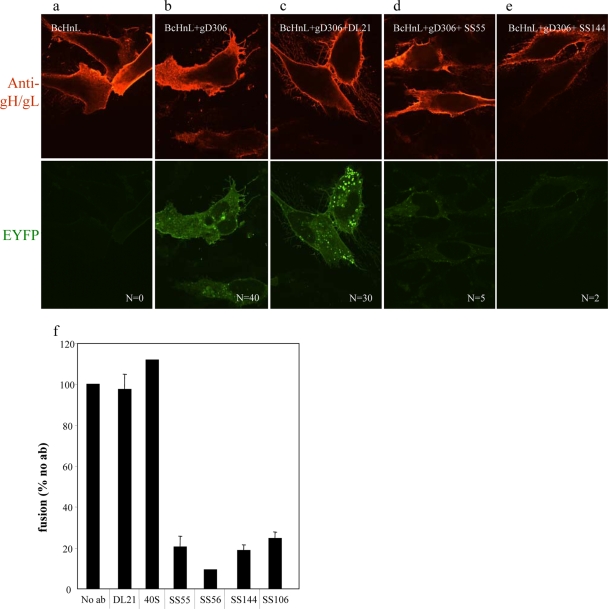
FIG. 2.
gB-gH/gL interaction can be blocked with gB-neutralizing antibodies from FR1. Immunofluorescence of nectin-expressing cells transfected with gL and EYFP-tagged gB (Bc) and gH (Hn). Twenty hours posttransfection, cells were incubated 1 h with 100 μg/ml MAb. Fusion was triggered with 250 μg/ml soluble gD306. Cells were fixed and examined with a confocal microscope at 100× magnification. In the absence of gD, no fusion and no BiMC was detected. Soluble gD306 triggers fusion and BiMC in the absence (b) or in the presence (c) of nonneutralizing antibody DL21. Neutralizing antibodies (d and e) block EYFP restoration and fusion. N, number of syncytia from a representative experiment; numbers are from one coverslip per sample. (f) Quantification of fusion (luciferase assay). No ab, no antibodies.
As a negative control for BiMC, C10 cells were transfected for 20 h with plasmids for gL and YFP-tagged gB (Bc) and gH (Hn). We readily detected gH/gL (red) on the surface of the transfected cells, and as expected, we observed no syncytia, nor did we detect the green fluorescence that is indicative of BiMC (Fig. 2a) (1, 2). When cells were similarly transfected and then incubated with gD306 for 1 h, 40 syncytia per coverslip were observed, and all exhibited BiMC at the plasma membrane (Fig. 2b). When a third monolayer of transfected cells was incubated with the nonneutralizing MAbs DL21 (Fig. 2c) or 40S (not shown) and then incubated with gD306, we again observed fusion (30 to 40 per coverslip) and BiMC. Both of these MAbs map to FR1 (6). In contrast, when the transfected cells were incubated with FR1-neutralizing MAb SS55 or SS144 prior to gD306 addition, we observed few syncytia (2 to 5 per coverslip), and importantly, we also found no evidence of BiMC (Fig. 2d and e). Similar results were obtained for the neutralizing MAbs SS56 and SS106 (data not shown). Thus, all of these neutralizing MAbs to FR1 blocked the gB-gH/gL interaction. Using a quantitative fusion assay, we found that MAbs SS55, SS56, and SS144 (all mapping to FR1) reduced overall fusion by 80% or more, while the two nonneutralizing MAbs 40S and DL21 had no effect on fusion compared to the control sample that was not pretreated with anti-gB antibody (Fig. 2f).
We conclude that residues between 670 to 725 that comprise FR1 (6) are involved either directly or indirectly in the interaction between gB and gH/gL as well as fusion. One possibility is that the fusion loops of gB must be inserted into a target membrane prior to the interaction of the protein with gH/gL. Alternatively, residues in this region could directly contact gH/gL, and antibody binding to them might block both BiMC and fusion.
Mutations in gB fusion loops prevent bimolecular complementation and fusion.
