Abstract
Interaction between pUL34 and pUL31 is essential for targeting both proteins to the inner nuclear membrane (INM). Sequences mediating the targeting interaction have been mapped by others with both proteins. We have previously reported identification of charge cluster mutants of herpes simplex virus type 1 UL34 that localize properly to the inner nuclear membrane, indicating interaction with UL31, but fail to complement a UL34 deletion. We have characterized one mutation (CL04) that alters a charge cluster near the N terminus of pUL34 and observed the following. (i) The CL04 mutant has a dominant-negative effect on pUL34 function, indicating disruption of some critical interaction. (ii) In infections with CL04 pUL34, capsids accumulate in close association with the INM, but no perinuclear enveloped viruses, cytoplasmic capsids, or virions or cell surface virions were observed, suggesting that CL04 UL34 does not support INM curvature around the capsid. (iii) Passage of UL34-null virus on a stable cell line that expresses CL04 resulted in selection of extragenic suppressor mutants that grew efficiently using the mutant pUL34. (iv) All extragenic suppressors contained an R229→L mutation in pUL31 that was sufficient to suppress the CL04 phenotype. (v) Immunolocalization and coimmunoprecipitation experiments with truncated forms of pUL34 and pUL31 confirm that N-terminal sequences of pUL34 and a C-terminal domain of pUL31 mediate interaction but not nuclear membrane targeting. pUL34 and pUL31 may make two essential interactions—one for the targeting of the complex to the nuclear envelope and another for nuclear membrane curvature around capsids.
Egress of herpesvirus capsids from the nucleus occurs by envelopment of capsids at the inner nuclear membrane (INM) and is followed by de-envelopment at the outer nuclear membrane (ONM). This process can be broken down into a pathway of discrete steps that begin with recruitment of the viral envelopment apparatus to the INM. Herpes simplex virus type 1 (HSV-1) UL34 and UL31 and their homologs in other herpesviruses are required for efficient envelopment at the INM (7, 13, 22, 23, 29). HSV-1 pUL31 and pUL34 are targeted specifically to the INM by a mechanism that requires their interaction with each other (27, 28), and this mutual dependence is a conserved feature of herpesvirus envelopment (9, 14, 27, 28, 32, 33, 39). Localization of these two proteins at the INM results in the recruitment of other proteins, including protein kinase C delta and pUS3, to the nuclear membrane (22, 24, 30). The sequences in HSV-1 pUL34 that mediate interaction with UL31 and that lead to nuclear envelope targeting were mapped to amino acids (aa) 137 to 181 (16). The sequences in the murine cytomegalovirus (MCMV) homolog of UL31, M53, that mediate the nuclear envelope targeting interaction with the UL34 homolog, M50, were mapped to the N-terminal third of the protein in the first of four conserved regions (17), and Schnee et al. subsequently showed that this same region of pUL31 homologs from other families of herpesviruses mediates interaction with the corresponding pUL34 homologs (33).
After the targeting of the pUL34/pUL31 complex to the INM, subsequent steps in nuclear egress include, it is thought, (i) local disruption of the nuclear lamina to allow capsid access to the INM, (ii) recognition and docking of capsids by the envelopment apparatus at the INM, (iii) curvature of the inner and outer nuclear membranes around the capsid, (iv) scission of the INM to create an enveloped virion in the space between the INM and ONM, (v) fusion of the virion envelope with the outer nuclear membrane, and (vi) capsid release into the cytoplasm.
At least some of the viral and cellular factors critical for nuclear lamina disruption and for de-envelopment fusion have been identified. pUL34, pUL31, and pUS3 of HSV-1 have all been implicated in changes in localization, interaction, and phosphorylation of nuclear lamina components, including lamins A/C and B and the lamina-associated protein, emerin (3, 15, 19, 20, 24, 26, 34, 35). pUS3, pUL31, and glycoproteins B and H have been implicated in de-envelopment of primary virions at the ONM (8, 21, 28, 30, 38).
pUL34 and pUL31 are thought to be involved in steps between lamina disruption and de-envelopment, but genetic evidence in infected cells has so far been lacking. Klupp et al. have shown that overexpression of alphaherpesvirus pUL31 and pUL34 in the absence of other viral proteins can induce formation of small vesicles derived from the INM, suggesting a role for these two proteins in membrane curvature around the capsid (12). Tight membrane curvature is an energetically unfavorable event and is thought to be accomplished by coupling curvature to energetically favorable interactions between membrane-bound proteins or protein complexes (reviewed in reference 40). The data of Klupp et al. suggest the possibility that upon recognition of a capsid, pUL31 and pUL34 may interact in a way that induces tight curvature of the INM. Here we present data in support of this hypothesis, showing that a specific point mutation in UL34 induces accumulation of docked capsids at the INM, extragenic suppression of the mutant phenotype is associated with a mutation in UL31, and pUL31 and pUL34 can interact via sequences that are not involved in their INM targeting interaction.
We previously published a characterization of a library of 19 charge cluster mutants of pUL34. In each of these mutants, one charge cluster (defined as a group of five consecutive amino acids in which two or more of the residues have charged side chains) was mutated such that the charged residues were replaced by alanine. Six of the 19 charge cluster mutants tested failed to complement replication of UL34-null virus, indicating that they disrupt essential functions of pUL34. Interestingly, five of the six noncomplementing mutants were synthesized at levels comparable to that of wild-type UL34 and localized normally to the nuclear envelope, suggesting that they were unimpaired in their ability to make a nuclear envelope targeting interaction with UL31. In order to identify essential functions of pUL34 downstream of nuclear envelope targeting, we have undertaken a detailed study of the behavior and interactions of these mutants.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 and Vero cells were maintained as previously described (29). The properties of HSV-1(F) and vRR1072 (TK+), referred to here as UL34-null virus, have been described previously (6, 29).
Plasmids and cell lines.
pRR1072, pRR1072Rep, and pRR1162, which contains the CL04 charge cluster mutation in UL34 on the pRR1072Rep background, were previously described (2, 29).
pRR1340, used for construction of an infection-inducible CL04-expressing cell line, was constructed by ligation of the 1,820-bp XbaI-PmlI fragment of pRR1162 that contains the CL04 UL34 gene between the XbaI and NruI sites of the pcDNA3 vector. An otherwise identical plasmid for expression of wild-type (wt) pUL34 was constructed using pRR1072Rep as the source of the insert. Clonal cell lines RepAC (expressing wt pUL34) and CL04AI (expressing CL04) were constructed by transfection of the corresponding plasmid into Vero cells, selection with G418, and isolation of clones by limiting dilution. Expressing cell clones were identified by an immunofluorescence (IF) assay for pUL34 expression 20 h after infection with the UL34-null virus vRR1072(TK+).
pRR1238 for constitutive expression of wt pUL34 was constructed by digestion of pRR1072Rep with NcoI and BspEI, treatment with Klenow enzyme in the presence of deoxynucleoside triphosphates (dNTPs) to create blunt ends, and ligation of the UL34-containing fragment into EcoRV-digested pcDNA3. pRR1328 for constitutive expression of CL04 pUL34 was constructed by InFusion (Clontech) cloning of a PCR fragment containing the CL04 pUL34 coding sequence amplified from pRR1162 between the HindIII and XhoI sites of pcDNA3. PCR primers used for amplification of the CL04 pUL34 coding sequence were 5′-AGGGAGACCCAAGCTCCATGGCGGGACTGGGCAAG-3′ and 5′-TAGATGCATGCTCGATTATAGGCGCGCGCCAGCAC-3′.
pRR1330, in which the coding sequences for amino acids 91 to 228 are deleted from the UL34 gene (Fig. 1, line 3), was constructed by digestion of pRR1238 with SgrAI and HpaI, treatment with Klenow enzyme in the presence of dNTPs to create blunt ends, and religation of the large fragment of the plasmid. pRR1331, in which the same coding sequences are deleted from the CL04 gene (Fig. 1, line 4), was created in the same way as pRR1330, except that the parent plasmid was pRR1328.
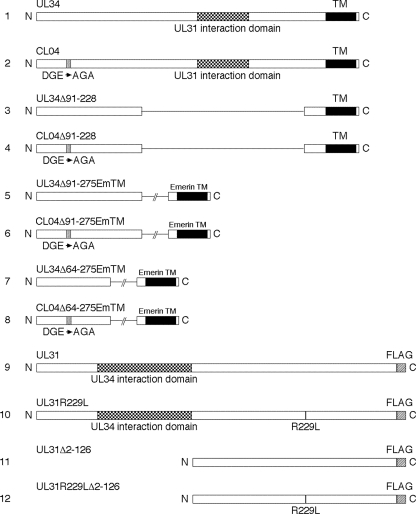
FIG. 1.
