Abstract
Herpesviruses have evolved numerous strategies to evade detection by the immune system. Notably, most of the herpesviruses interfere with viral antigen presentation to cytotoxic T lymphocytes (CTLs) by removing class I major histocompatibility complex (MHC) molecules from the infected cell surface. Clearly, since the herpesviruses have evolved an extensive array of mechanisms to remove class I MHC molecules from the cell surface, this strategy serves them well. However, class I MHC molecules often serve as inhibitory ligands for NK cells, so viral downregulation of all class I MHC molecules should leave the infected cell open to NK cell attack. Some viruses solve this problem by selectively downregulating certain class I MHC products, leaving other class I products at the cell surface to serve as inhibitory NK cell ligands. Here, we show that human herpesvirus 7 (HHV-7) U21 binds to and downregulates all of the human class I MHC gene products, as well as the murine class I molecule H-2Kb. HHV-7-infected cells must therefore possess other means of escaping NK cell detection.
Human herpesvirus-7 (HHV-7) is a betaherpesvirus that infects over 90% of the population by the age of 3 (for a review, see reference 58). Like all other herpesviruses, HHV-7 establishes a latent or persistent infection, lasting for the lifetime of its host. Primary infection is usually accompanied by febrile illness, but long-term infection with the virus is asymptomatic (3, 53). HHV-7 is T-lymphotrophic, but it has also been found in salivary epithelial cells (30, 62).
As viruses that remain latent or persistent throughout the life of their hosts, the herpesviruses must interact continually with the host immune system. In so doing, all herpesviruses have evolved mechanisms to interfere with viral antigen presentation by class I major histocompatibility complex (MHC) molecules as a means to escape detection by cytotoxic T lymphocytes (CTLs). Some herpesvirus gene products interfere with proteolysis of antigens or peptide transport into the endoplasmic reticulum (ER) (1, 20, 56, 61). Others retain or destroy class I molecules (2, 26, 59, 64), enhance the internalization of class I molecules, or divert class I molecules to lysosomes for degradation (11, 23, 25, 44). Judging from the number and molecular diversity of these strategies, the removal of MHC class I-peptide complexes from the cell surface must be evolutionarily advantageous to these viruses as a means of escaping immune detection. We have described one such immunoevasin, U21, from HHV-7. HHV-7 U21 binds to class I MHC molecules in the ER and diverts them to a lysosomal compartment, where they are degraded, effectively removing them from the cell surface (23). The mechanism of U21-mediated diversion of class I molecules to lysosomes is not known, but the relocalization of class I MHC molecules is specific—U21 does not cause the rerouting of either the transferrin receptor or CD4 to lysosomes (22, 23).
Since the herpesviruses have evolved such an extensive array of mechanisms to remove class I MHC molecules from the cell surface of infected cells, this strategy must serve them well. However, when natural killer (NK) cells detect an absence of class I MHC molecules on the surface of a cell (i.e., “missing self”), they become activated to kill that cell. NK cells detect the absence of class I MHC molecules through interaction of NK cell receptors with NK cell receptor ligands present on the surface of the target cell (for a review, see references 6 and 7). When an NK cell surveys a potential target, it integrates the number and strength of the activating and inhibitory signals it receives; after weighing the balance, it either remains indifferent to the target or becomes activated to kill it.
Class I MHC molecules are ligands for inhibitory NK cell receptors. Thus, when a virus removes class I MHC molecules from the cell surface to escape detection by CTLs, it simultaneously renders the cell vulnerable to NK cell attack. Not surprisingly, viruses have evolved counterstrategies to protect their host cells from NK cell-mediated attack. The class I MHC locus contains three classical class I gene products, HLA-A, -B, and -C, as well as other “nonclassical” products, including HLA-E and HLA-G. As a strategy to avoid both CTL and NK cell attack, some viral immunoevasins selectively downregulate HLA-A and HLA-B locus products, while leaving HLA-C, -E, and other inhibitory class I-like molecules at the plasma membrane (10, 16, 35). It has therefore been speculated that HLA-A and -B may be more effective at antigen presentation to CTLs than HLA-C (15, 40). The nonclassical class I molecule HLA-E, on the other hand, functions primarily to inhibit NK cell activation and does not present foreign antigen to CTLs (33). As such, its expression at the cell surface is even promoted by at least one immunoevasin, UL40 from human cytomegalovirus (HCMV) (54, 57).
We do not know how HHV-7 responds to the selective pressures exerted by NK cells. We have shown previously that U21 can associate with and downregulate HLA-A and -B, but we do not yet know the full extent of its promiscuity (23). For this reason, we now examine the ability of U21 to bind to and downregulate the various classical and nonclassical class I MHC gene products. We find that, unlike many other viral immunoevasins, HHV-7 U21 can associate with and downregulate HLA-C, -E, and -G and even murine class I MHC molecules. In an infection, this would shift the balance of inhibitory NK cell ligands on the cell surface to favor NK cell attack, suggesting that HHV-7 might compensate for such an imbalance through other means of NK cell evasion.
U21 is 55-kDa type I membrane protein with a short (50-amino-acid [aa]) cytoplasmic tail. We have shown that its transmembrane domain and cytoplasmic tail are not involved in its association with the lumenal domain of the class I molecule (22). In addition to gaining information about U21's potential influence on CTL and NK cell detection of HHV-7-infected cells, we also hoped that a survey of its ability to associate with various class I MHC gene products might help to illuminate regions of the class I molecule important for association with U21.
MATERIALS AND METHODS
Cell lines.
B6/WT3 mouse fibroblast (generously provided by T. Hansen, Washington University, St. Louis, MO), U373 astrocytoma, and JEG-3 choriocarcinoma cell lines were cultured in Dulbecco's modified Eagle medium (DMEM)-5% fetal bovine serum-5% newborn calf serum in the presence or absence of puromycin (375 ng/ml final concentration; Sigma-Aldrich, St. Louis, MO) or hygromycin B (final concentrations, 250 μg/ml for B6/WT3 and 50 μg/ml for JEG-3; Invitrogen, Carlsbad, CA). C1R B-lymphoblastoid cells (generously provided by J. Gumperz, University of Wisconsin, Madison, WI) were cultured in RPMI 1640-5% fetal bovine serum-5% newborn calf serum in the presence or absence of 400 μg/ml hygromycin B. 721.221 Epstein-Barr virus (EBV)-transformed B cells (generously provided by J. Gumperz, University of Wisconsin, Madison, WI) were cultured in RPMI 1640 with 10% iron-supplemented calf serum in the presence or absence of 200 ng/ml puromycin. Expression of the viral protein U21 in U373 cells was carried out via retrovirus-mediated gene transfer, using the vector pLNCX, which carries the selectable marker for neomycin resistance. The U373 cells uniformly express levels of U21 similar to that seen for HHV-7 infection of SupT1 T cells (23). Expression of U21 in B6/WT3, JEG-3, and C1R cells was carried out via lentivirus-mediated gene transfer, using the vector pHAGE-Hygro, which carries the selectable marker for hygromycin resistance (37). Expression of CD1d, HA-tagged m144 (m144HA), or murine MHC class I H-2Kb in U373 cells and of U21 in 721.221 cells was carried out via lentivirus-mediated gene transfer, using the vector pHAGE-puro, which carries the selectable marker for puromycin resistance (37). U21 expression is uniform within each cell population (data not shown).
Antibodies.
