Abstract
The aim of this study was to analyze trends in adult invasive pneumococcal disease (IPD) due to macrolide-resistant strains and to study the evolution of serotypes, genotypes, and macrolide-resistant determinants of strains collected in a prospective study between 1999 and 2007 in Barcelona, Spain. IPD due to macrolide-resistant strains of serotypes included in the 7-valent conjugate vaccine (PCV7) decreased from 2.16/100,000 (pre-PCV7 period, 1999 to 2001) to 0.80/100,000 (late-PCV7 period, 2005 to 2007) (P = 0.001), whereas IPD due to macrolide-resistant strains of non-PCV7 serotypes increased from 1.08/100,000 to 2.83/100,000 (P < 0.001). These changes were related to a fall of clones of PCV7 serotypes (ST81 [P < 0.05], ST90, ST315, and ST17) and an increase in new clones of serotypes 19A and 24F (ST230) and 33F (ST717) in the late-PCV7 period. The most common phenotype was MLSB (90.9%), related to the erm(B) gene. The frequent association between MLSB phenotype and tetracycline resistance [tet(M) gene], was related to transposons of the Tn916-family such as Tn6002 or Tn3872. In conclusion, overall adult IPD rates due to macrolide-resistant pneumococci stabilized between 1999 and 2007 in Barcelona. The decrease in macrolide-resistant PCV7 pneumococci was balanced by the increase in macrolide-resistant non-PCV7 pneumococci.
The worldwide increase in the prevalence of macrolide resistance (MR) in Streptococcus pneumoniae is a matter for concern, because of the risk of treatment failure in infections caused by these microorganisms (16). A global international surveillance project (PROTEKT, 2003 to 2004) found that the overall rate of erythromycin resistance was 37.2%, although significant geographical differences in these rates were observed (11). MR in S. pneumoniae is mediated mainly by two major mechanisms, namely, target site modification [encoded by the erm(B) gene and related to the MLSB phenotype] and an efflux pump (encoded by mef genes and related to the M phenotype). Other, less-common resistance mechanisms are mutations in the 23S rRNA and/or alterations in ribosomal proteins (L4 and L22) (18). Most macrolide-resistant S. pneumoniae strains are also resistant to tetracycline. This association is due to the insertion of the erm(B) gene into conjugative and composite transposons of the Tn916 family (Tn1545, Tn3872, Tn6002, and Tn6003) which carry the tet(M) gene (32).
Since the introduction of the pediatric heptavalent pneumococcal conjugate vaccine (PCV7), major changes in the epidemiology of invasive pneumococcal disease have been described. In the United States, the incidence of invasive pneumococcal disease (IPD) has decreased since PCV7 introduction, even in the nontarget population due to herd immunity (33). Since most PCV7 serotypes (6B, 9V, 14, 19F, and 23F) are usually penicillin and multidrug resistant, the rates of penicillin-resistant disease have decreased (17). Moreover, a decrease in macrolide-resistant IPD was observed in the United States (29), mainly due to a reduction of serotype 14 pneumococci harboring mef(E).
In Spain, since the introduction of voluntary PCV7 vaccination in June 2001, the rates of IPD due to PCV7 serotypes disease have decreased in both children and adults (13). However, an overall increase in IPD due to non-PCV7 serotypes was observed between 2005 and 2007 in our geographical area (2, 24).
The aim of the present study was to analyze trends in macrolide-resistant IPD and their relationship to serotypes, genotypes, and MR genes.
MATERIALS AND METHODS
Population and invasive disease surveillance.
The Hospital Universitari de Bellvitge is located in the south of Barcelona. The methods by which surveillance data were obtained have been described elsewhere (2).
MR IPD episodes were defined as the isolation of MR S. pneumoniae (erythromycin MIC, ≥1 μg/ml) from blood or other normally sterile sites. Three periods were considered: pre-PCV7 (1999 to 2001, baseline), early PCV7 (2002 to 2004), and late PCV7 (2005 to 2007).
Bacterial strains.
A total of 853 episodes of IPD were detected in 845 adult patients from 1999 to 2007. Of these, 187 episodes (22.1%) were caused by MR pneumococci isolated in 186 patients (one isolate per episode was considered). The sources of the MR strains included blood (n = 141), cerebrospinal fluid (n = 17), pleural fluid (n = 15), ascitic fluid (n = 12), joint fluid (n = 1), and aqueous humor (n = 1). A total of 185 (98.9%) of 187 MR strains were available for typing and PCR studies.
S. pneumoniae strains were identified by bile solubility and optochin susceptibility. Serotyping was carried out by using the Quellung reaction and/or a dot blot assay, with the use of antisera provided by the Statens Serum Institute (Copenhagen, Denmark) as described previously at the Spanish Reference Laboratory (12).
Antimicrobial susceptibility.
Antimicrobial susceptibility was tested by a microdilution method according to Clinical and Laboratory Standards Institute methods and criteria (6, 7). S. pneumoniae ATCC 6303 and S. pneumoniae ATCC 49619 were used as control strains.
Phenotypic characterization of MR was performed by using a double disk diffusion method with standard disks of erythromycin (15 μg) and clindamycin (2 μg) (7).
Detection of resistance genes.
MR genes [erm(B), erm(A) subclass erm(TR), mef(A), and mef(E)] were detected by PCR using primers and conditions described previously (5, 14, 30). The PCR products of the mef gene were digested with BamHI (Invitrogen) in order to differentiate between the mef(A) and mef(E) gene subclasses (23).