Wild-type and mutant UL31 and UL34 pcDNA3 expression constructs. Schematic drawings of expression constructs are shown. All constructs were expressed from the human cytomegalovirus immediate-early promoter in pcDNA3 and were constructed as described in Materials and Methods. The plasmids encoding each gene are the following: line 1, ppRRR1238; line 2, pRR1328; line 3, pRR1330; line 4, pRR1331; line 5, pRR1344; line 6, pRR1345; line 7, pRR1346; line 8, pRR1347; line 9, pRR1334; line 10; pRR1335; line 11, pRR1327; line 12, pRR1328.
pRR1344, in which amino acids 91 to 275 of wt pUL34 expressed from pcDNA3 are deleted and replaced by the emerin transmembrane domain (Fig. 1, line 5), was constructed by PCR amplification of sequences coding for amino acids 219 to 254 of emerin and ligation of the BspEI- and XbaI-digested PCR product between the SgrAI and XbaI sites of pRR1238. The primers used for amplification of the emerin TM sequence were 5′-GATCTCCGGACAGGATCGCCAGGTCCCGC-3′ and 5′-TAGCTCTAGACTAGAAGGGGTTGCCTTCTTCAGCCTG-3′. pRR1345, which is the equivalent construct for CL04 UL34 (Fig. 1, line 6), was made in the same way, except that the parent plasmid was pRR1328.
pRR1346, in which amino acids 64 to 275 of wt pUL34 expressed from pcDNA3 are deleted and replaced by the emerin transmembrane domain (Fig. 1, line 7) was made by insertion of an overlap PCR product into pcDNA3. For the first step in insert construction, sequences encoding amino acids 1 to 63 of UL34 fused to 19 nucleotides (nt) of the emerin sequence were amplified using pRR1238 as the template and the primers HSV1-63LEFT (5′-AGCTCTCGAGATGGCGGGACTGGGCAAGCCC-3′) and HSV1-63RIGHT (5′ GCGGGACCTGGCGATCCTGAAACGACTCGTCGGACCCGTCATG-3′). Next, sequences encoding amino acids 219 to 254 of emerin were amplified using the primers EmTMLEFT (5′-CAGGATCGCCAGGTCCCGC-3′) and EmTMRIGHT (5′-GGCCTCTAGACTAGAAGGGGTTGCCTTCTTCAGCC-3′). The resulting PCR products were gel purified and then combined as the template for a PCR using HSV1-63LEFT and EmTMRIGHT as primers. The resulting PCR product was digested with XbaI and XhoI and cloned between the XbaI and XhoI sites of pcDNA3. pRR1347, which is equivalent to pRR1346 except for the presence of the CL04 mutation in UL34 sequences (Fig. 1, line 8), was constructed in the same way, except that the template for amplification of UL34 sequences was pRR1328.
Plasmids for marker transfer of tagged UL31 wild-type or mutant sequences to the viral genome were constructed in several steps. First, a parent plasmid (pRR1336) containing UL31 wild-type sequences and flanking sequences from the UL30 and UL32 genes was constructed by reaction of pRR1060 (29) with BstEII and HindIII and with Klenow enzyme and dNTPs and by religation of the 8.64-kb fragment containing the vector, UL30, UL31, and UL32 sequences. Next, pRR1337, in which the 3′ end of the UL31 coding sequence was modified to include tandem FLAG epitope 8×His tags was made by insertion of an overlap PCR product into pRR1336. For the first step in insert construction, sequences between the AvrII site in UL31 and the 3′ end of the UL31 coding sequence and containing the 5′ part of the tandem tag were amplified using pRR1336 as the template and the primers TagUL31#1 (5′-CCGGGATGGGCTACTACCTAGGC-3′) and TagUL31#2 (5′ GTGATGGTGATGGTGCGCTGCTTTGTCGTCGTCCTTATAGTCCGGCGGAGGAAACTCGTCGAATGTTG-3′). For the second step in insert construction, sequences encoding the 3′ part of the tandem tag and sequences between the 3′ end of UL31 and the PstI site in UL30 were amplified using the primers TagUL31#3 (5′-GACGACGACAAAGCAGCGCACCATCACCATCACCATCACCATTAGTCATGCTAGAGTATCAAAG GCTCTATGCAACATTCG-3′) and TagUL31#4 (5′-CCTGCCCGAGGGACTGCAG-3′). The resulting overlapping PCR products were gel purified and then combined as the template for a PCR using the TagUL31#1 and TagUL31#4 primers. The resulting PCR product was digested with AvrII and PstI and cloned between the AvrII and PstI sites of pRR1336. pRR1338, which encodes the UL31 R229L mutation in the context of pRR1337, was made by insertion of an overlap PCR product into pRR1337. For the first step in insert construction, sequences between the AvrII site in UL31 and the site of the R229L mutation were amplified using pRR1337 as the template and the primers TagUL31#1 and UL31R229L#1 (5′-GCATCCGGTCTATtaataaGTAGTGCAGGTGGGCGGACG-3′) (lowercase indicates nucleotides altered to create the R229L mutation and an AseI restriction enzyme cleavage site). For the second step in insert construction, sequences from the site of the R229L mutation in UL31 to the PstI site in UL30 were amplified using the primers UL31R229L#2 (5′-CACCTGCACTACttattaATAGACCGGATGCTCACCGCGTG-3′) and TagUL31#4. The design of the primers created an overlap between the two PCR products that included the mutated sequences. The engineered mutated sequences included an AseI restriction site introduced at the site of the R229L mutation. The resulting overlapping PCR products were gel purified and then combined as the template for a PCR using the TagUL31#1 and TagUL31#4 primers. The resulting overlap PCR product was digested with AvrII and PstI and cloned between the AvrII and PstI sites of pRR1337.
pRR1334 and pRR1335, which contain the coding sequences for wt and R229L mutant tagged UL31, respectively, in pcDNA3 (Fig. 1, lines 9 and 10), were constructed by PCR amplification of the tagged UL31 coding sequences from pRR1337 and pRR1338 and cloning between the XhoI and XbaI sites of pcDNA3. pRR1327 and pRR1328, which code for tagged UL31 wt and R229L mutant amino acids 127 to 306, respectively (Fig. 1, lines 11 and 12), were constructed by PCR amplification of the corresponding sequences from pRR1337 and pRR1338 using the primers 5′-GATCCTCGAGATGGCGGGAGACGGGCG-3′ and 5′-GTACTCTAGACTAATGGTGATGGTGATGGTGATGGTGC-3′. PCR products were digested with XhoI and XbaI and cloned into the XhoI and XbaI sites of pcDNA3.
For complementation assays using combinations of wt and CL04 mutant UL34 and wt and R229L mutant UL31, plasmids were constructed that carry both the UL34 and UL31 genes, each driven by its own promoter/regulatory sequences. pRR1348, which carries both wt UL34 and wt, tagged UL31, was constructed by ligation of the 2.76-kb BstBI-PvuII fragment of pRR1337 into pRR1072Rep that had been cut with BstBI and PmlI. pRR1349, which carries wt UL34 and mutant, tagged UL34 was constructed in the same way, except that the UL31-containing insert was derived from pRR1338. pRR1350 and pRR1351, which carry CL04 UL34 and wt or mutant tagged UL31, respectively, were constructed in the same way as pRR1348 and pRR1349, except that pRR1162 was used as the parent plasmid.
Single-step growth measurement.
Measurement of replication of HSV-1(F), vRR1072(TK+), and CL04Rev viruses on Vero, RepAC, and CL04AI cells after infection at high multiplicity was performed as previously described (29).
Complementation assays.
Twelve-well cultures of Vero cells containing 400,000 cells were transfected with a total of 650 ng of plasmid mixtures using 5 μl of Lipofectamine according to the manufacturer's instructions. Complementation assays were performed on transfected cultures as previously described (2).
Marker transfer assays.
Genomes of UL34-null virus vRR1072(TK+) for marker transfer transfection were prepared in the form of a detergent-extracted, sonicated nuclear lysate from infected cells. A T25 culture of Vero cells was infected for 24 h with 5 PFU/cell of vRR1072(TK+), washed once with phosphate-buffered saline (PBS), and scraped into 1 ml of PBS. Cells were pelleted at 6,000 rpm in a microcentrifuge, resuspended in 1 ml PBS containing 0.2% NP-40, and placed on ice for 1 min. Nuclei were pelleted at 6,000 rpm for 2 min and then washed twice by resuspension in PBS without detergent and centrifugation at 6,000 rpm in the microcentrifuge. The pellet of washed nuclei was resuspended in 100 μl of PBS, lysed by one freeze-thaw cycle, and sonicated. Debris was pelleted by centrifugation at 14,000 rpm in the microcentrifuge for 2 min, and 2 μl of the supernatant was used for each transfection experiment.
For mapping of the extragenic suppressor, gel-purified restriction fragments of the CL04Rev genome (200 ng) were cotransfected with sonicated nuclear lysate from cells infected with vRR1072(TK+) into 12-well cultures, each containing 400,000 RepAC cells using 5 μl of Lipofectamine according to the manufacturer's protocol. Four days after transfection, when cytopathic effect was observed with most cells, virus stock was prepared and the titer was determined by plaque assay on RepAC cells. One thousand PFU (as measured on RepAC) cells were then plated onto confluent monolayers of CL04AI cells in 100-mm dishes. Four days after infection, cell monolayers were fixed with methanol for 10 min and then air dried. Fixed monolayers were stained with a solution of 0.04% amido black in 40% methanol and 10% acetic acid in water and then destained with water. Wild-type and recombinant R229L mutant UL31 sequences were tested in the same way, except that 200 ng of either PstI-linearized pRR1337 or PstI-linearized pRR1338 was used instead of genome restriction fragments.