The monoclonal antibody (MAb) W6/32 recognizes assembled, β2m-associated HLA-A, -B, -C, -E, and -G molecules; MAb HC10 recognizes unfolded free MHC class I heavy chain; MAb HCA2 recognizes unfolded HLA-G heavy chain; and MAb 12CA5 recognizes hemagglutinin (HA) epitope (each generously provided by H. Ploegh, Whitehead Institute, Cambridge, MA). MAb L31 recognizes free HLA-C heavy chains (generously provided by P. Giacomini, Regina Elena Cancer Institute, Rome, Italy). MAb Y-3 recognizes H-2Kb (generously provided by P. Cresswell, Yale University, New Haven, CT). The CD1d42 and CD1d51 MAbs recognize properly folded CD1d (generously provided by J. Gumperz, UW-Madison, WI). The MAb HA11 recognizes the HA epitope (Covance, San Diego, CA). The MAb H4B4 recognizes lamp2 (generously provided by T. August, Johns Hopkins Medical School, Baltimore, MD). The following rabbit polyclonal antibodies (PAb) were used: PAb MCW50 was raised against the cytoplasmic tail of HHV-7 U21 (Q. Wang and A. W. Hudson, unpublished data), PAb P8 recognizes unfolded H-2Kb heavy chain (generously provided by H. Ploegh, Whitehead Institute, Cambridge, MA), and “Mingus” is an anti-lgp120 PAb (generously provided by I. Mellman, Yale University, New Haven, CT). Alexa Fluor 488-, 594-, and 647-conjugated secondary antibodies were used in immunofluorescence studies (Molecular Probes, Eugene, OR). A phycoerythrin (PE)-conjugated anti-mouse MAb (BD Pharmingen, San Jose, CA) was used in flow cytometry experiments. A horseradish peroxidase (HRP)-conjugated anti-mouse MAb (Bio-Rad, Hercules, CA) was used in immunoblot assays.
Pulse-label experiments.
721.221 cells were incubated in methionine- and cysteine-free DMEM for 30 min at 37°C. The cells were labeled with 500 μCi/ml of 35S-Express label (1,175 Ci/mmol; PerkinElmer, Boston, MA) at 37°C for 1 h. Cells were lysed in 0.5% Nonidet P-40 lysis buffer (50 mM Tris-HCl [pH 7.4], 0.5% NP-40, 5 mM MgCl2, 150 mM NaCl). U373 and B6/WT3 cells were incubated in methionine- and cysteine-free DMEM containing 180 μCi/ml of 35S-Express label for 40 min at 37°C. Cells were lysed in 1% Nonidet P-40 lysis buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 5 mM MgCl2).
All lysates were centrifuged for 8 min at 16,000 × g at 4°C in an Eppendorf centrifuge to pellet nuclei and debris, followed by immunoprecipitation with specific antiserum and protein A agarose (RepliGen Corporation, Waltham, MA). Immunoprecipitates were normalized to equal trichloroacetic acid-precipitable 35S-labeled protein. The immunoprecipitates were washed twice with NP-40 wash buffer (50 mM Tris-HCl [pH 7.4], 0.5% NP-40, 5 mM EDTA, 150 mM NaCl), twice with the same buffer supplemented with 0.1% SDS, and subjected to SDS-PAGE.
Immunofluorescence microscopy.
Cells on coverslips were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 20 min, permeablized with 0.5% saponin in PBS and 3% bovine serum albumin (BSA), incubated with primary antibodies for 30 min, washed, and incubated with Alexa 488-, 594-, or 647-conjugated secondary antibodies. Coverslips were first treated with 0.01% poly-l-lysine (Sigma-Aldrich) for visualization of suspension cells.
Flow cytometry.
U373 cells were removed from plates, washed with ice-cold PBS, incubated with CD1d51 anti-CD1d antibody in PBS and 1% BSA for 30 min on ice, washed, and incubated in PE-conjugated anti-mouse IgG (R&D Systems, Minneapolis, MN). 721.221 (suspension) cells were similarly washed and incubated with W6/32 anti-MHC class I antibody followed by secondary antibody. Cell surface expression of class I MHC and CD1d molecules was then evaluated using a Beckman FACScalibur flow cytometer.
Immunoblots.
Immunoblots were performed with cell lysates or crude membrane preparations. To prepare lysates, cells were washed with ice-cold PBS and lysed in 1% NP-40 lysis buffer, and nuclei and debris were pelleted. For crude membrane preparations, cells were washed with ice-cold PBS, resuspended in scraping buffer (100 mM NaPi [pH 7.4], 5 mM EDTA, 250 mM sucrose, 50 mM NaF, 1 mM PMSF), sonicated twice for 15 s on ice, and centrifuged to pellet nuclei and debris, and the supernatant was centrifuged for 30 min at 100,000 × g at 4°C to pellet crude membranes. Membrane pellets were resuspended in membrane buffer (5 mM NaPi [pH 7.4], 1 mM EDTA, 250 mM sucrose). Protein concentrations of lysates and crude membranes were determined using a bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL). Protein assays were performed in triplicate and lysates of equal protein concentration were loaded on SDS gels, except where protein concentrations are specifically noted. The separated proteins were transferred to BA-85 nitrocellulose membranes (Whatman, Florham Park, NJ). To further verify the loading of equal protein concentrations, membranes were stained with Ponceau S to detect total protein. Membranes were probed with P8, HC10, HCA2, or L31 antibodies to free class I heavy chains or MCW50 antibody to U21, followed by an HRP-conjugated secondary antibody (Bio-Rad). Bands were visualized using Pierce SuperSignal Chemiluminescence reagents (Pierce) and quantitated with an Alpha Imager (AlphaInnotech, San Leandro, CA). For treatment with lysosomal protease inhibitors, cells were incubated either in medium containing 200 μM leupeptin (EMD Biosciences, Inc., San Diego, CA) and 20 μM folimycin (concanamycin A; EMD Biosciences, Inc.) or in medium containing dimethyl sulfoxide (DMSO) for 14 h. Culture medium was then replaced with fresh medium containing inhibitors or a DMSO control for 4 additional hours of incubation prior to lysis.
RESULTS
U21 binds to and affects trafficking of all classical class I MHC molecules.
Our initial assessment of the binding specificity of U21 for classical class I MHC gene products (HLA-A, -B, and -C) was performed using isoelectric focusing (IEF) gel analysis of immunoprecipitated class I MHC molecules (23). We recovered class I MHC molecules that coprecipitated with U21 and compared them to the IEF profile of class I MHC molecules recovered with W6/32, a conformation-specific anti-class I antibody that recognizes all β2m-bound HLAs. We found the two IEF profiles to be essentially identical, suggesting that U21 associates with the same array of class I gene products as W6/32 (23). Although W6/32 recognizes HLA-C, some of the HLA bands were underrepresented in this preliminary experiment, and we could not be certain that HLA-C alleles were represented among the polypeptides recovered and separated on the IEF gel. The idea that some viral immunoevasins will downregulate HLA-A and -B, but not HLA-C, prompted a more careful characterization of U21's ability to bind to and redirect HLA-C molecules.
As we have shown previously, in control U373 cells, class I molecules are localized mainly on the plasma membrane, while in cells expressing U21, class I MHC molecules are dramatically redistributed to the lysosomal compartment (Fig. 1, compare panels a and b) (17, 22, 23). To specifically examine the diversion of HLA-C by U21, we turned to the B-lymphoblastoid cell line C1R which, due to mutagenesis and negative selection with anti-HLA antibodies, expresses the HLA-Cw4 gene product, and not HLA-A or -B (52, 63). The C1R cells are nonadherent, and we observe relatively little plasma membrane localization of HLA-C in these cells, consistent with reports that 75 to 90% of HLA-C is retained intracellularly (36, 40, 50). Instead, we visualize HLA-C primarily in the biosynthetic compartment (Fig. 1, panel c). Expression of U21 in these cells resulted in a more punctate distribution of HLA-C, suggesting that U21 can redirect HLA-C molecules to the lysosomal compartment (Fig. 1, compare panels c and d). We note that the steady-state level of class I MHC expression in control U373 cells is ∼40-fold higher than in cells expressing U21 (Fig. 2a, compare lanes 1 and 2), yet W6/32-reactive class I molecules seen in U21-expressing cells appear disproportionately intense, perhaps because they are concentrated in the lysosomal compartment (Fig. 1b). We observe a similar phenomenon in UL21-expressing C1R cells (Fig. 1d).
FIG. 1.
U21 diverts class I MHC molecules to a perinuclear region. Control U373 cells (a), U21-expressing U373 cells (b), control C1R cells (c), and U21-expressing C1R cells (d) were labeled with W6/32, an antibody directed against properly folded class I MHC molecules (α-MHC-I), followed by Alexa 488-conjugated secondary antibody (Ab). Scale bars = 10 μm.
FIG. 2.