The Tn916 and Tn917 transposon-related genes (int, xis, tndX, tnpA, and tnpR), the tetracycline resistance gene [tet(M)] and the promoter of the aph3′-III gene were detected by PCR using primers and conditions described previously (5). The resistance gene combinations related to the different presumed transposons were: Tn6002 [erm(B), tet(M), int, and xis], Tn3872 [erm(B), tet(M), tnpA, and tnpR], Tn1545, or Tn6003 [erm(B), tet(M), int, xis, and aph3′-III] (4, 9). Strains with negative PCR results for all of the genes related to Tn916 or Tn917 were tested for the presence of the resolvase gene (tndX) of Tn5397. (4).
Molecular typing.
Molecular typing was performed by pulsed-field gel electrophoresis (PFGE) after restriction with SmaI. PFGE patterns were compared to international pneumococcal clones of the Pneumococcal Molecular Epidemiology Network (21).
Representative isolates of each PFGE pattern were studied by multilocus sequence typing (MLST; n = 73) as described previously (10). The allele number and sequence types (STs) were assigned using the pneumococcal MLST website. STs were divided into clonal complexes using eBURST, which is available at the MLST database. Clonal complexes were assigned by comparing our macrolide-resistant strains MLST data with the whole MLST database with the eBURST algorithm. Strains were grouped in the same clonal complex when six or more of the seven loci were identical. When an unusual association between serotype and sequence type was found, the serotype was confirmed by PCR, using previously described methodology (26).
Statistical analysis.
Statistical analyses were carried out by using SPSS for Windows (version 14.0) and EpiInfo (version 6.0; Centers for Disease Control and Prevention). We used chi-square or Fisher exact tests to compare proportions. Incidence rates of IPD, defined as the number of episodes per 100,000 inhabitants, and 95% confidence intervals (CI) were calculated. Two-sided P values of <0.05 were considered statistically significant.
RESULTS
Macrolide resistance in invasive pneumococcal disease.
During the study period, 187 episodes of IPD due to macrolide-resistant strains occurred in 186 patients. Just over two-thirds of patients (68%) were men, and the mean age of the patients was 61.2 years (from 18 to 93 years).
The overall rates of MR IPD remained stable during the study: 3.25 episodes/100,000 inhabitants in the pre-PCV7 period, 3.64 episodes/100,000 in the early-PCV7 period (P = 0.54), and 3.63/100,000 in the late-PCV7 period (P = 0.54). When the pre-PCV7 period and the early-PCV7 period were compared, no significant changes in MR IPD due to serotypes included in PCV7 (2.16/100,000 versus 2.13/100,000 [P = 0.94]) or non-PCV7 serotypes (1.08/100,000 versus 1.51/100,000 [P = 0.27]) were observed. However, when the pre-PCV7 period and the late-PCV7 period were compared, the rates of MR IPD due to PCV7 serotypes decreased by 63% (from 2.16/100,000 to 0.80/100,000; 95% CI = −79.7 to −32.4% [P = 0.001]) and those due to non-PCV7 serotypes increased by 161.7% (from 1.08/100,000 to 2.83/100,000; 95% CI = 53.3 to 346.7% [P < 0.001]).
Antimicrobial susceptibility.
Table 1 shows the antimicrobial susceptibility rates of MR isolates. In the late-PCV7 period a significant decrease in resistance to penicillin, cefotaxime, chloramphenicol, and cotrimoxazole was observed. The rates of macrolide resistance were higher among pneumococci isolated from patients older than 65 years (25.3% [101/400]) than those isolated from adults aged from 18 to 64 years (19.3% [86/446]; P = 0.03).
TABLE 1.
Antibiotic susceptibility of 187 invasive macrolide-resistant S. pneumoniae strains
| Antibiotic | Pre-PCV7 (1999-2001) (n = 54) |
Early-PCV7 (2002-2004) (n = 65) |
Late-PCV7 (2005-2007) (n = 68) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) |
%I | %R | I+R | MIC (μg/ml) |
%I | %R | I+R | Pa | MIC (μg/ml) |
%I | %R | I+R | Pb | |||||||
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | ||||||||||||
| Penicillin | ||||||||||||||||||||
| Old CLSI breakpointsc | 0.5 | 2 | ≤0.016-2 | 48.2 | 27.8 | 76.0 | 0.5 | 2 | ≤0.016-2 | 50.7 | 23.1 | 73.8 | 0.79 | 0.12 | 2 | ≤0.016-2 | 44.1 | 10.3 | 54.4 | 0.01 |
| New CLSI breakpointsd | ||||||||||||||||||||