Indirect IF.
IF was performed as previously described, with some variations (3, 27). Cells were fixed with 4% formaldehyde for 20 min and then washed with PBS. Cells were permeabilized and blocked in the same step by incubating in 10% Blokhen (Aves Labs) in IF buffer. Primary antibodies were diluted as follows in IF buffer: chicken anti-UL34 (1:1,000) and mouse monoclonal IgG anti-FLAG (1:1,000) (Sigma). Secondary antibodies were also diluted in IF buffer as follows: Alexa Fluor 594 goat anti-chicken IgG (1:1,000) was used to detect pUL34, and Alexa Fluor 488 goat anti-mouse IgG (1:400) was used to detect FLAG-UL31 fusions. Slo-fade II (Molecular Probes) was used to mount coverslips on glass slides. All confocal microscopy work was done with a Zeiss 510 confocal microscope. All images shown are representative of experiments performed a minimum of three times.
Immunoblotting.
Nitrocellulose sheets bearing proteins of interest were blocked in 5% nonfat milk plus 0.2% Tween 20 for at least 2 h. The membranes were probed with a previously described chicken polyclonal antibody directed against pUL34 (1:1,000) (27) and subjected to reaction with alkaline phosphatase-conjugated anti-chicken secondary antibody (Aves Laboratories), with mouse monoclonal antibody directed against the HSV-1 scaffolding protein (1:2,000) (Serotec) and reacted with alkaline phosphatase-conjugated anti-mouse secondary antibody (Sigma), or with anti-FLAG M2 mouse monoclonal antibody (Sigma) and reacted with alkaline-phosphatase-conjugated anti-mouse.
Transmission electron microscopy (EM) (TEM) of infected cells.
Confluent monolayers of Vero or CL04AI cells were infected with vRRR1072(TK+) at a multiplicity of infection (MOI) of 10 for 20 h and then fixed by incubation in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 2 h. Cells were postfixed in 1% osmium tetroxide, washed in cacodylate buffer, embedded in Spurr's resin, and cut into 95-nm sections. Sections were mounted on grids, stained with uranyl acetate and lead citrate, and examined with a JEOL 1250 transmission electron microscope.
BAC construction.
An HSV-1 bacterial artificial chromosome (BAC) genome carrying a UL34 deletion and a mutation in UL31 creating an R229L substitution was engineered in two steps using Red recombineering as described in reference 37. In the first step, the UL34 gene was disrupted by insertion of a kanamycin resistance (Kanr) cassette. This construction deletes exactly the same amino acids deleted in vRR1072(TK+). In the second step, the resulting UL34-null virus was mutagenized at the UL31 locus by insertion and scarless excision of a gentamicin resistance (Gmr) cassette.
The Kanr construct for insertion into the UL34 locus was generated by PCR from a pEPKan-S2 template using the primers KanUL34 Fwd and KanUL34 Rev (Table 1). The resulting PCR product was recombined into the HSV-1 BAC pYE-BAC102 (kind gift of Y. Kawaguchi [36]) as described previously (37). Kan-resistant recombinants were picked, and genomes were tested for insertion of the Kanr cassette by diagnostic PCR using the flanking primers UL34 test Fwd and UL34 test Rev (Table 1). Correct insertion of the Kanr cassette was confirmed by direct sequencing of the BAC DNA.
TABLE 1.
Primer used for construction and analysis of mutant BACs
| Primer name | Primer sequence |
|---|---|
| KanUL34 Fwda | 5′-CTTTGGTGGGTTTACGCGGGCACGCACGCTCCCATCGCGGGCGCCCGATTTATTCAACAAAGCCACGTTGTGTCTC-3′ |
| KanUL34 Reva | 5′-AGACCCCGCAGGGCCTGGTGCCACGGGCGGGAGGGCCCTTGGGTTGCCAGTGTTACAACCAATTAACCAATTCTGATTAG-3′ |
| UL34 test Fwd | 5′-CAGCGAACTTTACGGGACACAATCC-3′ |
| UL34 test Rev | 5′-ATAGGCTGCGGGGTAAACGTGT-3′ |
| Sce-GmR Forb | 5′-GACATGGATCCTAGGGATAACAGGGTAATAATTGACATAAGCCTGTTCGGTTCG-3′ |
| Sce-GmR Revb | 5′-GTAGCTCTAGAGGCCGCGGCGTTGTGAC-3′ |
| R229L Gm Fwdc | 5′- GTACGTCATCTTTCCCGGCACGTCCGCCCACCTGCACTACttatTaATAGACCGGATGCTCACCGCGCTCGGATCCTAGGGATAACAGGG-3′ |
| R229L Gm Revc | 5′-CGAACCGGTACCCCGGGCACGCGGTGAGCATCCGGTCTATtAataaGTAGTGCAGGTGGGCGGACGTCTAGAGGCCGCGGCGTTG-3′ |
| Gm mid For | 5′-GGTCGTGAGTTCGGAGACGTAGC-3′ |
| Gm mid Rev | 5′-CACTACGCGGCTGCTCAAACC-3′ |
| UL31 unique For | 5′-GTACGTCATCTTTCCCGGCACG-3′ |
| UL31 unique Rev | 5′-CGAACCGGTACCCCGGGCAC-3′ |
| UL31 test Fwd | 5′-TGCCCCTGGTGAAGACCAC-3′ |
| UL31 test Rev | 5′-GCTACGGCGGAGGAAACTCG-3′ |
Underlined sequences have homology to the Kanr cassette. Sequences not underlined have homology to the UL34 gene.
Underlined sequences have homology to the Gmr cassette.
Underlined sequences have homology to the UL31 gene. Lowercase indicates nucleotides altered to create the R229L mutation and an AseI restriction enzyme cleavage site. Sequences not underlined have homology to the Sce-Gmr cassette.
The Gm resistance cassette with the mutant UL31 flanking sequence was constructed in several steps. First, a Gm resistance cassette containing the Gmr promoter, protein coding sequence, and terminator flanked at the 5′ end with an SceI homing nuclease site was amplified from a pFastBac-1 template (Invitrogen) using primers Sce-GmR For and Sce-GmR Rev (Table 1). Second, PCR products containing the 5′ and 3′ halves of the Gm resistance gene were amplified from the Gmr cassette template using the primers R229L-Gm For and Gm mid Rev and the primers Gm mid For and R229L-Gm Rev, respectively. The two resulting PCR products overlap in the Gm coding sequence. The complete Gm resistance cassette with the UL31 flanking sequence was then assembled in a PCR using the overlapping partial genes and the primers UL31 unique For and UL31 unique Rev (Table 1). The resulting PCR product was recombined into the UL34-null BAC, Gm-resistant recombinants were picked, and genomes were tested for insertion of the Gm cassette by diagnostic PCR using the flanking primers UL31 test Fwd and UL31 test Rev (Table 1). Correct insertion of the Gm cassette was confirmed by direct sequencing of the BAC DNA. Scarless excision of the Gm cassette, leaving an intact UL31 gene carrying the R229L mutation, was carried out as described, and Gm-sensitive, Kan-resistant clones were tested for correct structure at both the UL31 and UL34 loci by diagnostic PCR using the UL34 test For, UL34 test Rev, UL31test For, and UL31test Rev primers. Correct structure was confirmed by direct sequencing of the BAC DNA at both loci.
Viruses were rescued from the wt BAC, UL34-null BAC, and UL31 R229L mutant BAC by transfection into UL34-expressing complementing cells.
Coimmunoprecipitation.
293T cells in 6-well culture were transfected with plasmids encoding portions of UL34 and UL31 using Lipofectamine 2000 according to the manufacturer's instructions. Two days after transfection, transfected monolayers were washed once with PBS, and then cells were lysed by addition of 0.5 ml immunoprecipitation (IP) buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) per well and sonicated for 10 s with a Fisher Sonic dismembrator at a power level of 2. Insoluble material was pelleted at top speed in a microcentrifuge for 2 min, and the supernatant was transferred to a fresh tube. Ten percent of each sample was removed and stored frozen as the input sample, and the remainder was immunoprecipitated using anti-FLAG M2 resin (Sigma) according to the manufacturer's instructions, using an overnight binding step and elution with 3×FLAG peptide. Immunoprecipitated samples were separated on 15% SDS-PAGE gels, blotted to nitrocellulose, and probed with antibodies directed against either FLAG or UL34.
RESULTS
CL04 mutant UL34 has a dominant-negative effect on wild-type protein function.