Binding of U21 to HLA-C is stabilized by lysosomal protease inhibitors. (a) Equal protein concentrations from lysates of U373- and U21-expressing U373 cells treated with lysosomal protease inhibitors folimycin (Foli) and leupeptin (Leu) were immunoblotted with HC10 to detect class I heavy-chain molecules. (b) Lysates and U21 immunoprecipitations (IP) of control C1R- and U21-expressing C1R cells treated with lysosomal protease inhibitors were immunoblotted with HC10 to detect class I MHC heavy chains. The upper band marked by the asterisk is due to reactivity of the HRP-conjugated secondary antibody directly with the anti-U21 antibody. (c) Samples from control and U21-expressing U373 cells treated with lysosomal protease inhibitors were blotted with L31 to detect HLA-C. The fold increase in signal intensity between bands is indicated by arrows. (d) Crude membrane preparations of U373, C1R, and U373 and C1R cells expressing U21 were immunoblotted with MCW50 antibody to U21, as indicated. Three micrograms of U373 crude membranes and 20 μg of C1R crude membranes were blotted, as indicated. The upper band marked by the asterisk is a cross-reacting background band.
To determine whether expression of U21 also affected the steady-state levels of HLA-C, we immunoblotted equal concentrations of total protein from control C1R cells and U21-expressing C1R cells using HC10, a monoclonal antibody reactive with free HLA-A, -B, and -C heavy chains (51). The total amount of HLA-C was reduced 2.2-fold in the C1R cells expressing U21, suggesting that the presence of U21 results in destabilization of HLA-C molecules (Fig. 2b, lanes 1 and 2). To determine whether U21 associates with HLA-C, we next performed coimmunoprecipitation experiments from control C1R and U21-expressing C1R cells, immunoprecipitating U21 and immunoblotting with HC10 to detect free HLA-C heavy chains. As shown in Fig. 2b, lane 4, class I MHC molecules coprecipitate with U21.
To further demonstrate the interaction between U21 and HLA-C, we performed the same experiment with U373 cells, which contain all of the classical class I gene products. In this experiment, the immunoblots were performed using L31, an antibody specific for free HLA-C heavy chains (48). Here, too, in U373 cells, 4.5-fold-lower steady-state levels of HLA-C were detected when U21 was present, further suggesting that U21 expression results in the degradation of HLA-C (Fig. 2c, lanes 1 and 2). For comparison, we also performed the experiment with U373 cells, detecting all free classical class I heavy chains using HC10. In U373 cells, we observed a larger (∼40-fold) reduction, likely because HLA-A and -B comprise 80 to 90% of the total surface class I (Fig. 2a, lanes 1 and 2).
U21 binds to class I molecules and redirects them to lysosomes, where they are degraded (17, 23). Predictably, inhibition of lysosomal proteases results in the stabilization of both U21 and class I MHC molecules (Fig. 2a, compare lane 4 with lane 2) (17, 23). Likewise, when U21-expressing C1R cells were treated with lysosomal protease inhibitors, we observed an 11-fold increase in the HLA-C that coimmunoprecipitates with U21 (Fig. 2b, compare lane 6 to lane 4), demonstrating that HLA-C, too, associates with U21 and is redirected to the lysosomal compartment for degradation. A 2.5-fold increase in recovery of coprecipitated HLA-C molecules following treatment with lysosomal protease inhibitors was also observed for U373 cells (Fig. 2c, compare lane 6 to lane 4).
The stable U21-expressing cell lines we describe were generated using retrovirus-mediated gene transfer and selected in puromycin or another selection agent. This mode of transduction generally yields uniform populations of cells that do not express toxic levels of protein, because the cells must continue to grow. We note that the levels of U21 expressed in U373 cells are similar (within 10-fold) to the levels of U21 expressed during HHV-7 infection of T cells in culture (23). The stable cell lines are quite uniform in their expression of U21 within each cell population, and the cells depicted in immunofluorescence images are representative of >80% of the cells examined. However, the steady-state levels of U21 expression vary considerably from cell type to cell type. To assess the steady-state U21 expression levels in the C1R cells compared with that in the U373-U21 cells, we performed immunoblotting experiments on crude membrane preparations from each cell line. Notably, all of the nonadherent cell lines we have tested express significantly lower levels of U21 than U373 cells (Fig. 2d, compare lanes 4 and 2).
U21 interacts with properly folded HLA-G molecules.
To further assess the ability of U21 to associate with the HLA molecules, we next examined whether U21 could affect the nonclassical class I molecule HLA-G. HLA-G is expressed only in placental tissue and is thought to be involved in protecting the fetus from its mother's NK cells (12, 13).
For these experiments, we expressed U21 in JEG-3 choriocarcinoma cells, which express HLA-G and HLA-C, but not HLA-A or HLA-B (46). We first examined the localization of class I MHC molecules. In control JEG-3 cells, the class I MHC (W6/32) immunofluorescence labeling pattern is somewhat different from that in U373 cells: the HLA-C and -G molecules in these cells label the cell surface with a grainy appearance, and the Golgi complex is more intensely visible (Fig. 3a). This pattern likely reflects the absence of HLA-A and -B in these cells and the relatively poor expression of HLA-C on the cell surface (36, 40, 50). In JEG-3 cells expressing U21, however, we observe striking redistribution of class I MHC molecules to a punctate perinuclear compartment similar to that observed for U373 U21 cells, demonstrating that U21 diverts W6/32-reactive class I MHC molecules in these cells (Fig. 3, compare panel b to panel a). Although this redistribution is dramatic, and the absence of HLA-A and HLA-B in these cells suggests that both HLA-C and HLA-G are relocalized, this experiment does not rule out the possibility that U21 excludes HLA-G and that this redistribution is attributable solely to HLA-C relocalization.
FIG. 3.
U21 associates with and relocalizes HLA-G. Control JEG-3 cells (a) and U21-expressing JEG-3 cells (b) were labeled with W6/32, an antibody directed against properly folded class I MHC molecules, followed by an Alexa 488-conjugated secondary antibody. Scale bars = 10 μm. (c) Lysates and U21 immunoprecipitations of control and U21-expressing JEG-3 cells treated with lysosomal protease inhibitors folimycin and leupeptin were immunoblotted with HCA2 to detect HLA-G. The fold increase in signal intensity between bands is indicated by arrows. (d) Crude membrane preparations of U373, JEG-3, and U373 and JEG-3 cells expressing U21 were immunoblotted with MCW50 antibody to U21, as indicated. Three micrograms of U373 crude membranes and 20 μg of JEG-3 crude membranes were immunoblotted, as indicated. The lower band marked by the asterisk is a cross-reacting background band.
Therefore, to assess whether U21 can associate with HLA-G, we first examined steady-state levels of HLA-G in the absence and presence of U21. Just as for HLA-C, the amount of HLA-G in cells expressing U21 was reduced (Fig. 3c, lanes 1 and 2). We next performed coimmunoprecipitation experiments (as described above for HLA-C) in which we performed immunoprecipitations with U21 to recover all U21-associated class I products, and we immunoblotted with HCA2, an antibody recognizing free HLA-G heavy chains (Fig. 3c, lanes 3 and 4). Although the HCA2 antibody was raised against HLA-A, it cross-reacts with HLA-G, and since JEG-3 cells lack HLA-A, HCA2 allows specific detection of HLA-G (47). In addition, since its cytoplasmic tail is shorter, HLA-G migrates faster than the classical class I molecules in SDS-polyacrylamide gels, so its detection is unmistakable. Like HLA-C, HLA-G coimmunoprecipitated with U21, and incubation in the presence of lysosomal protease inhibitors enhanced the amount of HLA-G recovered ∼4-fold, suggesting that U21 redirects HLA-G to the lysosomal compartment as well (Fig. 3c, compare lanes 4 and 6).
Similar to results for U21-expressing C1R cells, an immunoblot analysis of crude membranes showed steady-state expression of U21 in JEG-3 cells to be much lower than that in U373-U21 cells (Fig. 3d, compare lanes 4 and 2).
U21 associates with and reduces surface expression of HLA-E molecules.
