Abstract
Real-time detection of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) in cases of clinical bacteremia may promote appropriate antimicrobial therapy and infection control. Expense inherent to molecular diagnostics may prevent laboratories from utilizing real-time PCR for this purpose. BD GeneOhm StaphSR assay master mix was reconstituted and aliquoted into SmartCycler tubes in 25-μl volumes (freshly reconstituted master mix), with a portion being frozen at −70°C (frozen master mix). Incubation of 40 previously analyzed lysates from positive BacT/Alert SA and SN blood culture bottles (identified as 10 MRSA strains, 10 MSSA strains, 12 coagulase-negative Staphylococcus strains, and 8 Micrococcus strains) in freshly reconstituted master mix and master mix frozen between 1 week and 6 months generated the expected results in all PCRs. Similarly, positive- and negative-control reagents stored frozen at −70°C for up to 18 weeks yielded the expected reactions. Prospective analysis of 244 positive blood culture samples utilizing 1-week-frozen master mix and freshly reconstituted master mix yielded a 1.2% discordant rate upon initial testing due to three unresolved results (two unresolved results for freshly reconstituted master mix and one unresolved result for frozen master mix). Repeat testing produced a final 100% concordance rate between the two master mix preparations. Use of master mix that was frozen up to 6 months did not compromise performance of the BD GeneOhm StaphSR assay. This modification, resulting in less reagent waste, may allow a greater number of laboratories to consider real-time PCR methodology for detection of bacteremia caused by MRSA and MSSA.
Detection and proper identification of pathogens from the bloodstream are arguably two of the most important aims of diagnostic assays performed in the clinical microbiology laboratory. Estimates of the number of patients with bloodstream infections per year in the United States are commonly cited at 200,000 patients (17), with mortality rates approaching 30% (24). Staphylococcus aureus alone is responsible for approximately 20% of the cases of bloodstream infection (6, 24), and up to 57% of S. aureus isolates recovered from blood cultures demonstrate resistance to methicillin (7, 24). Methicillin-resistant S. aureus (MRSA) bloodstream infection not only generates additional financial and service burdens for health care facilities to a greater extent than those caused by methicillin-susceptible S. aureus (MSSA) (3, 18, 21), but it also demonstrates a significantly higher mortality rate (4, 18). Taken together, these data suggest that rapid identification and methicillin resistance profiling of S. aureus bloodstream infection may be of benefit to patients and allied health care professionals.
While nucleic acid hybridization assays (10) have been shown to directly identify S. aureus from positive blood cultures, only nucleic acid amplification modalities have taken the approach of delineating MRSA and MSSA from other Gram-positive coccus etiologies of bloodstream infection (16). In spite of these advances, some clinical microbiology laboratories may be apprehensive to implement these technologies due to financial considerations. Stamper et al. (20) reported on the utility of the BD GeneOhm StaphSR assay for direct identification and methicillin resistance profiling of coagulase-positive staphylococci from positive blood culture bottles, citing sensitivity and specificity values of ≥98.4% for MRSA and ≥96.7% for MSSA. In this report, we describe a modification of the protocol for reconstitution and storage of BD GeneOhm StaphSR assay master mix that not only allows for efficient detection of MRSA and MSSA but also promotes greater economy in reagent utilization.
(The results of this work were presented in part at the 109th General Meeting of the American Society for Microbiology, Philadelphia, PA, 17 to 21 May 2009 [15].)
In an institutional review board-approved protocol, one positive BacT/Alert SA or SN bottle (bioMérieux, Durham, NC) from consecutive septic episodes with a Gram stain morphology of Gram-positive cocci in pairs or clusters was processed to a lysate form for the BD GeneOhm StaphSR assay (BD Diagnostics, Ste-Foy, Quebec, Canada) per the manufacturer's specifications. Vials containing lyophilized master mix were reconstituted with 225 μl of kit-provided diluent according to the manufacturer's instructions; 25-μl aliquots were distributed into empty SmartCycler reaction tubes (Cepheid, Sunnyvale, CA) that were placed into a 4°C cooling block (freshly reconstituted master mix). In an experimental protocol, vials containing lyophilized master mix were reconstituted with 225 μl of kit-provided diluent; 25-μl aliquots were distributed into empty SmartCycler reaction tubes that were placed into a 4°C cooling block. The tubes were closed tightly, removed from the cooling block, and allowed to stand upright at −70°C for long-term storage protected from light (frozen master mix). Prior to storage, 3.0-μl aliquots of kit-provided nucleic acid (positive control) or sample buffer (negative control) were added to a subset of master mix aliquots.