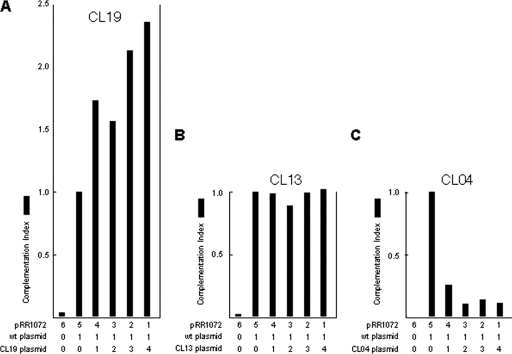
We previously described construction and characterization of a library of charge cluster mutants of UL34 (2). Initial attempts to study mutant UL34 function focused on construction of recombinant viruses expressing a mutant pUL34 in place of wt pUL34. Such a recombinant virus could be isolated for only one of the nonfunctional charge cluster mutants (CL13). For most of the UL34 charge cluster mutants, attempts were unsuccessful due to the inability to amplify the recombinant viruses on UL34-complementing cells. This failure during amplification suggested that the mutant pUL34 expressed from the viral genome might interfere with the function of the wt pUL34 expressed from the complementing cell genome. To test this hypothesis for the CL04 mutant, replicate cultures of Vero cells were transfected with a constant amount of wt pUL34-expressing plasmid pRR1072Rep and with increasing amounts of test plasmids that expressed a fully functional charge cluster mutant (CL19), the charge cluster mutant for which recombinant virus isolation was possible (CL13), or the putative dominant-negative mutant (CL04). Cultures were also transfected with pRR1072, which carries the enhanced green fluorescent protein (EGFP) gene in place of UL34 in the 1072Rep background, so that the total amount of UL34 promoter-containing plasmid was constant in each culture. Transfected cultures were infected with the UL34-null virus, and production of infectivity 20 h after infection was measured by a plaque assay using complementing cells. As shown in Fig. 2, expression of increasing amounts of functional pUL34 (CL19) along with wt pUL34 results in increasingly efficient complementation of the UL34-null virus (Fig. 2A). Expression of increasing amounts of nonfunctional but apparently noninterfering mutant CL13 does not interfere with the ability of wt pUL34 to complement the UL34-null virus (Fig. 2B). Increasing expression of the CL04 mutant, however, results in a dose-dependent decrease in the ability of wt pUL34 to complement, suggesting that the CL04 mutation exhibits a dominant-negative phenotype (Fig. 2C).
FIG. 2.
CL04 is a dominant-negative mutant of UL34. Graphs of complementation indices for combinations of cotransfected plasmids are shown. The complementation index for each condition was calculated by dividing the infectivity produced in each culture by the infectivity produced when the wt UL34-expressing plasmid is transfected in the absence of mutant UL34. The numbers below each bar indicate the number of ng × 100 of the corresponding plasmid transfected into the culture. In addition to the transfected plasmids indicated below each graph, all cultures were transfected with pCMVβ and assayed for β-galactosidase activity to control for differences in transfection efficiency. Transfection efficiencies were similar (within 30%) in all samples. One representative of three independent experiments is shown.
UL34 expression by stable cell lines.
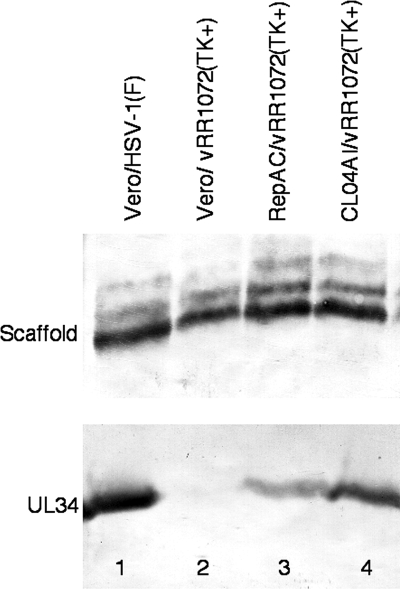
To better study CL04 function during infection, clonal cell lines expressing either wt pUL34 or CL04 pUL34 under the control of its own promoter-regulatory sequences were constructed by stable transfection of Vero cells. Two stable lines called RepAC and CL04AI, expressing wt and CL04 UL34, respectively, were isolated. To compare their pUL34 expression relative to each other and to wild-type virus infection, Vero cells were infected with 10 PFU/cell HSV-1(F), and Vero, wtUL34-expressing, and CL04 UL34-expressing cells were infected with 10 PFU/cell UL34-null virus for 18 h. Total protein was determined for each extract, and equivalent amounts were separated on an SDS-PAGE gel, transferred to nitrocellulose, and probed for pUL34. HSV-1 scaffolding protein was used as a loading control (Fig. 3). Neither of the clonal cell lines infected with UL34-null virus expressed as much pUL34 as wt virus-infected cells, but CL04 pUL34 expression was higher than that of wt pUL34 expression. This ensures that any putative CL04 phenotype observed in comparison of these two cell lines could not be ascribed to lower levels of CL04 protein expression.
FIG. 3.
Expression of wild-type and CL04 mutant UL34 by stable cell lines. Digital images of Western blots are shown. Vero cells (lanes 1 and 2) or cells stably expressing wt UL34 (lane 3) or CL04 UL34 (lane 4) were infected with wt HSV-1(F) (lane 1) or UL34-null vRRR1072(TK+) virus (lanes 2 to 4). Blotted infected cell proteins were probed for either scaffolding protein (top) or UL34 (bottom).
Characterization of the CL04 mutation effect on virus growth.
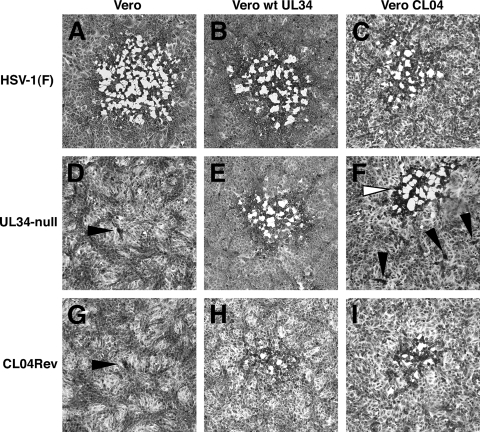
To test the ability of CL04 pUL34-expressing cells to support plaque formation, serial dilutions of wt HSV-1(F) and UL34-null virus were plated on Vero cells, wt pUL34-expressing cells, and CL04 pUL34-expressing cells. Plates were examined 48 h after infection, and digital images of representative plaques are shown in Fig. 4. HSV-1(F) forms robust plaques with similar efficiencies on all three of the cell lines tested (Fig. 4A to C). In contrast, UL34-null virus formed only tiny foci of one or a few infected cells on noncomplementing Vero cells (Fig. 4D, black arrowhead) but formed large plaques on cells expressing wt pUL34 (Fig. 4E). UL34-null virus formed slightly smaller plaques on wild-type UL34-expressing cells than on wild type virus (compare Fig. 4B and E), probably due to lower expression of pUL34 from the cell line genome than from the viral genome. UL34-null virus formed two populations of plaques on CL04-expressing cells. At dilutions for which plaques were easily visible on wt UL34-expressing cells, only small aggregations of a few infected cells could be seen on CL04-expressing cells (examples are indicated with black arrowheads in Fig. 4F). However, at high virus inputs (10,000 PFU/well), a few robust plaques developed on CL04-expressing cells (an example is indicated with the white arrowhead in Fig. 4F), suggesting the existence of a small subpopulation of the UL34-null virus that could replicate using CL04 mutant pUL34. After infection of CL04-expressing cells with UL34-null virus, robust plaques were purified by three rounds of plaque picking and amplifying. One of the amplified viruses, called CL04Rev, was selected for further analysis. Like its parent, vRR1072(TK+), CL04Rev could form only tiny foci of infection on noncomplementing Vero cells (Fig. 4G, black arrowhead) but efficiently formed large plaques on both wild type- and CL04-expressing cells (Fig. 4H and I), indicating that the UL34 deficiency in this virus could be complemented by either wt or CL04 mutant UL34.
FIG. 4.
Formation of plaques on wt UL34 or CL04 mutant UL34-expressing cell lines. Digital micrographs of infected cell monolayers stained with amido black are shown. The cell line infected is indicated above each column of panels, and the infecting virus is indicated to the left of each row. Black arrowheads indicate minute plaques comprised of a few cells. The white arrowhead (F) indicates a robust plaque formed by an extragenic suppressor virus.
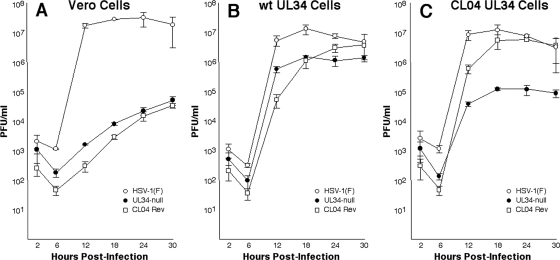
The single-step growth kinetics of HSV-1(F), vRR1072(TK+), and CL04Rev were measured with Vero, wild-type UL34-expressing RepAC, and mutant UL34-expressing CL04AI cells (Fig. 5). As shown previously, the UL34-null virus vRR1072(TK+) replicates poorly on Vero cells (29). CL04Rev replicates no better than vRR1072(TK+), indicating that it is not different from its parent in ability to replicate in the absence of pUL34. All three viruses replicate efficiently on RepAC cells, indicating that both UL34 null and CL04Rev viruses can be complemented by wild-type pUL34. Both UL34-null viruses produce about 10-fold less virus than wt HSV-1(F), however, probably because of the low level of pUL34 expressed by the complementing cells. CL04Rev replicates efficiently on CL04-expressing cells, achieving a peak titer similar to that attained by wt virus, whereas vRR1072(TK+) achieves a peak titer about 100-fold less than that of wt virus. This result indicates that CL04Rev can be efficiently complemented by CL04 UL34, while its parent cannot.