| Meningitis | 0.5 | 2 | ≤0.016-2 | 76.0 | 76.0 | 0.5 | 2 | ≤0.016-2 | 73.8 | 73.8 | 0.79 | 0.5 | 2 | ≤0.016-2 | 54.4 | 54.4 | 0.01 | |||
| Nonmeningitis | 0.5 | 2 | ≤0.016-2 | 0 | 0 | 0 | 0.5 | 2 | ≤0.016-2 | 0 | 0 | 0 | 0.12 | 2 | ≤0.016-2 | 0 | 0 | 0 | ||
| Cefotaximee | ||||||||||||||||||||
| Meningitis | 0.5 | 1 | 0.01-2 | 33.3 | 3.7 | 37.0 | 0.25 | 1 | 0.01-1 | 27.7 | 0 | 27.7 | 0.28 | 0.12 | 1 | 0.01-2 | 13.2 | 1.5 | 14.7 | 0.004 |
| Nonmeningitis | 0.5 | 1 | 0.01-2 | 3.7 | 0 | 3.7 | 0.25 | 1 | 0.01-1 | 0 | 0 | 0 | 0.20 | 0.12 | 1 | 0.01-2 | 1.5 | 0 | 1.5 | 0.09 |
| Erythromycin | >256 | >256 | 4->256 | 0 | 100 | 100 | >256 | >256 | 4->256 | 0 | 100 | 100 | >256 | >256 | 1->256 | 0 | 100 | 100 | ||
| Clindamycin | >0.5 | >0.5 | ≤0.25->0.5 | 0 | 96.2 | 96.2 | >0.5 | >0.5 | ≤0.25->0.5 | 0 | 87.7 | 87.7 | 0.09 | >0.5 | >0.5 | ≤0.25->0.5 | 0 | 88.3 | 88.3 | 0.11 |
| Tetracycline | 32 | 64 | ≤0.25-64 | 1.9 | 88.9 | 90.8 | 16 | 64 | 0.25-128 | 1.5 | 75.3 | 76.8 | 0.05 | 8 | 32 | 0.25-64 | 2.9 | 77.9 | 80.8 | 0.13 |
| Chloramphenicol | 4 | 16 | ≤2-16 | 50.0 | 50.0 | 4 | 16 | ≤2-16 | 35.4 | 35.4 | 0.10 | ≤2 | 16 | ≤2-32 | 29.4 | 29.4 | 0.02 | |||
| Cotrimoxazole | >2 | >2 | ≤0.5->2 | 9.3 | 53.7 | 63.0 | >2 | >2 | ≤0.5->2 | 1.5 | 58.5 | 60 | 0.74 | ≤0.5 | >2 | ≤0.5->2 | 2.9 | 33.8 | 36.7 | <0.001 |
P value comparing pre-PCV7 to early-PCV7 periods.
P value comparing pre-PCV7 to late-PCV7 periods.
Old penicillin breakpoints: sensitive ≤ 0.06 μg/ml, intermediate = 0.12 to 1 μg/ml, and resistant ≥ 2 μg/ml.
New penicillin breakpoints: meningeal sensitive ≤ 0.06 μg/ml and resistant ≥ 0.12 μg/ml, and nonmeningeal sensitive ≤ 2 μg/ml; intermediate: 4 μg/ml, resistant ≥ 8 μg/ml.
Cefotaxime breakpoints: meningeal sensitive ≤ 0.5 μg/ml, intermediate = 1 μg/ml, and resistant ≥ 2 μg/ml, nonmeningeal sensitive ≤ 1 μg/ml, intermediate = 2 μg/ml, and resistant ≥ 4 μg/ml.
In the pre-PCV7 period, 76.0% (41/54) of MR strains were also nonsusceptible to penicillin (≥0.12 μg/ml), whereas in the late-PCV7 period this coresistance decreased to 54.4% (37/68) (P = 0.01). However, all of the MR strains showed a penicillin MIC of ≤2 μg/ml and were considered susceptible to penicillin using CLSI breakpoints for nonmeningeal infection (7). Of the 187 invasive episodes of MR pneumococci, only 20 episodes (10.6%) were cases of meningitis. Of these, 60% (12/20) were penicillin resistant according to the new CLSI criteria.
No significant changes in the frequency of multiresistance, defined as resistance to ≥3 different antimicrobial families (15, 19), were observed throughout the study (81.5% [44/54] in the pre-PCV7, 84.6% [55/65] in the early-PCV7 period [P = 0.65], and 73.5% [50/68] in the late-PCV7 period [P = 0.3]). However, the proportion of multiresistant strains with serotypes included in PCV7 decreased significantly, as follows: 70.5% (31/44) in the pre-PCV7 period, 61.8% (34/55) in the early-PCV7 period, and 24.0% (12/50) in the late-PCV7 period (P < 0.001). The opposite occurred in the proportion of multiresistant strains with non-PCV7 serotypes: 29.5% (13/44) in the pre-PCV7 period, 38.2% (21/55) in the early-PCV7 period, and 76% (38/50) in the late-PCV7 period (P < 0.001).
Serotype distribution.
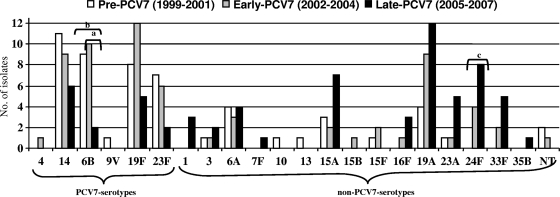
Figure 1 shows the evolution of serotype distribution of invasive MR strains. Twenty-one serotypes were found among the 187 invasive MR S. pneumoniae strains. The proportion of PCV7 serotypes decreased significantly from 66.7% (36/54) in the pre-PCV7 period to 22.1% (15/68) in the late-PCV7 period (P < 0.001), related to a decrease in MR strains of serotypes 6B, 9V, 14, 19F, and 23F. The frequency of non-PCV7 serotypes increased from 33.3% (18/54) in the pre-PCV7 period to 77.9% (53/68) in the late-PCV7 period associated with an increase in serotype (in order of increase): 24F, 19A, 33F, 15A, 23A, 16F, 7F, and 35B.
FIG. 1.
Distribution of serotypes of macrolide resistant invasive S. pneumoniae strains isolated from adult patients (1999 to 2007). Lettered brackets: a, significant decrease in MR IPD due to serotype 6B in early-PCV7 compared to late-PCV7 periods; b, significant decrease in MR IPD due to serotype 6B in pre-PCV7 compared to late-PCV7 periods; c, significant increase in MR IPD due to serotype 24F in pre-PCV7 compared to late-PCV7 periods.