We next investigated whether U21 could reduce the surface expression of HLA-E. For these experiments, we expressed U21 in 721.221 lymphoblastoid cells which, due to gamma ray-induced mutations in the HLA complex, express only HLA-E (29, 49).
Like most class I MHC molecules, stable assembly of HLA-E requires that peptide be bound within its peptide binding groove. Unlike other class I products, HLA-E's peptide binding groove accepts nonamer peptides derived from the cleavable signal sequences of other class I HLA molecules, to the exclusion of most conventionally processed antigens (4, 34). As such, it functions primarily as an inhibitory ligand for NK cells.
Since the 721.221 cells in these experiments express HLA-E in the absence of other class I products, class I-derived peptide antigens are unavailable; thus, assembly of HLA-E is impaired. Surface expression of endogenous HLA-E is therefore low (34). Still, when we expressed U21 in these cells and examined surface expression of HLA-E, the surface HLA-E was reduced to an almost undetectable level (Fig. 4a).
FIG. 4.
U21 associates with and downregulates surface expression of HLA-E. (a) Control 721.221 (shaded) and U21-expressing cells (bold) were incubated with W6/32 followed by secondary incubation with a PE-conjugated secondary antibody. Control cells incubated with secondary antibody only are indicated with the thin trace. (b) 721.221 control cells and U21-expressing cells were metabolically labeled for 1 h with [35S]cysteine and [35S]methionine, and HLA-E molecules were recovered with W6/32. (c) Crude membrane preparations of U373, 721.221, and U373 and 721.221 cells expressing U21 were immunoblotted with MCW50 antibody to U21, as indicated. Three micrograms of U373 crude membranes and 20 μg of 721.221 crude membranes were blotted, as indicated.
To determine whether U21 associates with HLA-E molecules, we performed coimmunoprecipitation experiments, using W6/32 to recover HLA-E from metabolically labeled U21-expressing 721.221 cells. When HLA-E molecules were immunoprecipitated with W6/32, U21 molecules were recovered as well, demonstrating that HLA-E, too, associates with U21 (Fig. 4b).
Similar to U21-expressing C1R and JEG-3 cells, immunoblot analysis of crude membranes showed steady-state expression of U21 in 721.221 cells to be much lower than that in U373-U21 cells (Fig. 4c, compare lanes 4 and 2).
U21 diverts the murine class I MHC molecule H-2Kb to lysosomes.
HHV-7 infection is species specific, and it does not infect mice. Because U21 was capable of association with all of the HLA gene products we examined, we next hypothesized that it may also be capable of interaction with the structurally similar murine class I MHC molecules, despite their more-divergent primary sequence (see Fig. 12). Our investigation into U21's association with murine class I MHC molecules was therefore performed not for physiological reasons, but to gain a deeper understanding of the breadth of its substrates.
FIG. 12.
Sequence alignments of HLA-A, -B, -C, -E, and -G, as well as murine H-2Kb. (a) Sequence comparison of the human HLAs. HLA-A through HLA-E share ∼58% identity and have a majority consensus at ∼94% of residues at the amino acid level. Identity is indicated by dark gray shading, while consensus is indicated by light gray shading. (b) Sequence comparison of HLA-A with murine H-2Kb, which share ∼73% similarity (light gray shading) and ∼64% amino acid identity (dark gray shading). Light gray lines over the sequences indicate α1 and α2 domains, and dark gray lines over the sequences indicate α3 domains. Boxes show predicted transmembrane domains, while black hash marks and squares indicate predicted signal peptide cleavage sites.
We first examined whether murine H-2Kb molecules could be relocalized by U21. When H-2Kb molecules are expressed in human U373 cells, they assemble with human β2m and are localized to the cell surface (Fig. 5a and c) (19). When we expressed U21 in H-2Kb-expressing U373 cells, the redistribution of H-2Kb was as striking as the redistribution of human class I molecules: H-2Kb molecules were localized to a perinuclear location that colocalized with lamp 2, a marker for the lysosomal compartment (Fig. 5, compare panels a and b, c and e, and e and f). To demonstrate that U21 expression results in the punctate distribution of class I molecules within the population of cells as a whole, smaller scale images are shown (Fig. 5a and b).
FIG. 5.
U21 diverts the murine class I MHC H-2Kb to the lysosomal compartment in human cells. U373 cells expressing H-2Kb with or without U21 were labeled with Y-3 anti-H-2Kb MAb and the anti-lamp2 lysosomal marker protein MAb H4B4. The antibodies used in each panel are indicated. Primary antibodies were detected with Alexa Fluor-conjugated secondary antibodies. Arrows indicate specific points of colocalization between H-2Kb and lamp 2 labeling. Scale bars = 10 μm.
U21 associates with H-2Kb in human astrocytoma cells.
To assess the association of U21 with H-2Kb, we performed coimmunoprecipitation experiments, recovering U21, H-2Kb, and endogenous class I MHC from metabolically labeled U373 cells expressing U21, H-2Kb, or both gene products together (Fig. 6a). Using control U373 cells, we recovered HLA heavy chains with W6/32 (Fig. 6a, lane 1). From U373 cells expressing H-2Kb, we recovered H-2Kb with the anti-H-2Kb antibody Y3 (Fig. 6a, lane 6). Of note, in U373 cells expressing H-2Kb, HLA class I heavy chains recovered with W6/32 were barely detectable (Fig. 6a, compare lanes 4 and 1). The decrease in W6/32-reactive human class I levels in these cells likely reflects a competition between orthologous class I molecules for the available β2m. The observation that murine H-2 molecules can associate with human β2m molecules is long known (19, 21); thus, the observation that murine H-2Kb appears to possess greater affinity than the human class I molecules for β2m, causing a reduction in W6/32-reactive β2m-bound human class I molecules, is consistent with the literature.
FIG. 6.
U21 associates with H-2Kb in human cells. (a) U373 control cells (lanes 1 to 3), along with U373 cells expressing U21 and/or H-2Kb (lanes 4 to 12), were labeled for 40 min with [35S]cysteine and [35S]methionine. Class I MHC molecules were recovered with W6/32 (lanes 1, 4, 7, and 10), U21 was recovered with PAb MCW50 anti-U21 (lanes 2, 5, 8, and 11), and H-2Kb was recovered with the anti-H-2Kb MAb Y-3 (lanes 3, 6, 9, and 12). Recovery of properly folded class I molecules with the W6/32 antibody was notably lessened in H-2Kb-expressing cells (lanes 4 and 10). The film exposure time for lanes 1 to 6 (72 h) was longer than that of lanes 7 to 12 (40 h). (b and c) Eight micrograms of the indicated lysates were immunoblotted with the HC10 antibody reactive with free HLA heavy chains (a) and P8 antibody to free H-2Kb heavy chains (b).
If indeed the HLA heavy-chain molecules “lose” the competition for available β2m, then although W6/32-reactive, properly folded class I MHC molecules are barely detectable in the H-2Kb-expressing cells, unfolded β2m-free HLA class I MHC molecules should be present at similar levels, regardless of H-2Kb-expression. This is indeed the case: U373 and U373-H-2Kb lysates show comparable levels of class I MHC heavy chains, when detected with a monoclonal antibody recognizing unfolded HLA heavy chains (Fig. 6b, compare lanes 1 and 3).
Immunoprecipitation of HLA class I alleles from U373 cells expressing U21 also results in recovery of fewer HLA heavy-chain molecules than from U373 control cells (Fig. 6a, compare lane 7 to lane 1). In this case, however, the reduced class I recovered is due to the expression of U21 in these cells, which results in degradation of class I molecules in lysosomes (Fig. 2a and 6b, lanes 1 and 2). Immunoprecipitation of U21 from these cells coprecipitates class I HLA heavy chains (Fig. 6a, lane 8).
In cells expressing both U21 and H-2Kb, we again observed very few labeled HLA heavy-chain molecules coimmunoprecipitating with W6/32 (Fig. 6a, lane 10), while immunoprecipitation with U21 resulted in the coprecipitation of H-2Kb and HLA molecules (Fig. 6a, compare lane 11 with lanes 10 and 12). The reciprocal immunoprecipitation with antibody to H-2Kb recovered U21 in addition to H-2Kb, demonstrating interaction between the two proteins (Fig. 6a, lane 12). Note that in metabolic labeling experiments, we visualize only 35S-labeled protein. Thus, the relative amounts of labeled protein in these experiments cannot be compared, since their steady-state levels, localization, and rates of synthesis are dissimilar. Lane 10 of Fig. 6a is particularly illustrative of this point; here, U21 appears more abundant than the class I MHC molecules that were immunoprecipitated.