FIG. 5.
Single-step growth of wild-type and mutant viruses on complementing and noncomplementing cells. Replicate cultures of Vero (A), RepAC (B), or CL04AI cells (C) were infected at an MOI of 5 with HSV-1(F) (open circles), vRR1072(TK+) (closed circles), or CL04Rev (open squares). Residual virus was removed or inactivated with a low-pH wash, and at the indicated times, total culture virus was titrated on RepAC cells. Virus yields are expressed as the number of PFU per milliliter. Each data point represents the mean of three independent experiments. Error bars indicate the range of values.
An extragenic suppressor of the CL04 mutation maps to UL31.
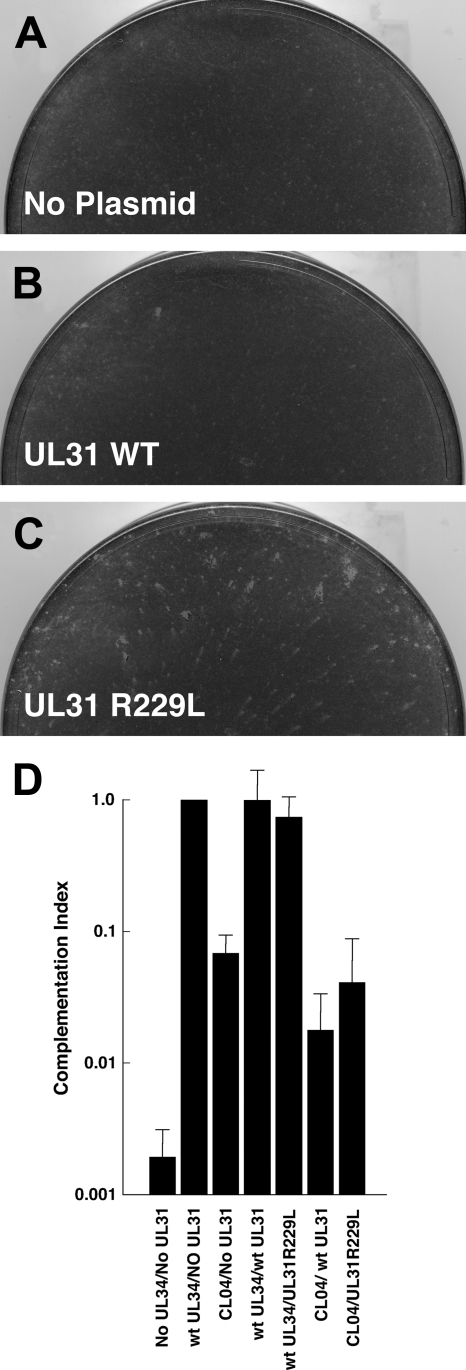
Marker transfer experiments were performed by cotransfection of vRR1072(TK+) nuclear lysate and gel-purified restriction fragments from the CL04Rev genome into RepAC cells. Viral progeny of cotransfection were plated on CL04-expressing cells and scored for formation of robust plaques. Only the 40.9-kb HindIII fragment, containing all of the UL20 through UL36 genes and a portion of the UL19 gene, reproducibly transferred the robust plaque phenotype. Genes having reported connections to DNA packaging or nuclear egress (UL19, UL20, UL25, UL26, UL28, UL31, UL33, UL35, and UL36) were then sequenced from the parental vRR1072(TK+) strain and from CL04Rev. Only UL31 showed a sequence difference compared to the parental virus—a single nucleotide substitution, changing the arginine at position 229 to leucine. Revertant viruses were isolated in six independent selections and purifications. All six carried the same amino acid substitution in UL31, suggesting that the R229L mutation was necessary for the revertant phenotype. Two approaches were used to determine whether the R229L mutation in UL31 might be sufficient for the revertant phenotype. For the first approach, marker transfer was performed by transfecting cloned plasmids containing either the wt UL31 sequence or containing an engineered R229L mutation and genomes of the UL34-null virus vRR1072(TK+) onto cells that express wt UL34. The viral progeny recovered were then tested by plating at low multiplicity on CL04-expressing cells and looking for the ability to form robust plaques. Only transfection with the mutant UL31 resulted in formation of robust plaques on CL04-expressing cells (compare Fig. 6C with A and B), suggesting that the UL31 R229L mutation was sufficient to suppress the CL04 phenotype. To confirm this result, a second approach using transient complementation of UL34-null virus was used. Replicate cultures of Vero cells were transfected with one of six plasmids that express either wt pUL34 alone, CL04 pUL34 alone, both wt pUL34 and wt pUL31, both wt pUL34 and pUL31R229L, both CL04 pUL34 and wt pUL31, or both CL04 pUL34 and pUL31R229L. One day after transfection, cultures were infected with UL34-null virus for 18 h, and viral infectivity was measured by plaque assay on wt pUL34-expressing RepAC cells (Fig. 6D). As previously shown, wt pUL34 complements replication of a UL34-null virus much better than CL04 pUL34. We expected that if the UL31 R229L mutant were sufficient to suppress the CL04 UL34 mutant phenotype, then complementation by the plasmid expressing both CL04 pUL34 and the pUL31 R229L mutant would be significantly better than that of the plasmid that expressed CL04 pUL34 and wt pUL31. Surprisingly, CL04 pUL34 complemented poorly, regardless of whether it was expressed with wt pUL31 or the pUL31 R229L mutant.
FIG. 6.
Marker transfer and complementation assays for UL31 R229L mutant function. Digital micrographs of CL04AI cell monolayers infected with the progeny of cotransfection of UL34-null vRR1072(TK+) viral genomes with either no plasmid (A), plasmid carrying wt UL31 (B), or plasmid carrying UL31R229L (C). (D) Graph of complementation indices for transfected plasmids is shown. Complementation index for each plasmid was calculated by dividing the infectivity produced in each culture by the infectivity produced when plasmid that expressed wt UL34 alone was transfected. Plasmids for each condition are the following: no UL34/no UL31, pRR1072; wt UL34/no UL31, pRR1072Rep; CL04/no UL31, pRR1162; wt UL34/wt UL31, pRR1348; wt UL34/UL31 R229L, pRR1349; CL04/wt UL31, pRR1350; CL04/UL31 R299L, pRR1351. Each bar represents the mean of five independent experiments. Error bars indicate the standard deviation.
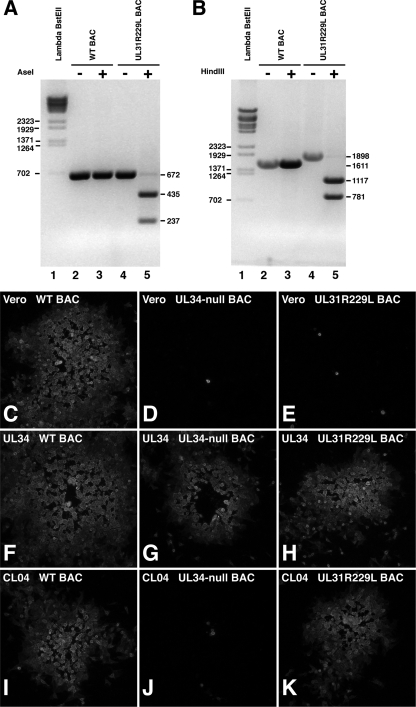
The conflicting results of the marker transfer and complementation experiments led us to test for sufficiency of the R229L mutation by characterization of a recombinant virus rescued from a BAC that had been mutagenized so that it was UL34 null and carried the UL31 R229L mutation. We constructed a UL34-null BAC with the same UL34 deletion as that in vRR1072(TK+) and then introduced the R229L mutation into the UL31 gene. Wild-type and UL34-null and UL34-null/UL31 R229L mutant viruses were rescued by transfection of the BAC clones into UL34-expressing complementing cells. Introduction of the R229L mutation into the UL31 locus was accompanied by introduction of an AseI restriction enzyme site. To confirm the presence of the mutation, the UL31 locus was PCR amplified from the wild-type and UL34-null/UL31 R229L mutant viruses, and the PCR products were digested with AseI (Fig. 7A). Amplification from both wt and mutant BAC DNAs resulted in a product of 672 bp, as expected (Fig. 7, lanes 2 and 4). The wild-type PCR product was not digested (Fig. 7, lane 3), whereas the UL34-null/UL31 R229L mutant PCR product was digested to release the expected fragments of 435 and 237 bp (Fig. 7, lane 5). To confirm the presence of the Kan replacement of UL34 sequences, the UL34 locus was PCR amplified from wild-type, UL34-null, and UL34-null/UL31 R229L mutant BAC genomes, and the PCR products were digested with HindIII (Fig. 7B). The wild-type UL34 locus does not contain a HindIII site, but the introduced Kanr cassette contains a single HindIII site. As expected, the wild-type sequence was not digested with HindIII (Fig. 7, lanes 2 and 3), whereas the UL34-null (not shown) and UL34-null/UL31 R229L mutant PCR products were digested into the expected fragments of 1,117 and 781 bp (Fig. 7, lane 5).