Aliquots (3.0 μl) of processed lysates were delivered to either frozen master mix tubes that were allowed to acclimate for 5 min in a 4°C cooling block or freshly reconstituted master mix. All reaction tubes were pulse centrifuged for 5 to 10 s and loaded onto a SmartCycler fluorescence thermal cycler (Cepheid) for 45 cycles of PCR. The lysates were subsequently frozen at −20°C. Potency studies involved incubation of archived positive (MRSA), reactive (MSSA), and negative lysates in both freshly reconstituted master mix and frozen master mix that had been stored for 1 week to 6 months at −70°C. In addition, control material frozen for 3, 9, and 18 weeks was subjected to PCR analysis. A prospective clinical study analyzed 244 lysates in tandem using freshly reconstituted master mix and 1-week-frozen master mix. All organisms from positive blood cultures were identified using standard microbiological techniques. Penicillinase-stable penicillin susceptibility results were determined by cefoxitin disk diffusion testing (22). Lysates demonstrating amplification inhibition (unresolved result) were thawed and repeat tested by PCR according to the manufacturer's instructions. Concordant results generated by PCR from freshly reconstituted and frozen master mix constituted reference results.
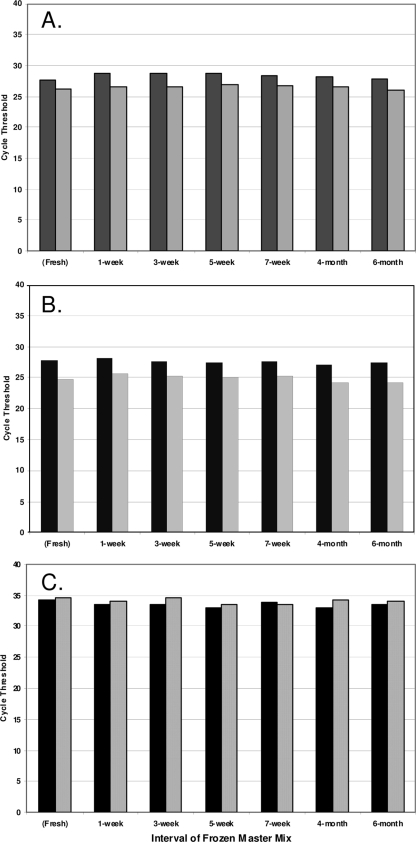
Forty previously analyzed lysates (10 MRSA strains, 10 MSSA strains, 12 coagulase-negative Staphylococcus [CNS] strains, and 8 Micrococcus strains) were incubated in freshly reconstituted master mix and in master mix frozen from 1 week to 6 months. The expected results were generated in all instances. Temporal cycle threshold (CT) profiles for two representative lysates from each classification are illustrated in Fig. 1. The CT values for the two MRSA lysates (Fig. 1A) ranged from 27.7 to 28.8 and from 26.1 to 27.0 and did not exhibit significant changes in frozen master mix potency (P = 0.58; one-way analysis of variance). Similarly, unchanged potency of frozen master mix over time was demonstrated with MSSA lysates (Fig. 1B), with CT value ranges of 27.1 to 28.2 and 24.1 to 25.6 (P = 0.11). Detection of internal-control nucleic acid (Fig. 1C) was not significantly affected by interval of frozen master mix, with CT value ranges of 32.9 to 34.3 and 33.5 to 34.6 (P = 0.73).
FIG. 1.
(A and B) Cycle threshold (CT) values for BD GeneOhm StaphSR assay master mix frozen at variable intervals prior to incubation with lysates derived from clinical blood cultures yielding methicillin-resistant Staphylococcus aureus (A) and methicillin-susceptible Staphylococcus aureus (B). Data from two representative lysates are exhibited per panel and are shaded accordingly. (C) Internal-control CT for frozen master mix incubated with representative coagulase-negative Staphylococcus lysates (solid black bars) and Micrococcus lysates (gray dotted bars).
Preparation of assay controls with reconstituted master mix and subsequent storage at −70°C for 3, 9, and 18 weeks (mean of 12 determinations per parameter) resulted in expected amplification of target staphylococcal and internal-control sequences (Table 1). A mean CT value increase was noted between 9- and 18-week-frozen prepared negative-control material (P = 0.003; t test for independent samples). In spite of this difference, the efficacy of 18-week-frozen master mix in amplification of internal-control nucleic acid (mean CT value of 34.59; Table 1) was largely unchanged from amplification of internal-control nucleic acid via freshly reconstituted master mix (mean CT value of 34.63).
TABLE 1.