To assess relative expression levels of class I MHC molecules, immunoblots were performed with lysates from these stable cell lines. As seen with U373 cells expressing U21 (Fig. 6b, lanes 1 and 2), the steady-state levels of class I HLA heavy-chain molecules are reduced when U21 is expressed in the U373 H-2Kb cells (Fig. 6b, lanes 3 and 4).
We note that there appears to be slightly more H-2Kb present in the U373 U21 H-2Kb cells than in the U373 H-2Kb cells (Fig. 6a, compare lanes 12 and 6). We suspect that these levels differ because the U373 and U373-U21 cells were independently transduced with an H-2Kb retrovirus, and as a result, the cells express slightly different levels of H-2Kb (Fig. 6c, compare lanes 3 and 4).
U21 associates with H-2Kb in murine fibroblast cells.
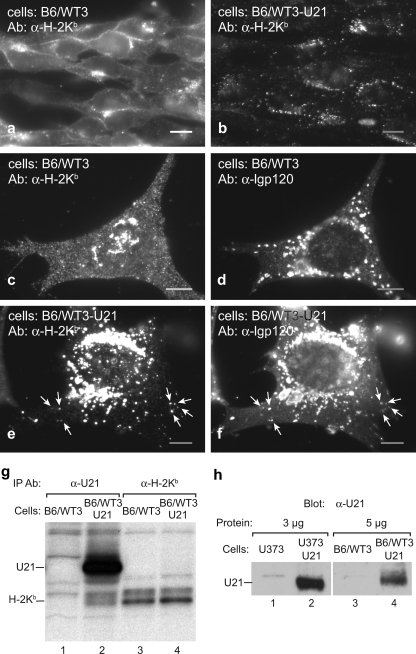
We next assessed whether U21 could affect endogenous murine class I MHC molecules. The mechanism of U21-mediated rerouting of class I MHC molecules is not known, but if its function depends upon cellular trafficking proteins, in order to affect class I H-2 molecules in murine cells, the murine versions of these auxiliary cellular proteins must substitute for their human counterparts. To determine whether U21 could bind to and divert H-2Kb molecules to a lysosomal compartment in murine cells, we expressed U21 in the b haplotype murine fibroblast line B6/WT3 and examined localization of H-2Kb by immunofluorescence microscopy. In B6/WT3 cells, the endogenous H-2Kb labels the cell surface and the biosynthetic compartments but has a more grainy appearance in these cells than it does when expressed in human U373 cells (compare Fig. 7c to Fig. 5c). Here, too, expression of U21 results in a dramatic change in localization of H-2Kb to the lysosomal compartment, showing significant colocalization with the murine lysosomal marker protein lgp120 (Fig. 7, compare panels c and e and panels e and f). To demonstrate that U21 expression results in the punctate distribution of class I molecules within the population of cells as a whole, smaller-scale images are shown (Fig. 7a and b).
FIG. 7.
U21 associates with endogenous H-2Kb and diverts it to a lysosomal compartment. B6/WT3 and U21-expressing cells were labeled with Y-3 anti-H-2Kb antibody along with anti-lgp120 antibody to murine lamp-1. The antibodies used in each panel are indicated. Primary antibodies were detected with Alexa Fluor-conjugated secondary antibodies. Arrows denote specific points of colocalization between H-2Kb and lgp-120. Scale bars = 10 μm. (g) Control B6/WT3 and U21-expressing cells were labeled for 40 min with [35S]cysteine and [35S]methionine. U21 was recovered with anti-U21 PAb, and H-2Kb was recovered with Y-3. (h) Crude membrane preparations of U373, B6/WT3, and U373 and B6/WT3 cells expressing U21 were immunoblotted with MCW50 antibody to U21, as indicated. Three micrograms of U373 crude membranes and 5 μg of B6/WT3 crude membranes were blotted, as indicated.
We also performed coimmunoprecipitation experiments with these cells, recovering U21 and H-2Kb molecules from metabolically labeled control and U21-expressing B6/WT3 cells. Immunoprecipitation with an antibody directed against the C terminus of U21 coprecipitated H-2Kb molecules (Fig. 7g, lane 2), and precipitation with an antibody to H-2Kb recovered H-2Kb from control cells as well as from U21-expressing cells (Fig. 7g, lanes 3 and 4). Strangely, however, although U21 coprecipitated with H-2Kb in H-2Kb-expressing U373 cells, U21 failed to coimmunoprecipitate with H-2Kb from U21-expressing B6/WT3 cells, despite identical experimental conditions (compare Fig. 7g, lane 4, to Fig. 6a, lane 12). The most obvious difference between this experiment and the one performed with U373 cells lies in the available β2m xenotype. In murine cells, H-2Kb folds in complex with endogenous murine β2m, whereas in human cells, it folds with human β2m. Although both complexes are recognized by the H-2Kb-specific antibody, and the β2m orthologs are 66% identical and 80% similar at the amino acid level, it is possible that a slight difference in the conformation achieved after binding of β2m may influence immunoprecipitation outcomes: the antibody binding to H-2Kb might somehow exclude U21 from immunoprecipitations when complexed with murine but not human β2m. Immunoblot analysis of crude membranes showed steady-state expression of U21 in B6/WT3 cells to be lower than that in U373-U21 cells (Fig. 7 h, compare lanes 4 and 2).
U21 does not alter CD1d localization.
In further examining the breadth of substrates that can be relocalized by U21, we next assessed the trafficking of CD1d. CD1d is a class I MHC-like molecule that presents lipid antigens to T-cell receptors on NKT cells. Although not encoded in the MHC, it assembles with β2m and has a three-dimensional structure nearly superimposable with class I MHC molecules (31; for a review, see reference 24). Because CD1d has been recently described as a target for downregulation by HCMV, Kaposi's sarcoma-associated virus, herpes simplex virus, and vesicular stomatitis virus (41-43, 45), we hypothesized that U21 might also cause relocalization of CD1d to lysosomes.
CD1d expression is confined to professional antigen-presenting cells, so to assess U21's ability to affect CD1d, we expressed exogenous CD1d using human U373 cells. When expressed in control U373 cells, CD1d localizes to the cell surface (Fig. 8c). When CD1d and U21 were coexpressed in U373 cells, no relocalization of CD1d was observed (Fig. 8e), despite reasonably high levels of U21 expression (Fig. 8, compare panel f to panel b).
FIG. 8.
U21 does not alter the localization of CD1d. U373 cells expressing CD1d, U21, or both were labeled with MAb CD1d42 anti-CD1d and with PAb anti-U21. The antibodies used in each panel are indicated. Primary antibodies were detected with Alexa Fluor-conjugated secondary antibodies. Scale bars = 10 μm.
To more quantitatively assess whether expression of U21 has an effect on surface CD1 localization, we performed flow cytometric analysis of the surface CD1d expression. Surface levels of CD1d were unaffected by the expression of U21 in the cells (Fig. 9a). The uniformity of CD1d expression within the cell populations is also illustrated in these flow cytometric histograms. Immunoblot analysis of crude membranes showed steady-state expression of U21 in U373 CD1d-expressing cells to be similar to that in U373-U21 cells (Fig. 9b, compare lanes 4 and 2).
FIG. 9.
U21 does not alter surface expression of CD1d. (a) Control CD1d-expressing U373 cells (shaded) and cells expressing both CD1d and U21 (bold) were incubated with CD1d51 anti-CD1d MAb followed by secondary incubation with a PE-conjugated secondary antibody. Control cells incubated with secondary antibody only are indicated by the thin trace. (b) Three micrograms of crude membrane preparations of U373 control cells and U373 cells expressing U21 and/or CD1d were immunoblotted with MCW50 antibody to U21.
U21 does not interact with m144.