FIG. 7.
Growth of BAC-derived UL34-null, UL31 R229L mutant virus on wt and mutant UL34-expressing cells. (A and B) Digital images show electrophoretically separated PCR products that are either digested with restriction enzyme (lanes 3 and 5) or undigested (lanes 2 and 4). The sizes of the undigested and digested products are indicated on the right of the gel. Lambda BstEII digest size standards are shown in lane 1, and the sizes of standard molecular weight bands are indicated on the left of the gel. (A) PCR products from the UL31 locus in virus rescued from wild-type HSV-1 BAC (lanes 2 and 3) or UL34-null/UL31 R229L mutant (A, lanes 4 and 5) are shown. (B) PCR products from the UL34 locus in the same viruses are shown. Digital micrographs of immunofluorescently stained plaques formed on Vero (C to E), wt UL34-expressing RepAC cells (F to H), or CL04-expressing CL04AI cells (I to K) by viruses rescued from wt HSV-1(F) BAC (C, F, and I), UL34-null/UL31 wt BAC (D, G, and J), or UL34-null/UL31 R229L mutant BAC (E, H, and K).
To determine whether the presence of the R229L mutation confers the ability to grow using CL04 UL34, Vero, wild-type UL34-expressing, and CL04-expressing cells were infected at low multiplicity with BAC-derived wild-type, UL34-null, and UL34-null/R229L mutant viruses. After two days, plaques were detected by indirect immunofluorescence using primary antibody directed against glycoprotein D. Representative plaques are shown in Fig. 7C to K. BAC-derived wild-type virus was able to form plaques on all three cell lines (Fig. 7C, F, and I), although the plaques observed with CL04-expressing cells (Fig. 7I) were slightly smaller than those seen with Vero or wild-type UL34-expressing cells. Like vRR1072(TK+), the UL34-null BAC-derived virus formed large plaques on wild-type UL34-expressing cells (Fig. 7G) but showed only single infected cells or minute plaques of only a few cells on either Vero or CL04-expressing cells (Fig. 7D and J, respectively). The UL34-null/UL31 R229L mutant BAC-derived virus was unable to form plaques on Vero cells (Fig. 7E) but efficiently formed large plaques on cells expressing either wild-type or CL04 UL34 (Fig. 7H and K, respectively), suggesting that in the context of a recombinant virus, the R229L mutation is sufficient to allow efficient viral replication using the CL04 mutant UL34.
Localization of the CL04 mutant UL34.
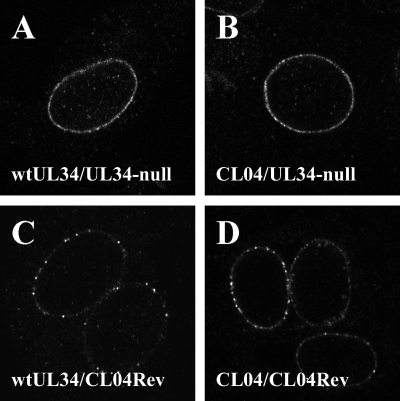
We previously reported that the CL04 mutant pUL34 localized normally to the nuclear envelope in transient transfection/infection assays. The mapping of an extragenic suppressor mutation to UL31 after selection of virus on a cell line stably expressing CL04 led us to examine the localization of the CL04 mutant protein in infected cells. CL04-expressing cells or wt UL34-expressing cells were infected with either vRR1072(TK+) (UL34-null mutant and UL31 wt) or CL04Rev (UL34-null mutant and UL31 R229L mutant) at an MOI of 10 for 16 h, fixed, and processed for immunofluorescence using antibody directed against pUL34 (Fig. 8). As reported previously, wt pUL34 in a wild-type virus-infected cell localizes almost exclusively to the nuclear envelope and is distributed evenly (Fig. 8A). Consistent with earlier transient transfection/infection assays, CL04 pUL34 expressed in the CL04AI cell line shows the same localization as wt pUL34 when expressed in the context of a wild-type infection (Fig. 7B). The localization of both wt and CL04 pUL34 changes when they are expressed in cells infected by the CL04Rev virus carrying the UL31 R229L mutation (Fig. 8C and D). In both cases, pUL34 is still found almost exclusively at the nuclear envelope, but it is distributed in discrete puncta.
FIG. 8.
Localization of wt UL34 and CL04 mutant UL34 in transfected, infected cells. Digital confocal images of cells transfected with pRR1072Rep expressing wt (A and C) or pRR1162 expressing CL04 mutant UL34 (B and D), subsequently infected with either vRR1072(TK+) (A and B) or CL04RevB (C and D), and immunofluorescently stained for UL34 are shown.
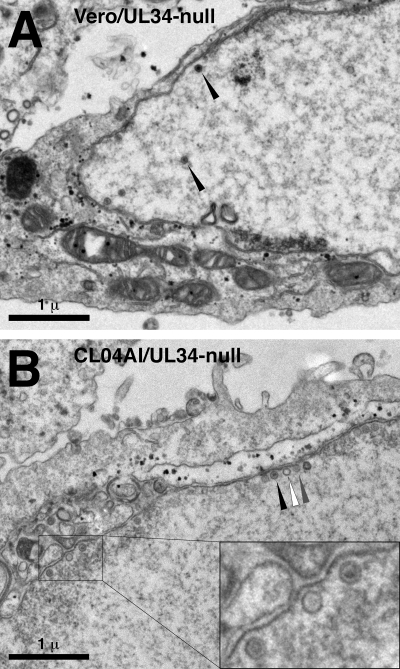
CL04 mutant UL34 envelopment.
In order to characterize the defect in CL04 UL34, Vero cells or CL04-expressing cells were infected with the UL34-null mutant virus at an MOI of 10 for 20 h and then prepared for analysis by transmission electron microscopy (Fig. 9). As shown previously, Vero cells infected with UL34-null virus do not produce cell surface or cytoplasmic virions or capsids, even though nuclear empty and DNA-containing capsids are present (Fig. 9A). These nuclear capsids are found distributed throughout the nucleoplasm, and though a few may be closely associated with the nuclear membrane, most capsids are not found in close association with the INM (Fig. 9A; Table 2). CL04-expressing cells infected with the UL34-null virus also produce neither cell surface nor cytoplasmic virions or capsids but differ from normal Vero cells, in that most accumulated nuclear capsids are found in close association with the INM (Fig. 9B; Table 2). In many cases, the association of capsids with INM was accompanied by slight curvature of the nuclear membrane (Fig. 9B, inset), a feature not seen with UL34-null-infected Vero cells. Interestingly, close association of capsids with the nuclear envelope in these cells shows no dependence upon packaged DNA. Three types of capsids may be distinguished in these preparations: (i) DNA-containing capsids (C capsids) appearing as capsids with an electron-dense center (black arrowhead in Fig. 9B), (ii) empty capsids having an electron-transparent interior (A capsids) (white arrowhead in Fig. 9B), and (iii) capsids with moderate electron density in the interior (B capsids) (gray arrowhead in Fig. 9B). For all capsid types, the frequency of such capsids in close association with the INM is equivalent to their frequency among total intranuclear capsids (Table 3).
FIG. 9.
TEM analysis of nuclear egress from cells that express CL04 UL34. Digital micrographs show Vero (A) or CL04AI (B) cells infected with the UL34-null virus vRR1072(TK+) for 20 h. Black arrowheads (A) point to examples of intranuclear capsids. Black arrowheads (B) point to examples of capsids docked at the INM. The white arrowhead (B) points to an instance of a capsid docked with slight curvature of the membrane. Scale bars are shown at the lower left of each panel.
TABLE 2.
Percentage of INM-associated capsids in UL34-infected Vero and CL04AI cells
| Cells | Docked capsids (%)a | Nucleoplasmic capsids (%) |
|---|---|---|
| Vero | 26.7 | 73.3 |
| CL04AI | 58.1 | 41.9 |
INM-associated capsids are defined as capsids having one side no more than one-fourth of a capsid diameter from the inner nuclear membrane.
TABLE 3.
Percentage of docked A, B, and C capsids
| Capsid type | % of total nuclear | % of docked nuclear |
|---|---|---|
| A | 26.6 | 26.4 |
| B | 52.7 | 55.8 |
| C | 20.7 | 17.8 |
Interactions between deletion mutants of UL34 and UL31.