Mean cycle threshold (CT) values for replicates of prepared BD GeneOhm StaphSR assay positive- and negative-control reagents frozen at −70°C for 3 weeks, 9 weeks, and 18 weeksa
| Reconstituted reagent | Mean CT ± SD following −70°C storage for: |
||
|---|---|---|---|
| 3 wks | 9 wks | 18 wks | |
| Positive controlb | 35.15 ± 0.29 | 35.20 ± 0.24 | 35.40 ± 0.22 |
| Negative controlc | 33.87 ± 0.43 | 34.10 ± 0.40 | 34.59 ± 0.16 |
CT values for negative-control material are indicative of internal-control amplification.
The mean CT value ± standard deviation for freshly prepared positive-control reagent (22 determinations) was 34.67 ± 0.48.
The mean CT value ± standard deviation for freshly prepared negative-control reagent (22 determinations) was 34.63 ± 0.32.
Analysis of 244 lysates derived from positive blood cultures in freshly reconstituted master mix and frozen master mix yielded three discordant results attributable to unresolved results (two unresolved results for freshly reconstituted master mix and one unresolved result for frozen master mix). After repeat testing of frozen lysates, 100% final concordance between freshly reconstituted and frozen master mix was demonstrated. Final microbiology results included 160 (65.6%) monobacteremic instances of CNS, 35 (14.3%) of MSSA, and 25 (10.2%) of MRSA. Three discrepant results were observed in which the BD GeneOhm StaphSR assay was positive for detection of MRSA and microbial identification was consistent with MSSA. These discrepant results were confirmed by positive MRSA results derived from off-label testing of the blood culture lysates with the BD GeneOhm MRSA assay (targeting the same staphylococcal cassette chromosome mec [SCCmec]/orfX junction as the BD GeneOhm StaphSR assay for MRSA detection) as well as positive MRSA results from testing the S. aureus isolates via the BD GeneOhm StaphSR assay. Further mecA characterization was not performed to resolve or determine the cause of the discordant results; however, utilization of frozen master mix made no contribution to the discrepancies. Studies have demonstrated that mecA dropouts (5, 12) and partial deletions (8) yield such PCR/culture discrepancies in the context of the BD GeneOhm MRSA assay.
While accurate identification of MRSA in bloodstream infection is critical for effective clinical management of patients (2, 4, 18), other data suggest that real-time detection of MSSA may prove to be of equal importance. Small and Chambers (19) have shown decreased in vitro bactericidal activity of vancomycin against recent MSSA strains compared to nafcillin. This translated into a vancomycin clinical failure rate of 38% for endocarditis caused by MSSA among intravenous drug abusers. In contrast, the failure rate for monotherapy with antistaphylococcal β-lactam agents was <3%, as derived from a meta-analysis of MSSA endocarditis within the same demographic (19). Gentry et al. (9) demonstrated complete response in 74% and 50% of patients who were administered nafcillin and vancomycin, respectively, for MSSA endocarditis. Chang et al. (1) reported higher treatment failure rates in patients with MSSA bloodstream infection who were managed with vancomycin (20%) than those who were managed with nafcillin (4%). Furthermore, these studies document a longer persistence of bacteremia following vancomycin therapy.
Although PCR-based assays can play an important role in selection of therapy for bloodstream infections due to MRSA or MSSA, the cost of PCR technology may be prohibitive to a number of health care institutions. Relating this issue of economy to a specific commercial MRSA/MSSA PCR assay, the BD GeneOhm StaphSR assay provides lyophilized master mix in approximate 25-cm3 volumes in individual vials and directs the user to reconstitute each vial with 225 μl of kit-provided diluent. Although the final reconstitution volume exceeds 200 μl (actually approximating 250 μl), it is suggested by the manufacturer that eight 25-μl aliquots are available for PCR assays involving patient specimens and appropriate positive and negative controls. Because freshly reconstituted master mix is stable for only 3 h in the original vial at 2 to 8°C per package insert, laboratories may be unable to utilize all potential aliquots for PCR testing. As an example, when eight potential aliquots are yielded from reconstitution of a lyophilized master mix vial for a patient lysate batch size of three (with controls bringing the total batch size to five), three of the eight aliquots (37.5%) may be unused (Table 2). If it is further assumed that laboratories contribute to diagnosis of staphylococcal bloodstream infection in a real-time fashion, then a random distribution of batch sizes may be realized over a continuum. Based on these assumptions, on average, 31.3% of master mix would go unused if eight aliquots were consistently generated upon lyophilized master mix reconstitution. That figure would increase to 45% if 10 aliquots were consistently generated from lyophilized master mix reconstitution.
TABLE 2.