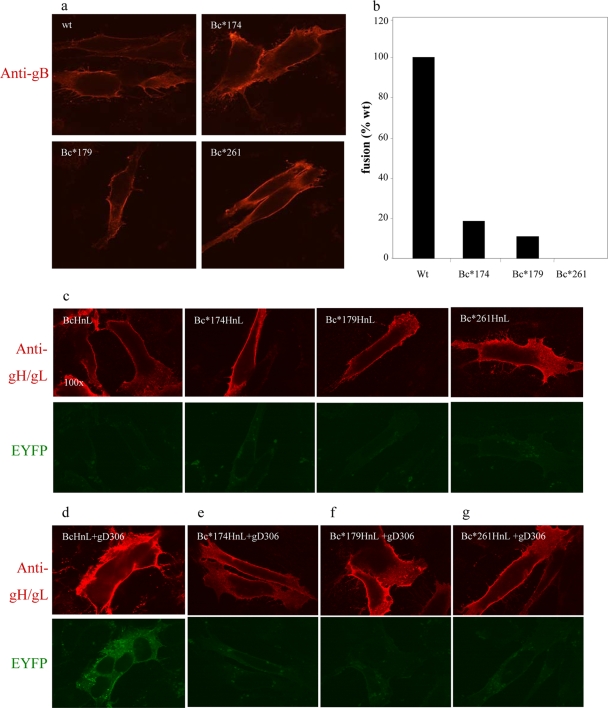
To determine how FR1 contributes to BiMC, we employed three gB mutants, W174R, Y179S, and A261D, with changes in the fusion loops that render them null for fusion, virus complementation, liposome binding, and virus egress (16, 17, 51). We constructed gB expression plasmids, appending the C-terminal portion of EYFP to the carboxy terminus of each to create gBc*174, gBc*179, and gBc*261. C10 cells were transfected with gHn, gL, and gBc(wt) or one of the gB-EYFP mutants. The mutant forms of gB were all expressed on the cell surface at levels comparable to that of wild-type gB (Fig. 3a). Each of these constructs was greatly impaired or null for cell-cell fusion, as measured by the luciferase assay (Fig. 3b) (16, 17). In the absence of gD, neither wild-type nor mutant gB interacted with gH/gL, as expected (Fig. 3c). When gD(306) was added, bright green syncytia, i.e., BiMC, and fusion were observed in cells transfected with wild-type gBc (Fig. 3d) but not in cells transfected with any of the fusion loop mutants, gBc*174, gBc*179, or gBc*261 (Fig. 3e to g). These results confirm those of our previous study, which showed that the fusion loops are essential for gB function (16, 17, 51), but further show that membrane insertion is essential for the interaction between gB and gH/gL leading to BiMC. Another possibility is that while insertion of the fusion loops precedes this interaction, it is not the cause of it, and mutations in this region block both events. These data also suggest that neutralizing MAbs to FR1 likely block BiMC in an indirect manner, by preventing gB from inserting its fusion loops into the cell membrane. However, we cannot exclude the possibility that some of the MAbs to FR1 block an additional gB function.
FIG. 3.
Characterization of EYFP-tagged fusion loop mutants. (a) Surface expression of EYFP-tagged gB fusion loop mutants in fixed, nonpermeabilized cells. (b) Quantification of fusion levels in cells triggered with soluble gD306 (luciferase assay). (c) gB fusion loop mutants and wild-type gB do not interact with gH/gL in the absence of gD. (d) Wild-type gB interacts with gH/gL, an interaction that leads to fusion. (e to g) Fusion loop mutants are negative for both BiMC and fusion.
Effect of gB FR2 antibodies on BiMC and fusion.