In an effort to evade detection from NK cells, some viruses encode proteins that act as decoy class I MHC molecules, masking the absence of class I MHC that is downregulated by the virus (27). The murine cytomegalovirus (MCMV) m144 protein is one such immunoevasin (9, 32, 39). Because m144 assembles with β2m and possesses a three-dimensional structure similar to that of class I MHC molecules, as part of our investigation into the structural and sequence requirements for association with U21, we examined whether m144 may also be a substrate for U21.
To determine if U21 could alter the localization of m144, we expressed C-terminally HA-tagged m144 in control U373 cells and in U373 cells expressing U21 (Fig. 10, compare panel a to panels c and e). Despite expression of U21, m144 remained on the cell surface and in the biosynthetic compartment (Fig. 10, compare panels c and e.) To demonstrate that U21 expression has no effect upon the distribution of m144 molecules within the population of cells as a whole, smaller-scale images are also shown (Fig. 10g and h).
FIG. 10.
U21 does not alter the localization of m144HA. U373 cells expressing HA-tagged m144, U21, or both were labeled with HA11 anti-HA MAb and with anti-U21 PAb. The antibodies used in each panel are indicated. Primary antibodies were detected with Alexa Fluor-conjugated secondary antibodies. Scale bars = 10 μm.
The cytoplasmic HA tag on m144 negated the option to quantitatively assess its surface expression by flow cytometry, but to determine whether U21 could interact with m144, we performed coimmunoprecipitation experiments, recovering U21 and HA-tagged m144 from metabolically labeled cells. Because both U21 and m144HA migrate similarly in SDS-PAGE (data not shown), to resolve the two proteins, immunoprecipitates were first treated with PNGase F to remove N-linked glycans from the molecules. Immunoprecipitation with an anti-HA antibody recovered m144HA from m144HA-expressing cells (Fig. 11a, lane 4). Similarly, immunoprecipitation with antibody to U21 recovered U21 from U21-expressing cells (Fig. 11a, lane 5). In cells expressing both proteins, recovery of U21 did not coprecipitate m144HA, nor did the recovery of m144HA coprecipitate U21 (Fig. 11a, lanes 7 and 8), suggesting that U21 does not associate with m144 despite its structural similarity to class I MHC. Immunoblot analysis of crude membranes showed steady-state expression of U21 in U373 m144-expressing cells to be similar to that in U373-U21 cells (Fig. 11b, compare lanes 4 and 2).
FIG. 11.
U21 does not associate with m144HA. (a) U373 control cells (lanes 1 to 2), along with U373 cells expressing U21 and/or HA-tagged m144 (lanes 3 to 8) were labeled for 40 min with [35S]cysteine and [35S]methionine. U21 molecules were recovered with PAb MCW50 anti-U21 (lanes 1, 3, 5, and 7), and m144HA was recovered with MAb HA11 anti-HA (lanes 2, 4, 6, and 8). Immunoprecipitations were treated with PNGase F prior to electrophoresis. (b) Three micrograms of crude membrane preparations of U373 control cells or U373 cells expressing U21 and/or m144HA (labeled m144) were blotted with MCW50 antibody to U21.
Amino acid sequence comparison of U21-sensitive MHC proteins.
U21 is notable among immunoevasins in its ability to bind to all human HLAs. To survey the primary sequence variation among U21's substrates, we aligned their amino acid sequences. The HLAs are highly similar (Fig. 12a). The α3 domains, marked by dark gray lines over the sequences, display the greatest identity. Not surprisingly, H2-Kb is least similar among the class I MHC sequences, having 64% identity to HLA-A (Fig. 12b). Still more divergence is seen with CD1d; despite sharing the same MHC fold, CD1d has only 17% identity (28% similarity) with HLA-A (sequence alignment not shown). The greatest degree of conservation among all of the proteins exists within the α3 domain.
DISCUSSION
The ability of viruses to downregulate certain class I MHC gene products, while leaving others at the cell surface, has been well documented (10, 16, 25, 35, 46). For example, human immunodeficiency virus Nef downregulates HLA-A and -B gene products, but not HLA-C or -E gene products (10). HCMV US2 affects HLA-A and most -B alleles, but not HLA-C, -E, or -G, and HCMV US11 affects HLA-A, -B, and some -C, but not HLA-E or -G (16, 35, 46). The selectivity of these viral immunoevasins for particular class I MHC products seems straightforward: viruses strive to downmodulate class I MHC molecules that act as effective antigen-presenting molecules for CTLs, while leaving molecules that serve as inhibitory ligands for NK cells at the cell surface. There are exceptions to this rule: HCMV US6 prevents assembly of all TAP-dependent HLA molecules, including HLA-C and -E (28, 35), and HCMV US3 prevents surface expression of all tapasin-dependent class I products, which include HLA-C, -E, and -G (28). Nonetheless, it seems that HCMV goes to great lengths to ensure that NK cell-inhibitory HLA-E is expressed at the cell surface: the cleavable signal peptide from the UL40 gene product serves as a canonical ligand for HLA-E's peptide binding groove, enhancing cell surface expression of HLA-E in a TAP-independent manner (54, 57).
Here, we demonstrate that HHV-7 U21 is not selective in its association with class I molecules; it binds to and affects the cell surface expression of classical, nonclassical, and even murine class I MHC molecules, but not of the structurally similar CD1d or MCMV m144 molecules. In downregulating the inhibitory NK cell ligand HLA-E, U21-expressing cells would seem to expose themselves to NK cell killing. Other viruses that downmodulate HLA-E along with the other classical class I molecules have evolved compensation mechanisms for this apparent oversight: HCMV, for example, encodes UL40 to bypass US6's effect upon HLA-E. In addition, HCMV itself encodes class I MHC homologs (UL18, UL142) that can act as NK cell-inhibitory ligands (8, 61; for a review, see reference 60). HCMV also encodes a microRNA that represses the activating NK cell ligand MICB (38), as well as several other protein products (UL16, UL141) that downregulate still more activating NK cell receptor ligands (8, 55). Thus, we hypothesize that HHV-6 and -7, betaherpesviruses that are most closely related to HCMV, must also encode other mechanisms of NK cell evasion.
In a previous work, we demonstrated that U21, a type I membrane protein with a 50-amino-acid cytoplasmic tail, does not rely on its transmembrane domain or cytoplasmic tail to mediate the trafficking of class I MHC molecules to lysosomes (22). From experiments performed with chimeric U21 molecules, we demonstrated that the association of U21 with class I molecules occurs within their lumenal domains and is β2m or conformation dependent (22). We believe that U21 utilizes as-yet-unknown cytoplasmic cellular sorting machinery for diversion of class I molecules to lysosomes; thus, the observation that U21 can reroute H-2Kb molecules when it is expressed in murine cells implies that any unidentified cellular proteins necessary to reroute class I molecules are functional and conserved in mice.
HHV-6 and -7 exhibit exquisite species specificity and do not infect mice. Thus, while U21's ability to affect murine H-2Kb may not be physiologically meaningful, the discovery that U21 can affect murine H-2Kb sheds unexpected light on U21's association with its substrates. Human class I molecules assume a slightly different conformation when bound to murine β2m than when complexed with human β2m and are no longer recognized by the conformation-specific antibody W6/32 (14, 19). Similar conformational changes occur when murine H-2K associates with human β2m, yet regardless of the source of β2m, U21 retains its ability to bind to and reroute class I molecules, suggesting that U21's association with class I/β2m heterodimers is independent of this conformation change.
HLA-G, a nonclassical class I molecule expressed in chorionic trophoblasts, is thought to provide the fetus protection from maternal NK cells (12, 13). However, the majority of HHV-7 infections are presumably acquired through contact with the saliva of adults (5), and congenital infection with HHV-7 has not been demonstrated (18); thus, U21-mediated downregulation of HLA-G in trophoblasts is an unlikely scenario. Nonetheless, U21's ability to reroute HLA-G to the lysosomal compartment contributed to our understanding of U21's ability to associate with a wide range of class I molecules. U21 associates exclusively with properly folded, β2m-bound class I molecules (22). Together with the information we have gained with respect to its association with such a wide range of class I molecules, our data suggest that U21 may have conformation-dependent binding determinants for class I binding, rather than a primary sequence consensus.