Molecular typing.
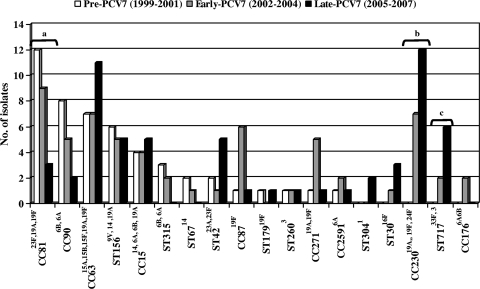
Table 2 shows the genotype and serotype distributions and MR gene detection by PCR of 185 available MR invasive pneumococci. Nine clonal groups accounted for 75% of MLSB strains (CC81, CC63, CC230, CC90, CC15 [ST9, ST17, and ST1201], CC87, CC156, CC311, and CC717). The predominant clone (52.9%) among 17M phenotype strains was the serotype 14 variant of ST156. Figure 2 shows the evolution of genotypes during the study period. When the pre-PCV7 and early-PCV7 periods were compared, only CC230 significantly increased (0% versus 9.2%, P = 0.03). More significant changes were observed when pre-PCV7 and late-PCV7 periods were compared. Six genotypes related to PCV7 serotypes (ST9 and ST17 of CC15, CC81, CC87, CC90, and CC315) decreased, but only descending rates of CC81 were statistically significant (23% versus 5.1%, P = 0.006). Genotypes related to non-PCV7 serotypes increased (CC63 and ST42 of CC311) or emerged (CC30, CC230, CC717, and ST1201 of CC15 and CC304) but only reached statistical significance in two cases: CC230 (0% versus 20.3%, P = 0.001) and CC717 (0% versus 10.2%, P = 0.02).
TABLE 2.
Characterization of 185 invasive macrolide-resistant S. pneumoniae strains
| Phenotype (no. of isolates) and STa | ST(s) found in this study | Related PNEM clone(s) | No. of isolates (%) | Gene(s) detected by PCRb | Presumed transposon(s) (no. of isolates)b | Serotype(s) (no. of isolates) |
|---|---|---|---|---|---|---|
| MLSB (168) | ||||||
| ST81 | ST81, ST3708 | Spain23F-1 | 24 (14.3) | erm(B), tet(M), int, xis | Tn6002 (17) | 23F (13), 19F (4) |
| erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (6) | 19A (5), 19F (1) | ||||
| erm(B), tet(M), tndX | Tn5397* (1) | 19A (1) | ||||
| ST63 | ST63, ST2100, ST3997 | Sweden15A-25 | 24 (14.3) | erm(B), tet(M), tndX | Tn5397 (23) | 15A (10), 19F (7) 15F (3), 19A (2), 15B (1) |
| erm(B)† | None (1) | 15A (1) | ||||
| ST230 | ST230, ST276, ST2307, ST3980 | Denmark14-32 | 19 (11.3) | erm(B), tet(M), int, xis | Tn6002 (14) | 24F (11), 19A (2), 19F(1) |
| erm(B), tet(M), aph3′, int, xis | Tn1545/Tn6003 (3) | 19A (3) | ||||
| erm(B), tet(M), tndX | Tn5397* (1) | 24F (1) | ||||
| erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (1) | 19A (1) | ||||
| ST90 | ST90, ST1624, ST3996 | Spain6B-2 | 15 (8.9) | erm(B), tet(M), int, xis | Tn6002 (12) | 6B (11), 6A (1) |
| erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (2) | 6B (2) | ||||
| erm(B), tet(M), aph3′, int, xis | Tn1545/Tn6003 (1) | 6B (1) | ||||
| ST15 | ST9, ST17, ST1201, ST1664, ST3988 | England14-9, Spain14-5 | 12 (7.1) | erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (8) | 14 (5), 19A (3) |
| erm(B), tet(M), int, xis | Tn6002 (2) | 6A (1), 19A (1) | ||||
| erm(B)† | None (2) | 6B (1), 14 (1) | ||||
| ST87 | ST88, ST87 | 8 (4.8) | erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (4) | 19F (4) | |
| erm(B), tet(M), int, xis | Tn6002 (3) | 19F (3) | ||||
| erm(B), tet(M), tndX | Tn5397* (1) | 19F (1) | ||||
| ST156 | ST156 | Spain9V-3 | 8 (4.8) | erm(B), tet(M), int, xis | Tn6002 (3) | 14 (2), 9V (1) |
| erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (3) | 14 (3) | ||||
| erm(B), tet(M), aph3′, int, xis | Tn1545/Tn6003 (1) | 14 (1) | ||||
| erm(B), mef(E), tet(M), tndX | MEGA+Tn5397* (1) | 19A (1) | ||||
| ST311 | ST42 | 8 (4.8) | erm(B), tet(M), int, xis | Tn6002 (6) | 23A (5), 23F (1) | |
| erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (1) | 23A (1) | ||||
| erm(B), tet(M), tndX | Tn5397* (1) | 23A (1) | ||||
| ST717 | ST717 | 8 (4.8) | erm(B), tet(M), int, xis | Tn6002 (8) | 33F (7), 3 (1) | |
| ST315 | ST315 | Poland 6B-20 | 5 (3.0) | erm(B), tet(M), int, xis | Tn6002 (5) | 6B (4), 6A (1) |
| ST271 | ST202, ST271, ST320 | 7 (4.2) | erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (5) | 19A (4), 19F (1) | |
| erm(B), mef(E), tet(M), int, xis | Tn2010 (2) | 19A (1), 19F (1) | ||||