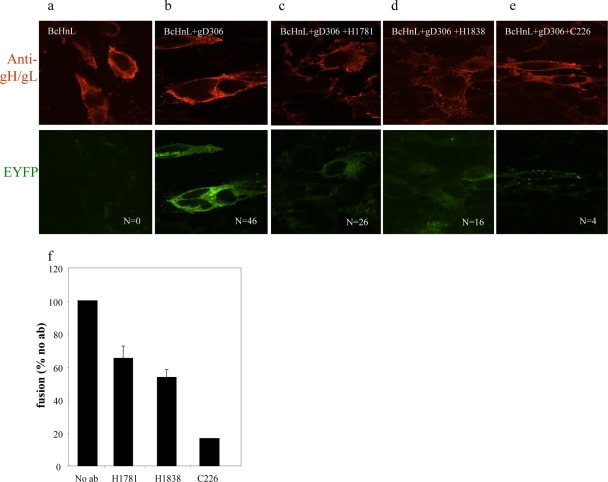
To determine the role of FR2 in BiMC and fusion, we used three neutralizing MAbs that map to this region (7). They are H1781 (linear epitope, residues 391 to 410), H1838 (linear epitope, residues 454 to 473), and C226 (conformation-dependent epitope, mapped to residues 234 to 472). Because C226 and H1838 compete with each other for gB binding, it has been suggested that much of the C226 epitope is within residues 454 to 473. Based on studies with multiple-antibiotic-resistant (mar) mutants, one residue, D419, has been identified as part of the C226 epitope (7). Of these, C226 is very powerful in its ability to block virus infection, whereas neutralization by H1781 and H1838 is much less effective (our unpublished data and Bender et al. [6]). The amino acids that constitute FR2 lie within the middle of structural domain II of gB (Fig. 1). Unfortunately, we have not identified nonneutralizing MAbs that map to this region for use as negative controls. Since neither C226 nor H1838 nor H1718 had any effect on gB730t insertion into liposomes (16), we hypothesize that these MAbs likely neutralize virus by interfering with a gB function that does not directly involve the fusion domain. We therefore asked whether these three FR2 antibodies would block BiMC. As in Fig. 2, we included both negative and positive controls for both BiMC and fusion (no gD or gD added, respectively) (Fig. 4a and b). We then preincubated the transfected cells with MAb H1781, H1838, or C226 prior to the addition of gD306 (Fig. 4c to e). MAbs H1781 and H1838 only partially inhibited fusion (Fig. 4c, d, and f), and the amount seen by luciferase correlated well with the numbers of syncytia (compare Fig. 4c and d with Fig. 4f). All of the syncytia exhibited BiMC, and some residual EYFP fluorescence was noted among the unfused cells. However, in most of the unfused cells, little or no BiMC occurred, particularly compared with the positive control in Fig. 4b. In contrast, when the cells were treated with C226, only four syncytia were seen on the coverslip, and the unfused cells exhibited no EYFP fluorescence (Fig. 4e). Fusion was inhibited more than 90% as measured using the luciferase assay (Fig. 4f). Thus, C226 efficiently blocks BiMC and cell-cell fusion as well as viral entry (data not shown). Taken together, the data suggest that FR2 plays a critical role in the interaction between gB and gH/gL as well as fusion.
FIG. 4.
gB-gH/gL interaction can be blocked with gB-neutralizing antibodies from FR2. Immunofluorescence of nectin-expressing cells transfected with gL and EYFP-tagged gB (Bc) and gH (Hn) and stained with R137 gH/gL polyclonal antibodies. In the absence of gD, no BiMC and no fusion occurs (a). In the presence of neutralizing MAbs (c to e), gD306 does not induce fusion and gB does not interact with gH/gL. N, number of syncytia found on each coverslip (one coverslip per sample). (f) Quantification of fusion by luciferase assay.
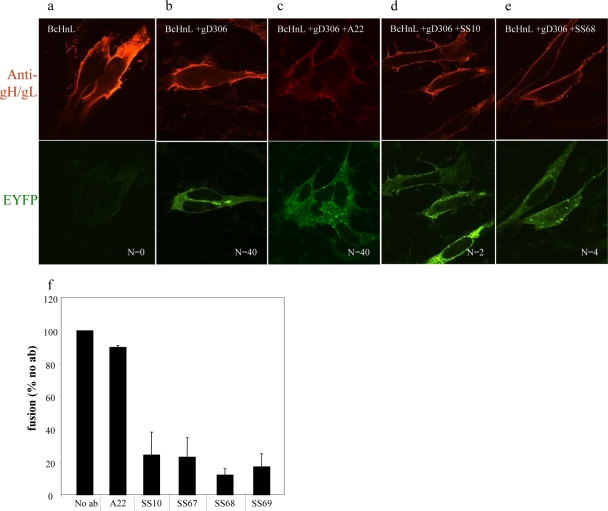
Neutralizing antibodies to FR3 of gB dissociate BiMC from fusion.
FR3 is located in the gB crown (domain IV) and is important for binding of virion gB to a putative cell receptor (7, 42). Several potent neutralizing antibodies map to overlapping antigenic sites within FR3, and it is presumed that their activity is associated, at least in part, with blocking the interaction of gB with the cell surface (6).