HHV-7 is an exceptionally successful virus, shed in the saliva of almost all healthy adults. Its persistence and prevalence within the population suggest that it must constantly interact with the immune system. U21 downregulates all of the classical and nonclassical class I MHC molecules, including the NK cell-inhibitory ligands. If U21 expression results in the efficient removal of NK cell-inhibitory ligands from the cell surface of HHV-7-infected cells, then HHV-7 has likely evolved other novel means of NK cell evasion.
Acknowledgments
We thank Jenny Gumperz for reagents and helpful discussions, Ted Hansen, Patrizio Giacomini, Dom Tortorella, Caroline Kulesza, and Richard Vile for generously supplying reagents, and Lisa Kimpler and Robert Fritz for critical readings of the manuscript.
This work was supported by NIH grant 1R01AI069099 (to A.W.H.).
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Ahn, K., A. Gruhler, B. Galocha, T. R. Jones, E. J. Wiertz, H. L. Ploegh, P. A. Peterson, Y. Yang, and K. Fruh. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613-621. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, K., T. H. Meyer, S. Uebel, P. Sempe, H. Djaballah, Y. Yang, P. A. Peterson, K. Fruh, and R. Tampe. 1996. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J. 15:3247-3255. [PMC free article] [PubMed] [Google Scholar]
- 3.Asano, Y., S. Suga, T. Yoshikawa, T. Yazaki, and T. Uchikawa. 1995. Clinical features and viral excretion in an infant with primary human herpesvirus 7 infection. Pediatrics 95:187-190. [PubMed] [Google Scholar]
- 4.Braud, V., E. Y. Jones, and A. McMichael. 1997. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 27:1164-1169. [DOI] [PubMed] [Google Scholar]
- 5.Caserta, M. T., M. P. McDermott, S. Dewhurst, K. Schnabel, J. A. Carnahan, L. Gilbert, G. Lathan, G. K. Lofthus, and C. B. Hall. 2004. Human herpesvirus 6 (HHV6) DNA persistence and reactivation in healthy children. J. Pediatr. 145:478-484. [DOI] [PubMed] [Google Scholar]
- 6.Cerwenka, A., and L. L. Lanier. 2001. Natural killer cells, viruses and cancer. Nat. Rev. Immunol. 1:41-49. [DOI] [PubMed] [Google Scholar]
- 7.Cerwenka, A., and L. L. Lanier. 2003. NKG2D ligands: unconventional MHC class I-like molecules exploited by viruses and cancer. Tissue Antigens 61:335-343. [DOI] [PubMed] [Google Scholar]
- 8.Chalupny, N. J., A. Rein-Weston, S. Dosch, and D. Cosman. 2006. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem. Biophys. Res. Commun. 346:175-181. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, T. L., and P. J. Bjorkman. 1998. Characterization of a murine cytomegalovirus class I major histocompatibility complex (MHC) homolog: comparison to MHC molecules and to the human cytomegalovirus MHC homolog. J. Virol. 72:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, G. B., R. T. Gandhi, D. M. Davis, O. Mandelboim, B. K. Chen, J. L. Strominger, and D. Baltimore. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661-671. [DOI] [PubMed] [Google Scholar]
- 11.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. U. S. A. 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis, S. A., M. S. Palmer, and A. J. McMichael. 1990. Human trophoblast and the choriocarcinoma cell line BeWo express a truncated HLA class I molecule. J. Immunol. 144:731-735. [PubMed] [Google Scholar]
- 13.Ellis, S. A., I. L. Sargent, C. W. Redman, and A. J. McMichael. 1986. Evidence for a novel HLA antigen found on human extravillous trophoblast and a choriocarcinoma cell line. Immunology 59:595-601. [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrier, P., C. Layet, D. H. Caillol, B. R. Jordan, and F. A. Lemonnier. 1985. The association between murine beta 2-microglobulin and HLA class I heavy chains results in serologically detectable conformational changes of both chains. J. Immunol. 135:1281-1287. [PubMed] [Google Scholar]
- 15.Furman, M. H., H. L. Ploegh, and D. J. Schust. 2000. Can. viruses help us to understand and classify the MHC class I molecules at the maternal-fetal interface? Hum. Immunol. 61:1169-1176. [DOI] [PubMed] [Google Scholar]
- 16.Gewurz, B. E., E. W. Wang, D. Tortorella, D. J. Schust, and H. L. Ploegh. 2001. Human cytomegalovirus US2 endoplasmic reticulum-lumenal domain dictates association with major histocompatibility complex class I in a locus-specific manner. J. Virol. 75:5197-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glosson, N. L., and A. W. Hudson. 2007. Human herpesvirus-6A and -6B encode viral immunoevasins that downregulate class I MHC molecules. Virology 365:125-135. [DOI] [PubMed] [Google Scholar]
- 18.Hall, C. B., M. T. Caserta, K. C. Schnabel, C. Boettrich, M. P. McDermott, G. K. Lofthus, J. A. Carnahan, and S. Dewhurst. 2004. Congenital infections with human herpesvirus 6 (HHV6) and human herpesvirus 7 (HHV7). J. Pediatr. 145:472-477. [DOI] [PubMed] [Google Scholar]
- 19.Hebert, A. M., J. Strohmaier, M. C. Whitman, T. Chen, E. Gubina, D. M. Hill, M. S. Lewis, and S. Kozlowski. 2001. Kinetics and thermodynamics of beta 2-microglobulin binding to the alpha 3 domain of major histocompatibility complex class I heavy chain. Biochemistry 40:5233-5242. [DOI] [PubMed] [Google Scholar]
- 20.Hill, A., P. Jugovic, I. York, G. Russ, J. Bennink, J. Yewdell, H. L. Ploegh, and D. Johnson. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411-415. [DOI] [PubMed] [Google Scholar]
- 21.Hochman, J. H., Y. Shimizu, R. DeMars, and M. Edidin. 1988. Specific associations of fluorescent beta-2-microglobulin with cell surfaces. The affinity of different H-2 and HLA antigens for beta-2-microglobulin. J. Immunol. 140:2322-2329. [PubMed] [Google Scholar]
- 22.Hudson, A. W., D. Blom, P. M. Howley, and H. L. Ploegh. 2003. The ER-lumenal domain of the HHV-7 immunoevasin U21 directs class I MHC molecules to lysosomes. Traffic 4:824-837. [DOI] [PubMed] [Google Scholar]
- 23.Hudson, A. W., P. M. Howley, and H. L. Ploegh. 2001. A human herpesvirus 7 glycoprotein, U21, diverts major histocompatibility complex class I molecules to lysosomes. J. Virol. 75:12347-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson, A. W., and H. L. Ploegh. 2002. The cell biology of antigen presentation. Exp. Cell Res. 272:1-7. [DOI] [PubMed] [Google Scholar]
- 25.Ishido, S., C. Wang, B. S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, T. R., E. J. Wiertz, L. Sun, K. N. Fish, J. A. Nelson, and H. L. Ploegh. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. U. S. A. 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonjic, S., M. Babic, B. Polic, and A. Krmpotic. 2008. Immune evasion of natural killer cells by viruses. Curr. Opin. Immunol. 20:30-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jun, Y., E. Kim, M. Jin, H. C. Sung, H. Han, D. E. Geraghty, and K. Ahn. 2000. Human cytomegalovirus gene products US3 and US6 down-regulate trophoblast class I MHC molecules. J. Immunol. 164:805-811. [DOI] [PubMed] [Google Scholar]
- 29.Kavathas, P., F. H. Bach, and R. DeMars. 1980. Gamma ray-induced loss of expression of HLA and glyoxalase I alleles in lymphoblastoid cells. Proc. Natl. Acad. Sci. U. S. A. 77:4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kempf, W., V. Adams, P. Mirandola, L. Menotti, D. Di Luca, N. Wey, B. Muller, and G. Campadelli-Fiume. 1998. Persistence of human herpesvirus 7 in normal tissues detected by expression of a structural antigen. J. Infect. Dis. 178:841-845. [DOI] [PubMed] [Google Scholar]