| ST2591 | ST3977, ST3987, ST3998 | 4 (2.4) | erm(B), tet(M), aph3′, int, xis | Tn1545/Tn6003 (2) | 6A (2) | |
| erm(B), tet(M), int, xis | Tn6002 (2) | 6A (2) | ||||
| ST260 | ST260 | 3 (1.8) | erm(B), tet(M), int, xis | Tn6002 (1) | 3 (1) | |
| erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (2) | 3 (2) | ||||
| ST66 | ST67 | Tennessee14-18 | 3 (1.8) | erm(B), tet(M), int, xis | Tn6002 (3) | 14 (3) |
| ST30 | ST30 | 3 (1.8) | erm(B), tet(M), int, xis | Tn6002 (3) | 16F (3) | |
| ST177 | ST179 | Portugal19F-21 | 2 (1.2) | erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (2) | 19F (2) |
| ST304 | ST304 | Sweden1-40 | 2 (1.2) | erm(B), tet(M), int, xis | Tn6002 (2) | 1 (2) |
| ST460 | ST97 | 1 (0.6) | erm(B), tet(M), int, xis | Tn6002 (1) | 10 (1) | |
| ST135 | ST135 | 1 (0.6) | erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (1) | 6B (1) | |
| ST191 | ST191 | Netherlands7F-39 | 1 (0.6) | erm(B), tet(M), int, xis | Tn6002 (1) | 7F (1) |
| ST344 | ST344 | NorwayNT-42 | 1 (0.6) | erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (1) | 19A (1) |
| ST306 | ST306 | Sweden1-28 | 1 (0.6) | erm(B)† | None (1) | 1 (1) |
| ST386 | ST386 | 1 (0.6) | erm(B), tet(M), int, xis | Tn6002 (1) | 6A (1) | |
| ST558 | ST558 | 1 (0.6) | erm(B)† | None (1) | 35B (1) | |
| Not founder | ST1866, ST3986, ST3989 | 3 (1.8) | erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (1) | 4 (1) | |
| erm(B), tet(M), int, xis | Tn6002 (2) | 6A (1), 23A (1) | ||||
| MLST not done | 3 (1.8) | erm(B), tet(M), int, xis | Tn6002 (2) | 15A (1), NT (1) | ||
| erm(B), tet(M), int, xis, tnpA, tnpR | Tn3872 (1) | NT (1) | ||||
| M (17) | ||||||
| ST156 | ST156 | Spain9V-3 | 9 (52.9) | mef(E)† | MEGA (9) | 14 (9) |
| ST176 | ST138, ST3979 | 2 (11.8) | mef(E)† | MEGA (1) | 6B (1) | |
| mef(A)† | Tn1207.1 (1) | 6A (1) | ||||
| ST15 | ST9 | England14-9 | 1 (5.9) | mef(A)† | Tn1207.1 (1) | 14 (1) |
| ST30 | ST30 | 1 (5.9) | mef(E)† | MEGA (1) | 16 (1) | |
| ST63 | ST3978 | Sweden15A-25 | 1 (5.9) | mef(E)† | MEGA (1) | 14 (1) |
| ST242 | ST242 | Taiwan23F-15 | 1 (5.9) | mef(E), tet(M), int, xis | Tn2009 (1) | 23F (1) |
| ST473 | ST473 | 1 (5.9) | mef(E)† | MEGA (1) | 6A (1) | |
| MLST not done | 1 (5.9) | mef(E)† | MEGA (1) | NT (1) |
ST, that is, the predicter founder in the global MLST database.
*, Tn5397-related element with expression of tet(M) gene; †, tetracycline-susceptible strains.
FIG. 2.
Distribution of major clones of macrolide resistant invasive S. pneumoniae strains isolated from adult patients (1999 to 2007). Lettered brackets: a, significant decrease of CC81 in pre-PCV7 compared to late-PCV7 periods; b, significant increase of CC230 in pre-PCV7 compared to late-PCV7 periods; c, significant increase of ST717 in pre-PCV7 compared to late-PCV7 periods.
Capsular switch events were observed in 11 genotypes: CC63, CC81, CC90, CC156, CC230, ST320, and ST202 of CC271, ST42 of CC311, CC315, CC344, and CC717. In three of them, the capsular switching is rarely described: one strain of serotype 3 belonged to ST717, previously related to serotype 33F strains, one strain of serotype 19A belonged to ST344 previously related to nontypeable pneumococci, and the remaining strain was a serotype 19A with ST156 related to serotype 9V.
Eleven new sequence types were found: ST3977 (serotype 6A), ST3978 (serotype 14), ST3979 (serotype 6B), ST3980 (serotype 24), ST3986 (serotype 6A), ST3987 (serotype 6A), ST3988 (serotype 6A), ST3989 (serotype 23A), ST3996 (serotype 6B), ST3997 (serotype 15F), and ST3998 (serotype 6A). Only two of them (ST3986 and ST 3989) were singletons as listed in the MLST database.
Gene detection by PCR.
Of 187 invasive MR strains, 170 (90.9%) showed an MLSB phenotype and 17 (9.1%) showed an M phenotype. The proportion of M phenotype strains increased from 3.7% (2/54) in the pre-PCV7 period to 10.3% (7/68) in the late-PCV7 period (P = 0.12).