The same negative and positive controls were included (Fig. 5a and b). When gBc-, gHn-, and gL-transfected cells were treated with the FR3 nonneutralizing MAb A22, followed by addition of gD, we observed numerous syncytia and bright plasma membrane EYFP fluorescence indicative of BiMC (Fig. 5c). In contrast, the neutralizing FR3 neutralizing MAbs SS10 and SS68 effectively inhibited cell-cell fusion (2 and 4 syncytia, respectively) but had no effect on the intensity or location of EYFP fluorescence on the many unfused cells (Fig. 5d and e). Additional neutralizing MAbs from the same functional region (SS67, SS69) also inhibited syncytia formation by 80% and also had no effect on BiMC in the unfused cells (data not shown). The inhibitory effect of SS10, SS67, SS68, and SS69 on fusion was confirmed by the luciferase assay (Fig. 5f). We conclude that residues recognized by the neutralizing antibodies within FR3 are essential for cell-cell fusion but are not involved in the interaction between gB and gH/gL. Thus, our data suggest that the interaction between gB and gH/gL occurs independently of fusion. Furthermore, our data suggest that the interaction occurs in the vicinity of where MAbs to FR2 bind on gB.
FIG. 5.
Antibodies from FR3 do not interfere with the interaction between gB and gH/gL, but they block cell-cell fusion. Nectin-1 cells transfected with gL and EYFP-tagged gB and gH/gL do not show BiMC in the absence of gD306 (a). In the presence of gD (b) and nonneutralizing MAbs (A22), BiMC and fusion occur (c). Neutralizing MAbs (d and e) do not affect the gB-gH/gL interaction. N, number of syncytia found on each coverslip (one coverslip per sample). (f) Syncytia formation quantification by luciferase assay. The extent of fusion was calculated as the percentage of no-antibody sample.
DISCUSSION
The basic working model of HSV entry and fusion is that gD binds to a receptor (e.g., nectin-1 or HVEM), and this leads to several conformational changes in gD structure that allow it to activate downstream events (18, 26). Using bimolecular complementation, we and others showed that one of these events is a gD/receptor-induced interaction between gB and gH/gL (1, 2). In our prior studies, we found that this interaction as well as cell-cell fusion could be triggered by adding the gD ectodomain to receptor-bearing cells expressing EYFP-tagged (on the C termini) forms of gB and gH along with untagged gL. The interaction between gB and gH/gL drives the two proteins within 6 Å of each other, resulting in restoration of EYFP (BiMC). Fusion occurred concurrently, and syncytia could be detected within 10 min of the addition of gD306 (1).
In the present study, we used a panel of well-characterized anti-gB MAbs (6) to determine which ones could block fusion and which of those block BiMC. As shown by others (32, 49), we found that many neutralizing MAbs to gB were able to block cell-cell fusion. No nonneutralizing MAbs were able to do this, nor did they block BiMC. Importantly, a subset of anti-gB MAbs that block fusion were also able to block BiMC, and these map to FR1 and FR2 (6). The key finding of this study is that neutralizing MAbs to FR3 blocked fusion but failed to block the interaction between gB and gH/gL, as evidenced by BiMC of unfused cells. Taken together, our data strongly suggest that the interaction between gB and gH/gL is triggered by binding of gD to receptor and precedes fusion as an essential step. Furthermore, the interaction appears to involve amino acids in FR1 and FR2, but not FR3. When we substituted wild-type-tagged EYFP-tagged gB with tagged gB mutants having lethal changes in the fusion loops, we found that neither fusion nor BiMC occurred. These results suggest that insertion of gB via its fusion loops into the cell membrane precedes the interaction between gB and gH/gL, unless the mutations affected both insertion into the membrane and the interaction with gH/gL. Had it occurred after the interaction, then we would have observed BiMC. Thus, we postulate that the order of the essential steps that occur by binding of gD to its receptor are (i) binding of gD to its receptor; (ii) insertion of gB fusion loops which could occur prior to or as a result of the gD-receptor interaction; (iii) interaction between gB and gH/gL triggered by gD binding to its receptor; (iv) fusion triggered by binding of gB to gH/gL; and (v) possible other intermediary steps, such as gB binding to a cell receptor, or hemifusion (47).
Some confounding data.