Reversion of a point mutation in the coding sequence of one protein by an extragenic suppressor in the coding sequence of another strongly suggests that the proteins encoded by the two mutant genes interact and that the interaction, or its functional consequence, is mediated at least in part by the altered amino acids (11). pUL34 and pUL31 are known to interact, but the previously described interacting domains have been mapped to the central part of pUL34 and the N-terminal half of pUL31 (summarized in Fig. 1). Both the CL04 mutation in UL34 and the R229L mutation in UL31 are outside these previously characterized interaction domains (Fig. 1, lines 1, 2, 9, and 10). In order to look for an additional interaction between pUL34 and pUL31, mediated by N-terminal sequences of UL34 and C-terminal sequences of UL31, deletion mutants were constructed that lack the previously characterized interaction domains. Internal deletions of wt and CL04 UL34 that lack amino acids 91 to 228 were constructed (Fig. 1, lines 3 and 4). In order to exclude a possible contribution of the UL34 transmembrane domain to this interaction, deletion mutants of UL34 and CL04 that lack amino acids 91 to 275 and amino acids 64 to 275 and in which the UL34 transmembrane domain is replaced by that of the cellular nuclear lamina-associated protein emerin were also constructed (Fig. 1, lines 5 to 8). wt and R229L mutant UL31 N-terminal truncations that delete amino acids 2 to 126 and that add a FLAG-His tag to the C terminus of the protein were constructed (Fig. 1, lines 11 and 12).
Plasmid transfection.
Transfection of the plasmids described above, alone and in combination, revealed the following.
(i) wt pUL34.
wt pUL34 lacking amino acids 91 to 229 or lacking amino acids 91 to 275 and carrying the emerin transmembrane domain when expressed alone localizes like full-length wt pUL34 (compare Fig. 10A through C). As shown previously, UL34 expressed in the absence of any UL31 construct is found mostly on cytoplasmic membranes, although some in a patchy distribution on the nuclear membrane can be found as well. All of the UL34 constructs tested here show very similar localizations, although pUL34 lacking amino acids 64 to 275 shows a greatly reduced tendency to localize in patches (Fig. 10D).
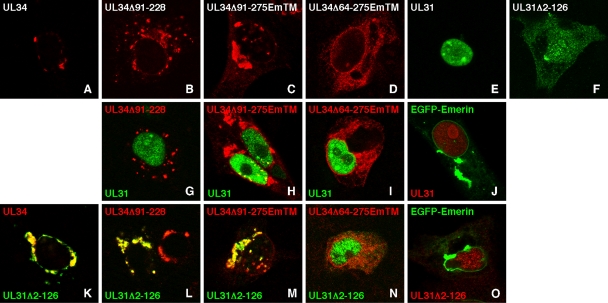
FIG. 10.
Interaction between wild-type and mutant UL34 deletion constructs and the C-terminal domain of UL31. Digital confocal micrographs of transfected Vero cells are shown. Cells were transfected either with a single plasmid (A to F) or with combinations of two plasmids (G to O) The plasmids used are those depicted in Fig. 1 and are indicated within each panel. All UL31 constructs were FLAG tagged and were detected with anti-FLAG primary antibody and either green (E-F, G to I, and K to N) or red (J and O) secondary antibody. All UL34 constructs were detected with anti-UL34 primary antibody and red secondary antibody. EGFP emerin was detected by GFP fluorescence in green (J and O). For combination transfections, the color of the text in the panel corresponds to the color of the antibody used to detect that construct. Only merged images are shown.
(ii) Full-length wt pUL31 and N-terminal truncation mutant.
Whereas full-length wt pUL31 localizes exclusively to the nucleoplasm when expressed alone (Fig. 10E), the N-terminal truncation mutant shows some concentration in the nucleus but also shows diffuse cytoplasmic localization (Fig. 10F), suggesting that sequences required for specific pUL31 nuclear targeting are located in the N-terminal 125 amino acids.
(iii) Colocalization of pUL34 constructs with N-terminally truncated pUL31.
pUL34 constructs lacking the previously identified UL31 interaction domain can colocalize with N-terminally truncated pUL31. Expression of the N-terminal deletion of wt pUL31 with full-length pUL34 (Fig. 10K), internally deleted pUL34 (Fig. 10L), or C-terminally truncated pUL34 lacking amino acids 91 to 275 (Fig. 10M) results in recruitment of the truncated pUL31 to sites of pUL34 localization and complete colocalization of the two proteins. While simple colocalization of two proteins may occur by coincidence, the complete recruitment of the UL31 construct from its normal site of localization to the site of UL34 localization is strongly suggestive of a stable, physical interaction between aa 1 to 90 of pUL34 and aa 126 to 305 of pUL31. Deletion of amino acids 65 to 275 from pUL34, however, resulted in loss of ability to recruit truncated pUL31 from the nucleus and loss of colocalization (Fig. 10N), suggesting that sequences of pUL34 between aa 65 and 90 are necessary for this interaction.
(iv) Failure of pUL34 deletion constructs to colocalize with full-length pUL31.
All pUL34 constructs lacking the previously identified UL31 interaction domain do not colocalize with full-length pUL31. As reported by Liang and Baines, internally deleted wt pUL34 fails to colocalize with full-length pUL31 and neither protein is properly targeted to the nuclear membrane (Fig. 10G) (16). Failure to colocalize with full-length pUL31 was also seen for the pUL34 truncation mutants that lack amino acids 91 to 275 (Fig. 10H) and 64 to 275 (Fig. 10I).
(v) Similar patterns of colocalization and recruitment.
The same patterns of colocalization and recruitment were seen regardless of whether wt pUL34, CL04 pUL34, wt pUL31, or the pUL31 R229L mutant was expressed (Table 4), suggesting that the CL04 mutation does not result in loss of interaction.
TABLE 4.
Interactions of UL34 and UL31 wild-type and mutant constructs
| Construct | UL34 |
CL04 |
EGFP-emerin | ||||
|---|---|---|---|---|---|---|---|
| Full length | Δ91-275EmTM | Δ64-275EmTM | Full length | Δ91-275EmTM | Δ64-275EmTM | ||
| UL31 | + | − | − | + | − | − | − |
| UL31 Δ2-126 mutant | + | + | − | + | + | − | − |
| UL31 R229L mutant | + | − | − | + | − | − | − |
| UL31 R229L Δ2-126 mutant | + | + | − | + | + | − | − |
As a specificity control, EGFP-emerin was cotransfected with full-length and truncated pUL31 to see whether these proteins could be relocalized by overexpression of any type II membrane protein or if the emerin transmembrane domain could mediate relocalization of UL31 constructs in the absence of UL34 sequences (Fig. 10J and O). Coexpression of emerin at levels high enough to induce formation of large cytoplasmic aggregates did not perturb the localization of either pUL31 construct.
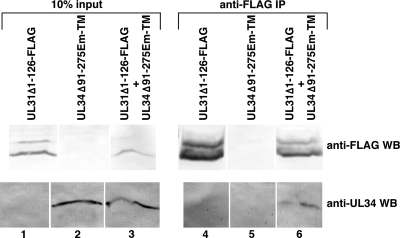
Recruitment of UL31Δ2-126 by UL34Δ91-275EmTM strongly suggests a stable physical interaction between these two proteins. To confirm this interaction, 293T cells were transfected with these two constructs alone and in combination and then immunoprecipitated with anti-FLAG antibody. Unimmunoprecipitated input samples and immunoprecipitated proteins were separated on SDS-PAGE gels, blotted to nitrocellulose, and then probed with antibodies directed against the FLAG epitope (for detection of the UL31 construct) or anti-UL34 (Fig. 11). Expression of both constructs was detected in transfected cells (Fig. 11, lanes 1 to 3), and UL31Δ2-126 expressed alone or in combination with UL34Δ91-275EmTM was efficiently immunoprecipitated using anti-FLAG (Fig. 11, lanes 4 to 6, top). UL34Δ91-275EmTM was not immunoprecipitated by anti-FLAG when expressed alone (Fig. 11, lane 5, bottom) but was coimmunoprecipitated when coexpressed with UL31Δ2-126.
FIG. 11.
Digital images of Western blots are shown. Unfractionated lysates (lanes 1 to 3) or proteins immunoprecipitated with anti-FLAG antibody (lanes 4 to 6) were separated by SDS-PAGE and probed with antibody directed against FLAG epitope to detect UL31 (top row) or with antibody directed against pUL34 (bottom row).
DISCUSSION
A dominant-negative phenotype for a mutation usually reflects the ability of the mutation to disrupt a subset of the essential interactions made by the wild-type form of the protein. The existence of a dominant-negative UL34 mutation suggests that wt UL34 makes at least two essential interactions, one of which is altered by the mutation. To this point, the only essential interaction known for the UL34 protein was the interaction with UL31 that mediates the targeting of both proteins to the nuclear membrane. This interaction is not disrupted by the CL04 mutation, inasmuch as targeting of CL04 pUL34 to the nuclear membrane occurs normally in the presence of wt pUL31 (Fig. 7). pUL34 must make another essential interaction.
Evidence presented here suggests that in addition to the pUL34/pUL31 interaction that mediates nuclear envelope targeting, pUL34 and pUL31 make another interaction that is required for a step in nuclear egress subsequent to targeting to the nuclear envelope. There are two lines of evidence in support of this hypothesis. First, the CL04 mutation can be extragenically suppressed, and all suppressor viruses carry a mutation in UL31 that converts the arginine at position 229 to a leucine, thereby converting a positive-charge side chain to a hydrophobic side chain of similar bulk. The CL04 mutation converts two negatively charged residues, aspartic acid 35 and glutamic acid 37, to alanine. One of these two, aspartic acid 35, is mutated to alanine in the fully functional charge cluster mutant CL03, suggesting that the critical mutated residue in CL04 is glutamic acid 37 (2). It is tempting to speculate that a charge interaction between glutamic acid 37 of UL34 and arginine 229 of UL31 is critical for the function of the pUL34/pUL31 complex. Second, the regions of pUL31 and pUL34 sequences that contain the mutations physically interact when expressed in constructs that lack the interaction domains that mediate nuclear envelope targeting.