Table 2 shows the PCR detection of macrolide and tetracycline-resistant genes of 168 available MLSB phenotype isolates and 17M phenotype isolates.
MLSB phenotype strains (n = 168).
The erm(B) gene was detected in all 168 MLSB strains. No erm(TR) gene was detected. Three strains (3/168, 1.8%) harbored both erm(B) and mef(E) genes (two strains were serotype 19A [ST156 and ST320, respectively], and one strain was serotype 19F [ST271]).
The association of tetracycline and erythromycin resistance was found in 89.3% (150/168) of MLSB strains and the tet(M) gene was detected in all of these strains.
Among 168 MLSB strains, 88 (52.4%) harbored erm(B), int, xis, and tet(M) genes, suggesting that the erm(B) gene was carried by Tn6002. Thirty-eight strains (22.6%) had erm(B), int, xis, tnpA, tnpR, and tet(M) genes, suggesting the presence of Tn3872. Twenty-seven strains (16.1%) harbored erm(B), tet(M), and tndX, indicating the presence of a related Tn5397 transposon. Seven strains (4.2%) harbored erm(B), int, xis, tet(M), and aph3′, suggesting the presence of Tn1545 or the recently described Tn6003 (9). Of the three strains harboring erm(B) and mef(E) genes, two also harbored the tet(M), int, and xis genes related to Tn2010.
M phenotype strains (n = 17).
Fifteen M strains (88.2%) harbored the mef(E) gene, whereas only two strains (11.8%) carried the mef(A) gene. Only two M phenotype strains (2/17, 12%) were tetracycline resistant and harbored the tet(M) gene. One of them also harbored the int and xis genes related to presumed Tn2009.
DISCUSSION
There have been major changes in the epidemiology of IPD in the United States since the introduction of PCV7 for children, the most important being the reduction in the incidence of IPD in children and adults and the decrease in penicillin resistance rates. Little is known about the impact of PCV7 introduction in the incidence rates of MR IPD. In Atlanta, GA, where PCV7 was introduced in 2000, the overall incidence of IPD due to MR S. pneumoniae fell from 7.7/100,000 in 2000 to 2.9/100,000 in 2002, coinciding with a significant decrease in macrolide-resistant 6B, 19F, and 23F serotypes, all included in PCV7. However, the incidence of macrolide resistance among non-PCV7 serotypes doubled within 2 years of the implementation of PCV7, from 0.30/100,000 in the 1994-to-1999 period to 0.7/100,000 in 2002 (P = 0.02) (29).
In Spain, PCV7 was introduced for children in June 2001, and the estimated uptake of PCV7 for children fluctuates from 22% in 2002 to 50% in 2006 (13). In spite of this low uptake, a significant decrease of IPD due to serotypes included in the PCV7 was observed in children (3, 24, 25) and adults (2) in the 2005 to 2007 period.
The present study demonstrates that the incidence of IPD due to MR pneumococci among adults remained stable throughout the 1999 to 2007 period in southern Barcelona. This stabilization was the result of a combination of a significant decrease in IPD due to MR serotypes included in PCV7 and a significant increase in IPD caused by MR serotypes not included in PCV7, such as 15A, 16F, 19A, 23A, 24F, and 33F.
Several observations should be made in view of these findings. First, the fall in IPD caused by PCV7 serotypes observed among adult patients in the late-PCV7 period was associated with a significant decrease in penicillin-, cefotaxime-, chloramphenicol-, and cotrimoxazole-resistant rates. However, the incidence of macrolide resistant pneumococci in our geographical area remained stable, because the decrease of multiresistant PCV7 serotypes was balanced by the increase of multiresistance among non-PCV7 serotypes.
Second, previous reports in the United States and our geographical area (17, 33) observed a decrease in IPD due to PCV7 serotypes observed in children related to PCV7 implementation (24). In agreement, a similar fall was also observed in our institution among adult patients with MR IPD due to PCV7 serotypes, suggesting a herd immunity phenomenon.
Third, it is well known that long-acting macrolides such as clarithromycin and azithromycin, which ensure a low serum concentration of the antibiotic for a long period of time, are associated with selection of coresistance to macrolides, penicillin, and cotrimoxazole (31). Although the overall use of macrolides in Spain decreased from 3.3 defined daily doses per 1,000 inhabitants and day (DID) in 1999 to 2.0 DID in 2006, the consumption of the long-acting macrolides clarithromycin and azithromycin remained stable during the study period, with an annual consumption of ∼1.4 DID for clarithromycin and 0.8 DID for azithromycin (1, 20).
Fourth, a phenomenon of serotype replacement has been observed in our geographical area in the late-PCV7 period among invasive MR pneumococci. The overall frequency of MR PCV7 serotypes (14, 6B, 19F, and 23F) decreased significantly from 66.7% in the pre-PCV7 period to 22.1% in the late-PCV7 period (P < 0.001), and this gap was filled by a significant increase in MR non-PCV7 serotypes (P < 0.001). The non-PCV7 serotypes 1, 16F, 24F, and 33F, which were macrolide susceptible in the pre-PCV7 period, have acquired macrolide resistant determinants under macrolide selective pressure in the PCV7 era. In addition, an expansion of classically macrolide-resistant serotypes 15A, 19A, and 23A was also observed in the late-PCV7 period in our geographical area.