A recent study, also employing BiMC, reported that gB and gH/gL can interact in the absence of gD (3). One of the problems with that study was the overexpression of the glycoproteins, and a second was the use of a mutant form of gB designed to send the protein to an abnormal cell compartment. These results are in contrast with those in an earlier publication from the same laboratory, which stated that gB interacts with gH/gL only in the presence of gD (2). We believe the reasons for the change in results may be due to a combination of technical considerations. T. Kerppola, who pioneered the assay, stresses that this technique is subject to a great deal of artifact (21, 23, 24). Among the pitfalls is the overexpression of proteins in cells, which can result in mislocalization of the proteins and formation of nonnative, nonspecific complexes. This problem was noted in a recent study of associations between the attachment and fusion proteins of a paramyxovirus (14). In our studies, we were careful to titrate plasmid concentrations and choose the lowest concentration that still allowed cell surface expression of the tagged glycoproteins (1). A second pitfall is the use of mutant forms of the protein that drive it to the wrong cell compartment (21, 23, 24). We believe that the designed excess accumulation of the two proteins, gB and gH/gL, could lead to a forced reformation of EYFP at a cellular site that is not relevant for the normal conditions necessary for virus entry or cell-cell fusion. Thus, we believe that in transfected cells, there is no significant interaction between gB and gH/gL on the cell surface in the absence of gD binding to its cell receptor.
Observations about the role of FR1.
This region consists of structural domains 1 and IV and contains two fusion loops per protomer and the structures needed to support fusion activity (6, 17, 19). Virus-neutralizing MAbs to either domain I (SS55) or domain IV (SS144) block insertion of the fusion domains of gB into lipid membranes (liposomes). These MAbs have a minimal effect on gB binding to cells (16). Here, we found that these antibodies block both BiMC and fusion (Fig. 2), suggesting that insertion of the fusion loops into the membrane is important for BiMC.
Site-directed mutagenesis of residues that form a hydrophobic ridge in the fusion loops revealed the importance of these amino acids in fusion (16, 17). When the mutants were expressed as soluble proteins, they were unable to bind liposomes, confirming the role of these amino acids in the fusion process (16). Here, we found that the full-length EYFP-tagged mutant forms of gB failed to participate in either BiMC or fusion. We suggest that the physical act of insertion of the gB fusion loops into the plasma membrane is a step in the process that enables gB to interact with gH.
Observations about the role of FR2.
MAb C226 recognizes a conformation-dependent epitope in FR2, whereas H1781 and H1838 recognize well-defined linear epitopes in the same region. Although these antibodies neutralize virus (6, 25, 37, 38), we found that only C226 significantly reduced cell-cell fusion to less than 20%. For H1781 and H1838, fusion levels were more modest, being 50 to 70% of those for the control sample. In addition, these results correlated well with the fact that C226 is a very potent neutralizer of virus infection, whereas H1781 and H1838 neutralize only to a partial extent (data not shown). C226 was also much more efficient than either H1781 or H1838 at blocking the interaction between gB and gH/gL on the cell surface. We postulate that FR2 contains the residues that are directly involved in gB binding to gH/gL. Further studies, e.g., with mutants, will be needed to test this hypothesis. Thus, we postulate that FR1 and FR2 serve two different functions: the first is involved in fusion loop insertion into target membranes, and the second may play a direct role in binding to gH/gL. We note, however, that residues of FR1 that are outside the fusion loops may play a functional role in BiMC and fusion. What then is the role of FR3?
Observations about the role of FR3.
FR3 is located in the crown of gB and contains amino acids that are important for viral entry, possibly ones that function in binding to a cell surface receptor (7, 16, 42). We tested four neutralizing MAbs (SS10, SS67, SS68, SS69) and found that none had any effect on the ability of gB to bind and insert into lipid membranes (7, 16). Neutralization by FR3 MAbs occurs by blocking virus attachment to a putative cell receptor, possibly pair immunoglobulin-like receptor (PILR) (42). Here, we found that neutralizing MAbs to FR3 were quite effective at blocking cell-cell fusion, but none of them interfered with BiMC associated with the gB-gH/gL interaction. Thus, these MAbs separate BiMC from fusion and suggest that binding of gB to a cellular receptor, presumably by residues in FR3, is not on the path to YFP reformation but occurs post-BiMC. We propose that this event occurs chronologically after the interaction between gB and gH/gL.
Thus, the functional region of gB plays a distinct role, and the antibodies to each region neutralize virus for a different reason, supporting the concept that there are at least three different activities that gB engages in during virus entry: binding to a receptor (FR3), binding to gH/gL (FR2), and fusion loop insertion (FR1).