Hypothetical number of unused aliquots of freshly reconstituted master mix, on the basis of lysate batch size, under current BD GeneOhm StaphSR assay specificationsa
| Lysate batch size (no. of lysates) [with controls included] | No. (%) of unused aliquots per master mix vial following reconstitution |
||
|---|---|---|---|
| 8 aliquots | 9 aliquots | 10 aliquots | |
| 1 [3] | 5 (62.5) | 6 (66.7) | 7 (70.0) |
| 2 [4] | 4 (50.0) | 5 (55.6) | 6 (60.0) |
| 3 [5] | 3 (37.5) | 4 (44.4) | 5 (50.0) |
| 4 [6] | 2 (25.0) | 3 (33.3) | 4 (40.0) |
| 5 [7] | 1 (12.5) | 2 (22.2) | 3 (30.0) |
| 6 [8] | 0 (0.0) | 1 (11.1) | 2 (20.0) |
| Mean % of unused aliquots per random distribution batch size | 31.3 | 38.9 | 45.0 |
Mandated controls are added to batch size prior to the calculation of unused reaction tubes.
With these rather substantial percentages of potential reagent waste, laboratories may be forced to batch patient lysates for PCR, thus not fully taking advantage of the real-time capacity of the assay. We hypothesized that reconstituting master mix as recommended and freezing at −70°C would maximize reagent utilization without compromising assay performance. Kofler and Klausegger (13) described an in-house PCR for detection of human cytomegalovirus from clinical specimens that utilized preformulated aliquots of master mix stored in 200-μl thin-walled tubes at −20°C. Hoorfar et al. (11) showed that reaction mixtures specific for Salmonella enterica 5′ nuclease PCR were stable for 3 months when stored in microwell plates at −20°C. West and Sawyer (23) reported that aliquots of preformulated master mix from four real-time PCR assays could be stored at −70°C without deleteriously affecting performance.
We also reported an effective master mix reconstitution and storage paradigm for a commercial MRSA PCR assay (14). Reconstituted BD GeneOhm MRSA master mix was frozen for up to 4 weeks at −70°C and retained potency, provided that prepared SmartCycler tubes were protected from light and allowed to acclimate in 4°C conditions. These same environmental conditions were employed in the current assessment of BD GeneOhm StaphSR frozen master mix, and a comparable efficacy was observed. Final concordance rates of ≥98.8% were noted in prospective studies of frozen and freshly reconstituted master mix with the BD GeneOhm MRSA (14) and StaphSR assays. Data presented in this study confirm and extend the frozen master mix paradigm. No change in CT data was detected when the potency of frozen BD GeneOhm StaphSR master mix was tested 6 months after reconstitution. This would potentially benefit smaller laboratories who may encounter fewer instances of bloodstream infection. Of further importance, we demonstrated that control reagents can be prepared in the same fashion as frozen master mix.
In conclusion, the kit-provided lyophilized master mix and control reagents that were reconstituted and stored for prolonged periods of time at −70°C retain sufficient potency for routine performance of the BD GeneOhm StaphSR assay. Freezing of the master mix results in cost-effective kit utilization and can assist in establishing efficient work flow practices. Smaller-volume laboratories can therefore more advantageously utilize PCR technology for a real-time contribution to clinical management of staphylococcal bloodstream infection.
Acknowledgments
We express our sincere gratitude to Janice Basile, Jason Burtch, Patricia Luedke, Cheryl Miller, Kimber Munson, Maureen Napierala, Robin Olson, and Maki Silberberg for excellent technical assistance.