Fifth, resistance to macrolides and other antimicrobials is associated with the spread of certain multiresistant international clones of S. pneumoniae (22, 27, 28). In the present study, the following 10 international clones in order of frequency were detected throughout 1999-2007 period: Spain23F-1 ST81, Sweden15A-25 ST63, ST 23019A,24F, Spain6B-2 ST90, ST8819F, Spain9V-3 ST156, ST4223A,23F, ST71733F, Poland6B-20 ST315, and Spain14-5 ST17, which accounted for 61.0% of macrolide-resistant pneumococci. However, after PCV7 introduction for children, MR was associated with the emergence of new MR clones (ST23024F,19A, ST27119A, ST71733F, and ST3016F), and the expansion of previously macrolide-susceptible clones that acquired MR determinants in the late-PCV7 period (ST120119A and ST3041).
Sixth, coresistance to macrolides and tetracycline is related to conjugative transposons, mobile genetic elements that can transfer resistance determinants in a wide variety of streptococci (32). There are some geographical differences in the prevalence and spread of transposons in pneumococci. In Italy and Spain, the most frequently reported transposons in S. pneumoniae strains are Tn6002 and Tn3872, whereas in Japan Tn917 is the most common (8, 9). The widespread occurrence of these transposons among new S. pneumoniae clones, related to non-PCV7 serotypes, could explain the stability in MR IPD rates that we observed in our area. Moreover, as previously reported, we found that different transposons could be carried by pneumococci sharing the same PFGE pattern (5, 9), such as Spain23F-ST81, a well-established clone in our country, which carried a large variety of transposons probably because of the possibility of different exchange events with other pneumococci or viridans group streptococci. In addition, new macrolide-resistant clones such as ST306, ST260, and ST191 were previously determined to be macrolide susceptible, supporting the role of horizontal transfer of transposons in the maintenance of MR rates. Moreover, the spread of new MR clones not previously detected in Spain, such as CC230, emphasizes the role of clonal dissemination in resistance.
In conclusion, the incidence of MR IPD among adults in Barcelona remained stable over the period studied because non-PCV7 serotypes filled the gap of macrolide-resistant PCV7 serotypes. This change was related to the acquisition of Tn916 family of transposons carrying macrolide-resistant determinants by emerging MR clones related to non-PCV7 serotypes; alternatively, the selection and spread of strains with these transposons may be due to serotype replacement. Further surveillance studies focused on clinical and molecular epidemiology of IPD caused by MR pneumococci and macrolide use are needed to know the impact of future conjugate vaccines on the serotype and clone distribution.
Acknowledgments
We thank the staff at the Microbiology Department of Hospital Universitari de Bellvitge for daily contributions to this project and Montserrat Alegre for excellent technical assistance.
We received support from the Fondo de Investigaciones Sanitarias de la Seguridad Social (PI060647 and PI02/0269), the Spanish Pneumococcal Infection Study Network G03/103 (Red Temática de Cooperación del FIS), the Ciber de Enfermedades Respiratorias (CB06/06/0037 Ministry of Health, Instituto de Salud Carlos III, Madrid, Spain), and the Institut d'Investigació Biomèdica de Bellvitge (to D.R.).
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Agencia Española de Medicamentos y Productos Sanitarios. 2006. Uso de antibióticos en España. Agencia Española de Medicamentos y Productos Sanitarios, Ministerio de Sanidad y Consumo, Barcelona, Spain. http://www.agemed.es/profHumana/observatorio/docs/uso-antibioticos-oct07.pdf.
- 2.Ardanuy, C., F. Tubau. R. Pallares, L. Calatayud, M. A. Domínguez, D. Rolo, I. Grau, R. Martín, and J. Liñares. 2009. Epidemiology of invasive pneumococcal disease among adult patients in Barcelona before and after pediatric 7-valent pneumococcal conjugate vaccine introduction, 1997-2007. Clin. Infect. Dis. 48:57-64. [DOI] [PubMed] [Google Scholar]
- 3.Aristegui, J., E. Bernaola, I. Pocheville, C. García, L. Arranz, G. Durán, L. Pérez, M. Bastida, C. Canduela, M. Herranz Aguirre, E. Garrote, M. A. Fletcher, and C. Pérez. 2007. Reduction in pediatric invasive pneumococcal disease in the Basque Country and Navarre, Spain, after introduction of the heptavalent pneumococcal conjugate vaccine. Eur. J. Clin. Microbiol. Infect. Dis. 26:303-310. [DOI] [PubMed] [Google Scholar]
- 4.Brenciani, A., A. Bacciaglia, M. Vecchi, L. A. Vitali, P. E. Varaldo, and E. Giovanetti. 2007. Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob. Agents Chemother. 51:1209-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calatayud, L., C. Ardanuy, E. Cercenado, A. Fenoll, E. Bouza, R. Pallares, R. Martín, and J. Liñares. 2007. Serotypes, clones, and mechanisms of resistance of erythromycin-resistant Streptococcus pneumoniae isolates collected in Spain. Antimicrob. Agents Chemother. 51:3240-32466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility test for bacteria that growth aerobically; approved standard, 7th ed. CLSI document M7-A6. CLSI, Wayne, PA.
- 7.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing: 18th informational supplement. CLSI document M100-S18. CLSI, Wayne, PA.