Which events are activated by binding of gD to receptor, and when do they occur?
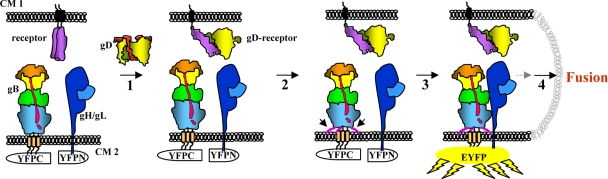
We propose the following model (Fig. 6). Twenty hours posttransfection, glycoproteins B, H, and L are expressed on the surface, poised for fusion. Once gD is added, it binds to its cell receptor (nectin-1) and undergoes conformational changes (10, 11, 28). That change then exposes a region or regions of gD which interact with the fusion components gB, gH/gL, or both (step 1). In vitro experiments with soluble proteins showed that association of wild-type gB with liposomes occurs independently of other proteins, including gD (16). An unknown stimulus dictates the insertion of gB fusion loops into the lipid membrane (step 2). This insertion presumably results in a structural change in gB that will facilitate an interaction with gD (data not shown). Once the fusion loops are inserted into the lipid membrane, gB then interacts with gH/gL (step 3), and fusion occurs (step 4).
FIG. 6.
Working model of events that lead to fusion. Cartoon representation of gB trimer and gD dimer with domains color coded as in the crystal structures previously published (19) and Fig. 1. Receptor and receptor bound to gD (HVEM is depicted) are shown in purple. Heterocomplex gH/gL is shown in blue, nonfluorescent YFPN and YFPC halves in white, and reconstituted YFP in yellow. CM, cell membrane. Arrowheads point to extended gB fusion loops (step 2).
Acknowledgments
Funding for this project was through NIH grants R01-AI076231 and R01-AI056045 to R.J.E. and G.H.C., respectively, from the National Institute of Allergy and Infectious Diseases and grant R37-AI18289 to G.H.C. and R.J.E. from the National Institute of Allergy and Infectious Diseases. Brian Hannah was supported by training grant AI-07234 from the National Institute of Allergy and Infectious Diseases.
We thank Florent Bender, Tina Cairns, and our laboratory colleagues for helpful advice.
Footnotes
Published ahead of print on 3 February 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Atanasiu, D., J. C. Whitbeck, T. M. Cairns, B. Reilly, G. H. Cohen, and R. J. Eisenberg. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 104:18718-18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81:11532-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2009. Cross talking among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J. Virol. 83:10752-10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backovic, M., and T. S. Jardetzky. 2009. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backovic, M., G. P. Leser, R. A. Lamb, R. Longnecker, and T. S. Jardetzky. 2007. Characterization of EBV gB indicates properties of both class I and class II viral fusion proteins. Virology 368:102-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender, F. C., M. Samanta, E. E. Heldwein, M. P. de Leon, E. Bilman, H. Lou, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J. Virol. 81:3827-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, F. C., J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2005. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J. Virol. 79:11588-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckmaster, E. A., U. Gompels, and A. Minson. 1984. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 × 103 molecular weight. Virology 139:408-413. [DOI] [PubMed] [Google Scholar]
- 9.Cairns, T. M., M. S. Shaner, Y. Zuo, M. Ponce-de-Leon, I. Baribaud, R. J. Eisenberg, G. H. Cohen, and J. C. Whitbeck. 2006. Epitope mapping of herpes simplex virus type 2 gH/gL defines distinct antigenic sites, including some associated with biological function. J. Virol. 80:2596-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 11.Cocchi, F., D. Fusco, L. Menotti, T. Gianni, R. J. Eisenberg, G. H. Cohen, and G. Campadelli-Fiume. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. U. S. A. 101:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J. Virol. 77:8127-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly, S. A., G. P. Leser, T. S. Jardetzky, and R. A. Lamb. 2009. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J. Virol. 83:10857-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 16.Hannah, B. P., T. M. Cairns, F. C. Bender, J. C. Whitbeck, H. Lou, R. J. Eisenberg, and G. H. Cohen. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83:6825-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannah, B. P., E. E. Heldwein, F. C. Bender, G. H. Cohen, and R. J. Eisenberg. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heldwein, E. E., and C. Krummenacher. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 20.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 21.Hu, C. D., and T. K. Kerppola. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadlec, J., S. Loureiro, N. G. Abrescia, D. I. Stuart, and I. M. Jones. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024-1030. [DOI] [PubMed] [Google Scholar]