Three experimental approaches designed to determine whether the R229L mutation in pUL31 is sufficient to suppress the CL04 mutation in pUL34 gave contradictory results (Fig. 5 and 6). The assays used differ, in that infected cells in the complementation assay may express both mutant suppressor pUL31 (from the complementing plasmid) and wt pUL31 (from the infecting virus), whereas in the marker transfer assay and assays with BAC recombinant viruses, only one type of pUL31 is expressed in each infected cell. It is possible that nonproductive interactions between mutant pUL34 and wild-type pUL31 will “poison” the system, even when otherwise productive mutant pUL34-suppressor pUL31 interactions can occur. Consistent with this possibility, the interaction assays shown in Fig. 9 suggest that the CL04 mutant pUL34 can interact with wt pUL31. This may suggest that the CL04 mutation does not disrupt the physical interaction between the N terminus of pUL34 and the C terminus of pUL31 but does prevent the functional consequence of that interaction in nuclear egress.
The interaction between the N-terminal domain of pUL34 and the C-terminal domain of UL31 occurs when amino acids 1 to 90 of UL34 are present, but not when amino acids 1 to 63 are present, suggesting that the C-terminal boundary of the interaction domain is located between amino acids 64 and 90. Interestingly, the pUL34 N terminus/pUL31 C terminus (UL34 N/UL31 C) interaction does not occur in the context of full-length pUL31 when pUL34 and pUL31 are the only viral proteins expressed (Fig. 9). For this interaction to function in nuclear egress, interaction with some other infected cell structure, perhaps the docking capsid, must change the conformation of the full-length pUL34 or pUL31 to allow the UL34 N/UL31 C interaction to take place.
The CL04 mutation in UL34 reveals a function for pUL34 after capsid docking. In Vero cells infected with UL34-null virus, virus capsids are unable to efficiently egress from the nucleus, and they accumulate in the nucleoplasm but not in close association with the INM. trans-complementation of the UL34-null virus with CL04 UL34 also results in the failure of nuclear egress but leads to the accumulation of a specific egress intermediate—capsids docked at the INM. This phenotype suggests two specific functions for UL34. (i) The observation that capsids cannot dock in the absence of pUL34 but can when a mutant pUL34 is present suggests that the presence of pUL34 is required for some step in envelopment leading to capsid docking. This step might be the docking step itself, it might be that pUL34-mediated nuclear lamina disruption is a necessary prerequisite to capsid docking, or it might be both. (ii) Accumulation of docked capsids in cells that express CL04 pUL34 suggests that the CL04 mutation disrupts a pUL34 function required for the step immediately following capsid docking at the INM, perhaps curvature of the nuclear membrane around the capsid. A function for a pUL34/pUL31 interaction in nuclear membrane curvature would be consistent with the results of Klupp et al., who observed that expression of pUL34 and pUL31 of HSV or pseudorabies virus (PRV) in the absence of other viral proteins can result in vesicularization of the INM into the perinuclear space in a membrane curvature event topologically similar to nuclear envelopment (12).
One model that accounts for observations reported here and those of Klupp et al. would include the following steps leading up to membrane curvature around the capsid. (i) Interaction between the previously mapped interaction/targeting domains of pUL34 and pUL31 mediates targeting to the nuclear membrane. At this stage, interaction between the pUL34 N domain and the pUL31 C domain is blocked by sequences in the N-terminal domain of pUL31. (ii) Following nuclear lamina disruption, capsids dock at the inner nuclear membrane in a process dependent upon pUL34 and perhaps on other viral or cellular factors. (iii) Capsid docking triggers a change in the structure of the pUL34/pUL31 complex allowing interaction between the pUL34 N domain and the pUL31 C domain. This interaction is not sensitive to the CL04 mutation in pUL34. (iv) The pUL34N/pUL31C interaction permits multimerization of pUL31/pUL34 complexes (perhaps by induction of a conformational change in one or both proteins), and geometry of these interactions drives curvature of the nuclear membrane around the capsid. This oligomerization-enabling step is disrupted by the CL04 mutation in pUL34 and can be suppressed by the R229L mutation on pUL31. This model accounts for several features of our observations. First, it is consistent with the dominant-negative character of the CL04 mutation. The presence of a noninteracting pUL34 could presumably poison the formation of multimers, even in the presence of wt pUL34. This model is also consistent with the failure of the pUL31 R229L mutant to suppress the CL04 phenotype when wild-type pUL31 is present, for the same reason.
One of the intriguing properties of the nuclear egress system is its selectivity. Primary envelopment apparently requires completion of the DNA packaging process. In wt virus-infected cells, empty capsids are enveloped more rarely than full capsids, and a variety of viral mutants that synthesize capsids but fail to complete the DNA packaging process do not efficiently envelop the capsids that are produced (1, 4, 5, 10, 18, 25, 31). The phenotype of the CL04 mutation was surprising, in that there was apparently no selectivity in the type of capsid that could dock at the INM. This suggests either that selection occurs subsequent to docking or that the CL04 mutation impairs docking selectivity. The first of these explanations seems unlikely since, if true, docked, empty capsids should accumulate at the INM in wild-type infection, and this has not been reported.
Acknowledgments
We thank the staff of the Central Microscopy Research Facility of the University of Iowa and especially Jean Ross for expertise and help with TEM analysis. We are grateful to Yasushi Kawaguchi for providing the HSV-1(F) BAC and to Klaus Osterrieder for other plasmids required for BAC mutagenesis.
These studies were supported by the University of Iowa and Public Health Service award AI 41478. S.L.B. was supported by the Washburn University Mary B. Sweet Sabbatical Program. S.H. was supported by NSF REU site grant DBI-0097361.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 2.Bjerke, S. L., J. M. Cowan, J. K. Kerr, A. E. Reynolds, J. D. Baines, and R. J. Roller. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601-7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjerke, S. L., and R. Roller. 2006. Roles for herpes simplex type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347:261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the U(L)31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church, G. A., and D. W. Wilson. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J. Virol. 71:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characteristics of herpes simplex virus strains differing in their effect on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 7.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H. J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 79:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 104:10187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarvik, J., and D. Botstein. 1975. Conditional-lethal mutations that suppress genetic defects in morphogenesis by altering structural proteins. Proc. Natl. Acad. Sci. U. S. A. 72:2738-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klupp, B. G., H. Granzow, W. Fuchs, G. M. Keil, S. Finke, and T. C. Mettenleiter. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. U. S. A. 104:7241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99-106. [DOI] [PubMed] [Google Scholar]
- 15.Leach, N., S. L. Bjerke, D. K. Christensen, J. M. Bouchard, F. Mou, R. Park, J. Baines, T. Haraguchi, and R. J. Roller. 2007. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 81:10792-10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang, L., and J. D. Baines. 2005. Identification of an essential domain in the herpes simplex virus 1 UL34 protein that is necessary and sufficient to interact with UL31 protein. J. Virol. 79:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lötzerich, M., Z. Ruzsics, and U. H. Koszinowski. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 80:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris, J. B., H. Hofemeister, and P. O'Hare. 2007. Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 81:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mou, F., T. Forest, and J. D. Baines. 11 April 2007. Us3 of herpes simplex type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. doi: 10.1128/JVI.00380-07. [DOI] [PMC free article] [PubMed]
- 21.Mou, F., E. Wills, and J. D. Baines. 2009. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 83:5181-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 23.Neubauer, A., J. Rudolph, C. Brandmuller, F. T. Just, and N. Osterrieder. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189-204. [DOI] [PubMed] [Google Scholar]
- 24.Park, R., and J. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon, A. P., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santarelli, R., A. Farina, M. Granato, R. Gonnella, S. Raffa, L. Leone, R. Bei, A. Modesti, L. Frati, M. R. Torrisi, and A. Faggioni. 2008. Identification and characterization of the product encoded by ORF69 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:4562-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnee, M., Z. Ruzsics, A. Bubeck, and U. H. Koszinowski. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 80:11658-11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott, E. S., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75:1818-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson-Holley, M., R. C. Colgrove, G. Nalepa, J. W. Harper, and D. M. Knipe. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79:12840-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191-197. [DOI] [PubMed] [Google Scholar]
- 38.Wisner, T. W., C. C. Wright, A. Kato, Y. Kawaguchi, F. Mou, J. D. Baines, R. J. Roller, and D. C. Johnson. 2009. Herpesvirus gB-induced fusion between the virion envelope and outer nuclear membrane during virus egress is regulated by the viral US3 kinase. J. Virol. 83:3115-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamauchi, Y., C. Shiba, F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82:1423-1428. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerberg, J., and M. M. Kozlov. 2006. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7:9-19. [DOI] [PubMed] [Google Scholar]