- 8.Cochetti, I., E. Tili, M. Vecchi, A. Manzin, M. Mingoia, P. E. Varaldo, and M. P. Montanari. 2007. New Tn916-related elements causing erm(B)-mediated erythromycin resistance in tetracycline-susceptible pneumococci. J. Antimicrob. Chemother. 60:127-131. [DOI] [PubMed] [Google Scholar]
- 9.Cochetti, I., E. Tili, M. Mingoia, P. E. Varaldo, and M. P. Montanari. 2008. erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob. Agents Chemother. 52:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 11.Farrell, D. J., C. Couturier, and W. Hryniewicz. 2008. Distribution and antibacterial susceptibility of macrolide resistance genotypes in Streptococcus pneumoniae: PROTEKT Year 5 (2003-2004). Int. J. Antimicrob. Agents 31:245-249. [DOI] [PubMed] [Google Scholar]
- 12.Fenoll, A., I. Jado, D. Vicioso, A. Pérez, and J. Casal. 1998. Evolution of Streptococcus pneumoniae serotypes and antibiotic resistance in Spain: update (1990 to 1996). J. Clin. Microbiol. 36:3447-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenoll, A., J. J. Granizo, L. Aguilar, M. J. Giménez, L. Aragoneses-Fenoll, G. Hanquet, J. Casal, and D. D. Tarragó. 2009. Temporal trends of invasive Streptococcus pneumoniae serotypes and antimicrobial resistance patterns in Spain from 1979 to 2007. J. Clin. Microbiol. 47:1012-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kataja, J., P. Huovinen, M. Skurnik, H. Seppälä, et al. 1999. Erythromycin resistance genes in group A streptococci in Finland. Antimicrob. Agents Chemother. 43:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klugman, K. P. 1990. Pneumococcal resistance to antibiotics. Clin. Microbiol. Rev. 3:171-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klugman, K. P., and J. R. Lonks. 2005. Hidden epidemic of macrolide-resistant pneumococci. Emerg. Infect. Dis. 11:802-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyaw, M. H., R. Lynfield, W. Schaffner, A. S. Craig, J. Hadler, A. Reingold, A. R. Thomas, L. H. Harrison, N. M. Bennett, M. M. Farley, R. R. Facklam, J. H. Jorgensen, J. Besser, E. R. Zell, A. Schuchat, C. G. Whitney, et al. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 6:1455-1463. [DOI] [PubMed] [Google Scholar]
- 18.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liñares, J., R. Pallares, T. Alonso, J. L. Perez, J. Ayats, F. Gudiol, P. F. Viladrich, and R. Martin. 1992. Trends in antimicrobial resistance of clinical isolates of Streptococcus pneumoniae in Bellvitge Hospital, Barcelona, Spain (1979-1990). Clin. Infect. Dis. 15:99-105. [DOI] [PubMed] [Google Scholar]
- 20.Llor, C., J. M. Cots, M. J. Gaspar, M. Alay, and N. Rams. 2009. Antibiotic prescribing over the last 16 years: fewer antibiotics but the spectrum is broadening. Eur. J. Clin. Microbiol. Infect. Dis. 28:893-897. [DOI] [PubMed] [Google Scholar]
- 21.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefévre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGee, L., K. P. Klugman, A. Wasas, T. Capper, and A. Brink. 2001. Serotype 19F multiresistant pneumococcal clone harboring two erythromycin resistance determinants [erm(B) and mef(A)] in South Africa. Antimicrob. Agents Chemother. 45:1595-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montanari, M. P., M. Mingoia, I. Cochetti, and P. E. Varaldo. 2003. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J. Clin. Microbiol. 41:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muñoz-Almagro, C., I. Jordan, A. Gene, C. Latorre, J. J. Garcia-Garcia, and R. Pallares. 2008. Emergence of invasive pneumococcal disease caused by nonvaccine serotypes in the era of 7-valent conjugate vaccine. Clin. Infect. Dis. 46:174-1782. [DOI] [PubMed] [Google Scholar]
- 25.Obando, I., L. A. Arroyo, D. Sánchez-Tatay, D. Tarragó, D. Moreno, W. P. Hausdorff, and A. B. Brueggemann. 2007. Molecular epidemiology of paediatric invasive pneumococcal disease in southern Spain after the introduction of heptavalent pneumococcal conjugate vaccine. Clin. Microbiol. Infect. 13:347-348. [DOI] [PubMed] [Google Scholar]
- 26.Pai, R., R. E. Gertz, and B. Beall. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44:124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinert, R. R., A. Ringelstein, M. van der Linden, M. Y. Cil, A. Al-Lahham, and F. J. Schmitz. 2005. Molecular epidemiology of macrolide-resistant Streptococcus pneumoniae isolates in Europe. J. Clin. Microbiol. 43:1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siira, L., M. Rantala, J. Jalava, A. J. Hakanen, P. Huovinen, T. Kaijalainen, O. Lyytikäinen, and A. Virolainen. 2009. Temporal trends of antimicrobial resistance and clonality of invasive Streptococcus pneumoniae isolates in Finland, 2002 to 2006. Antimicrob. Agents Chemother. 53:2066-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens, D. S., S. M. Zughaier, C. G. Whitney, W. S. Baughman, L. Barker, K. Gay, D. Jackson, W. A. Orenstein, K. Arnold, A. Schuchat, M. M. Farley, et al. 2005. Incidence of macrolide resistance in Streptococcus pneumoniae after introduction of the pneumococcal conjugate vaccine: population-based assessment. Lancet 365:855-863. [DOI] [PubMed] [Google Scholar]
- 30.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanderkooi, O. G., D. E. Low, K. Green, J. E. Powis, and A. McGeer. 2005. Toronto Invasive Bacterial Disease Network: predicting antimicrobial resistance in invasive pneumococcal infections. Clin. Infect. Dis. 40:1288-1297. [DOI] [PubMed] [Google Scholar]
- 32.Varaldo, P. E., M. P. Montanari, and E. E. Giovanetti. 2009. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, A. Schuchat, et al. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]