- 23.Kerppola, T. K. 2008. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol. 85:431-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerppola, T. K. 2006. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1:1278-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kousoulas, K. G., B. Huo, and L. Pereira. 1988. Antibody-resistant mutations in cross-reactive and type-specific epitopes of herpes simplex virus 1 glycoprotein B map in separate domains. Virology 166:423-431. [DOI] [PubMed] [Google Scholar]
- 26.Krummenacher, C., A. Carfi, R. J. Eisenberg, and G. H. Cohen. Entry of herpesviruses into cells: the enigma variations. In S. Pohlmann and G. Simmons (ed.), Viral entry into host cells. Landes Bioscience, in press. [DOI] [PubMed]
- 27.Krummenacher, C., A. V. Nicola, J. C. Whitbeck, H. Lou, W. Hou, J. D. Lambris, R. J. Geraghty, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 72:7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magliery, T. J., C. G. Wilson, W. Pan, D. Mishler, I. Ghosh, A. D. Hamilton, and L. Regan. 2005. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127:146-157. [DOI] [PubMed] [Google Scholar]
- 30.McShane, M. P., and R. Longnecker. 2004. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc. Natl. Acad. Sci. U. S. A. 101:17474-17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, C. G., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and N. W. Fraser. 2001. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol. Ther. 3:160-168. [DOI] [PubMed] [Google Scholar]
- 32.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 33.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 34.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 35.Peng, T., M. Ponce-de-Leon, H. Jiang, G. Dubin, J. M. Lubinski, R. J. Eisenberg, and G. H. Cohen. 1998. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J. Virol. 72:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng, T., M. Ponce de Leon, M. J. Novotny, H. Jiang, J. D. Lambris, G. Dubin, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J. Virol. 72:6092-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira, L. 1982. Use of monoclonal antibodies to HSV-1 and HSV-2 for serological analysis of the viral glycoproteins. Dev. Biol. Stand. 52:115-131. [PubMed] [Google Scholar]
- 38.Pereira, L., M. Ali, K. Kousoulas, B. Huo, and T. Banks. 1989. Domain structure of herpes simplex virus 1 glycoprotein B: neutralizing epitopes map in regions of continuous and discontinuous residues. Virology 172:11-24. [DOI] [PubMed] [Google Scholar]
- 39.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 40.Remy, I., and S. W. Michnick. 2004. Mapping biochemical networks with protein-fragment complementation assays. Methods Mol. Biol. 261:411-426. [DOI] [PubMed] [Google Scholar]
- 41.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 42.Satoh, T., J. Arii, T. Suenaga, J. Wang, A. Kogure, J. Uehori, N. Arase, I. Shiratori, S. Tanaka, Y. Kawaguchi, P. G. Spear, L. L. Lanier, and H. Arase. 2008. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sisk, W. P., J. D. Bradley, R. J. Leipold, A. M. Stoltzfus, M. Ponce de Leon, M. Hilf, C. Peng, G. H. Cohen, and R. J. Eisenberg. 1994. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J. Virol. 68:766-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell. Microbiol. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 46.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. Methods 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. U. S. A. 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanarsdall, A. L., B. J. Ryckman, M. C. Chase, and D. C. Johnson. 2008. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J. Virol. 82:11837-11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willis, S. H., A. H. Rux, C. Peng, J. C. Whitbeck, A. V. Nicola, H. Lou, W. Hou, L. Salvador, R. J. Eisenberg, and G. H. Cohen. 1998. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J. Virol. 72:5937-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright, C. C., T. W. Wisner, B. P. Hannah, R. J. Eisenberg, G. H. Cohen, and D. C. Johnson. 2009. Fusion between perinuclear virions and the outer nuclear membrane requires the fusogenic activity of herpes simplex virus gB. J. Virol. 83:11847-11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zago, A., C. R. Jogger, and P. G. Spear. 2004. Use of herpes simplex virus and pseudorabies virus chimeric glycoprotein D molecules to identify regions critical for membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 101:17498-17503. [DOI] [PMC free article] [PubMed] [Google Scholar]