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Chang, F. Y., J. E. Peacock, Jr., D. M. Musher, P. Triplett, B. D. MacDonald, J. M. Mylotte, A. O'Donnell, M. M. Wagener, and V. L. Yu. 2003. Staphylococcus aureus bacteremia: recurrence and the impact of antibiotic treatment in a prospective multicenter study. Medicine (Baltimore) 82:333-339. [DOI] [PubMed] [Google Scholar]
- 2.Cosgrove, S. E., and V. G. Fowler, Jr. 2008. Management of methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 46(Suppl. 5):S386-S393. [DOI] [PubMed] [Google Scholar]
- 3.Cosgrove, S. E., Y. Qi, K. S. Kaye, S. Harbarth, A. W. Karchmer, and Y. Carmeli. 2005. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect. Control Hosp. Epidemiol. 26:166-174. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 5.Desjardins, M., C. Guibord, B. Lalonde, B. Toye, and K. Ramotar. 2006. Evaluation of the IDI-MRSA assay for detection of methicillin-resistant Staphylococcus aureus from nasal and rectal specimens pooled in a selective broth. J. Clin. Microbiol. 44:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diekema, D. J., S. E. Beekmann, K. C. Chapin, K. A. Morel, E. Munson, and G. V. Doern. 2003. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J. Clin. Microbiol. 41:3655-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diekema, D. J., B. J. BootsMiller, T. E. Vaughn, R. F. Woolson, J. W. Yankey, E. J. Ernst, S. D. Flach, M. M. Ward, C. L. Franciscus, M. A. Pfaller, and B. N. Dobbeling. 2004. Antimicrobial resistance trends and outbreak frequency in United States hospitals. Clin. Infect. Dis. 38:78-85. [DOI] [PubMed] [Google Scholar]
- 8.Donnio, P.-Y., F. Février, P. Bifani, M. Dehem, C. Kervégant, N. Wilhelm, A.-L. Gautier-Lerestif, N. Lafforgue, M. Cormier, the MR-MSSA Study Group of the Collège de Bactériologie-Virologie-Hygiène des Hôpitaux de France, and A. Le Coustumier. 2007. Molecular and epidemiological evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob. Agents. Chemother. 51:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentry, C. A., K. A. Rodvold, R. M. Novak, R. C. Hershow, and O. J. Naderer. 1997. Retrospective evaluation of therapies for Staphylococcus aureus endocarditis. Pharmacotherapy 17:990-997. [PubMed] [Google Scholar]
- 10.Hensley, D. M., R. Tapia, and Y. Encina. 2009. An evaluation of the AdvanDx Staphylococcus aureus/CNS PNA FISH assay. Clin. Lab. Sci. 22:30-33. [PubMed] [Google Scholar]
- 11.Hoorfar, J., P. Ahrens, and P. Rådström. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huletsky, A., R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Bastien, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kofler, B., and A. Klausegger. 1999. Simplified PCR set-up using a frozen preformulated mix for the detection of cytomegalovirus. Diagn. Microbiol. Infect. Dis. 34:33-35. [DOI] [PubMed] [Google Scholar]
- 14.Munson, E., T. Block, J. T. Voegeli, J. E. Hryciuk, and R. F. Schell. 2009. Cost-effective frozen master mix modification of a commercial methicillin-resistant Staphylococcus aureus PCR assay. J. Clin. Microbiol. 47:1888-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munson, E., T. Kramme, and A. Culver. 2009. Frozen master mix modification of BD GeneOhm™ StaphSR assay, abstr. C-116, p. 117. Abstr. 109th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 16.Paule, S. M., A. C. Pasquariello, R. B. Thomson, Jr., K. L. Kaul, and L. R. Peterson. 2005. Real-time PCR can rapidly detect methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive blood culture bottles. Am. J. Clin. Pathol. 124:404-407. [DOI] [PubMed] [Google Scholar]
- 17.Pittet, D., N. Li, R. F. Woolson, and R. P. Wenzel. 1997. Microbiological factors influencing the outcome of nosocomial bloodstream infections: a 6-year validated, population-based model. Clin. Infect. Dis. 24:1068-1078. [DOI] [PubMed] [Google Scholar]
- 18.Reed, S. D., J. Y. Friedman, J. J. Engemann, R. I. Griffiths, K. J. Anstrom, K. S. Kaye, M. E. Stryjewski, L. A. Szczech, L. B. Reller, G. R. Corey, K. A. Schulman, and V. G. Fowler, Jr. 2005. Costs and outcomes among hemodialysis-dependent patients with methicillin-resistant or methicillin-susceptible Staphylococcus aureus bacteremia. Infect. Control Hosp. Epidemiol. 26:175-183. [DOI] [PubMed] [Google Scholar]
- 19.Small, P. M., and H. F. Chambers. 1990. Vancomycin for Staphylococcus aureus endocarditis in intravenous drug users. Antimicrob. Agents Chemother. 34:1227-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamper, P. D., M. Cai, T. Howard, S. Speser, and K. C. Carroll. 2007. Clinical validation of the molecular BD GeneOhm StaphSR assay for direct detection of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in positive blood cultures. J. Clin. Microbiol. 45:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone, P., E. Larson, and L. N. Kawar. 2002. A systematic audit of economic evidence linking nosocomial infections and infection control interventions: 1990-2000. Am. J. Infect. Control 30:145-152. [DOI] [PubMed] [Google Scholar]
- 22.Swenson, J. M., F. C. Tenover, and the Cefoxitin Disk Study Group. 2005. Results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus spp. J. Clin. Microbiol. 43:3818-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West, D. M., and J. Sawyer. 2006. Freezing complete polymerase chain reaction master mix reagents for routine molecular diagnostics. J. Vet. Diagn. Investig. 18:580-582. [DOI] [PubMed] [Google Scholar]
- 24.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]