- 31.Koch, M., V. S. Stronge, D. Shepherd, S. D. Gadola, B. Mathew, G. Ritter, A. R. Fersht, G. S. Besra, R. R. Schmidt, E. Y. Jones, and V. Cerundolo. 2005. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat. Immunol. 6:819-826. [DOI] [PubMed] [Google Scholar]
- 32.Kubota, A., S. Kubota, H. E. Farrell, N. Davis-Poynter, and F. Takei. 1999. Inhibition of NK cells by murine CMV-encoded class I MHC homologue m144. Cell. Immunol. 191:145-151. [DOI] [PubMed] [Google Scholar]
- 33.Lanier, L. L. 1998. Follow the leader: NK cell receptors for classical and nonclassical MHC class I. Cell 92:705-707. [DOI] [PubMed] [Google Scholar]
- 34.Lee, N., D. R. Goodlett, A. Ishitani, H. Marquardt, and D. E. Geraghty. 1998. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160:4951-4960. [PubMed] [Google Scholar]
- 35.Llano, M., M. Guma, M. Ortega, A. Angulo, and M. Lopez-Botet. 2003. Differential effects of US2, US6 and US11 human cytomegalovirus proteins on HLA class Ia and HLA-E expression: impact on target susceptibility to NK cell subsets. Eur. J. Immunol. 33:2744-2754. [DOI] [PubMed] [Google Scholar]
- 36.McCutcheon, J. A., J. Gumperz, K. D. Smith, C. T. Lutz, and P. Parham. 1995. Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA. J. Exp. Med. 181:2085-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mostoslavsky, G., A. J. Fabian, S. Rooney, F. W. Alt, and R. C. Mulligan. 2006. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc. Natl. Acad. Sci. U. S. A. 103:16406-16411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nachmani, D., N. Stern-Ginossar, R. Sarid, and O. Mandelboim. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376-385. [DOI] [PubMed] [Google Scholar]
- 39.Natarajan, K., A. Hicks, J. Mans, H. Robinson, R. Guan, R. A. Mariuzza, and D. H. Margulies. 2006. Crystal structure of the murine cytomegalovirus MHC-I homolog m144. J. Mol. Biol. 358:157-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neisig, A., C. J. Melief, and J. Neefjes. 1998. Reduced cell surface expression of HLA-C molecules correlates with restricted peptide binding and stable TAP interaction. J. Immunol. 160:171-179. [PubMed] [Google Scholar]
- 41.Raftery, M. J., M. Hitzler, F. Winau, T. Giese, B. Plachter, S. H. Kaufmann, and G. Schonrich. 2008. Inhibition of CD1 antigen presentation by human cytomegalovirus. J. Virol. 82:4308-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raftery, M. J., F. Winau, S. H. Kaufmann, U. E. Schaible, and G. Schonrich. 2006. CD1 antigen presentation by human dendritic cells as a target for herpes simplex virus immune evasion. J. Immunol. 177:6207-6214. [DOI] [PubMed] [Google Scholar]
- 43.Renukaradhya, G. J., M. A. Khan, D. Shaji, and R. R. Brutkiewicz. 2008. Vesicular stomatitis virus matrix protein impairs CD1d-mediated antigen presentation through activation of the p38 MAPK pathway. J. Virol. 82:12535-12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reusch, U., W. Muranyi, P. Lucin, H. G. Burgert, H. Hengel, and U. H. Koszinowski. 1999. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 18:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez, D. J., J. E. Gumperz, and D. Ganem. 2005. Regulation of CD1d expression and function by a herpesvirus infection. J. Clin. Invest. 115:1369-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schust, D. J., D. Tortorella, J. Seebach, C. Phan, and H. L. Ploegh. 1998. Trophoblast class I major histocompatibility complex (MHC) products are resistant to rapid degradation imposed by the human cytomegalovirus (HCMV) gene products US2 and US11. J. Exp. Med. 188:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sernee, M. F., H. L. Ploegh, and D. J. Schust. 1998. Why certain antibodies cross-react with HLA-A and HLA-G: epitope mapping of two common MHC class I reagents. Mol. Immunol. 35:177-188. [DOI] [PubMed] [Google Scholar]
- 48.Setini, A., A. Beretta, C. De Santis, R. Meneveri, A. Martayan, M. C. Mazzilli, E. Appella, A. G. Siccardi, P. G. Natali, and P. Giacomini. 1996. Distinctive features of the alpha 1-domain alpha helix of HLA-C heavy chains free of beta 2-microglobulin. Hum. Immunol. 46:69-81. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu, Y., and R. DeMars. 1989. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J. Immunol. 142:3320-3328. [PubMed] [Google Scholar]
- 50.Snary, D., C. J. Barnstable, W. F. Bodmer, and M. J. Crumpton. 1977. Molecular structure of human histocompatibility antigens: the HLA-C series. Eur. J. Immunol. 7:580-585. [DOI] [PubMed] [Google Scholar]
- 51.Stam, N. J., H. Spits, and H. L. Ploegh. 1986. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 137:2299-2306. [PubMed] [Google Scholar]
- 52.Storkus, W. J., D. N. Howell, R. D. Salter, J. R. Dawson, and P. Cresswell. 1987. NK susceptibility varies inversely with target cell class I HLA antigen expression. J. Immunol. 138:1657-1659. [PubMed] [Google Scholar]
- 53.Tanaka, K., T. Kondo, S. Torigoe, S. Okada, T. Mukai, and K. Yamanishi. 1994. Human herpesvirus 7: another causal agent for roseola (exanthem subitum). J. Pediatr. 125:1-5. [DOI] [PubMed] [Google Scholar]
- 54.Tomasec, P., V. M. Braud, C. Rickards, M. B. Powell, B. P. McSharry, S. Gadola, V. Cerundolo, L. K. Borysiewicz, A. J. McMichael, and G. W. Wilkinson. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287:1031. [DOI] [PubMed] [Google Scholar]
- 55.Tomasec, P., E. C. Wang, A. J. Davison, B. Vojtesek, M. Armstrong, C. Griffin, B. P. McSharry, R. J. Morris, S. Llewellyn-Lacey, C. Rickards, A. Nomoto, C. Sinzger, and G. W. Wilkinson. 2005. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 6:181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomazin, R., N. E. van Schoot, K. Goldsmith, P. Jugovic, P. Sempe, K. Fruh, and D. C. Johnson. 1998. Herpes simplex virus type 2 ICP47 inhibits human TAP but not mouse TAP. J. Virol. 72:2560-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ulbrecht, M., S. Martinozzi, M. Grzeschik, H. Hengel, J. W. Ellwart, M. Pla, and E. H. Weiss. 2000. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol. 164:5019-5022. [DOI] [PubMed] [Google Scholar]
- 58.Ward, K. N. 1998. Infections due to the herpesvirus group in immunocompromised patients. Curr. Opin. Infect. Dis. 11:425-430. [DOI] [PubMed] [Google Scholar]
- 59.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 60.Wilkinson, G. W., P. Tomasec, R. J. Stanton, M. Armstrong, V. Prod'homme, R. Aicheler, B. P. McSharry, C. R. Rickards, D. Cochrane, S. Llewellyn-Lacey, E. C. Wang, C. A. Griffin, and A. J. Davison. 2008. Modulation of natural killer cells by human cytomegalovirus. J. Clin. Virol. 41:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wills, M. R., O. Ashiru, M. B. Reeves, G. Okecha, J. Trowsdale, P. Tomasec, G. W. Wilkinson, J. Sinclair, and J. G. Sissons. 2005. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 175:7457-7465. [DOI] [PubMed] [Google Scholar]
- 62.Yadav, M., S. Nambiar, S. P. Khoo, and H. B. Yaacob. 1997. Detection of human herpesvirus 7 in salivary glands. Arch. Oral Biol. 42:559-567. [DOI] [PubMed] [Google Scholar]
- 63.Zemmour, J., A. M. Little, D. J. Schendel, and P. Parham. 1992. The HLA-A,B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J. Immunol. 148:1941-1948. [PubMed] [Google Scholar]
- 64.Ziegler, H., R. Thale, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U. H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6:57-66. [DOI] [PubMed] [Google Scholar]