Abstract
Globicatella sanguinis is a rare cause of acute meningitis. We demonstrated human carriage of Globicatella by identifying cefotaxime-resistant strains in groin and rectal specimens 9 months after invasive infection. The pathogenic strain isolated from the cerebrospinal fluid and the carriage strains were accurately identified by sodA gene sequence analysis.
CASE REPORT
A 56-year-old woman presented with vomiting and headache, associated with instability when walking, memory impairment, and clinical signs of meningitis. Lumbar puncture revealed turbid cerebrospinal fluid (CSF) with pleocytosis (11,800 × 106 white blood cells/liter with 94% polymorphonuclear cells), a glucose concentration of 0.59 g/liter, and a protein concentration of 2.32 g/liter. Gram-positive cocci were observed and subsequently identified as Globicatella sanguinis strain HDP50220. Based on the results of antibiotic susceptibility tests, the primary antibiotic treatment, which consisted of cefotaxime (100 mg/kg of body weight/day intravenously [i.v.]) combined with fosfomycin (200 mg/kg/day i.v.), was replaced by amoxicillin (200 mg/kg/day i.v.) for 3 weeks. Remission of meningitis was confirmed by the absence of bacterial growth from a follow-up CSF sample. Nine months later, Globicatella isolates HDP60616 and HDP60617 were retrieved from the groin and rectum, respectively.
The Globicatella strain from CSF grew as pinpoint, alpha-hemolytic colonies on blood and chocolate agars (Oxoid, Wesel, Germany) incubated for 18 h at 37°C under an atmosphere enriched with 5% CO2; Gram-positive, catalase-negative cocci organized into short chains and clusters were observed. Biochemical characteristics of the strain determined by the rapid ID 32 Strep system corresponded to those of G. sanguinis (profile 623.761.733.50; certainty of 99.9%), whereas those determined by the API 20 Strep system corresponded to those of Aerococcus viridans (profile 7306773; certainty of 88.9%) (both systems were obtained from bioMérieux, Marcy l'Etoile, France) (Table 1).
TABLE 1.
Phenotypic characteristics of Globicatella isolates compared to those described previouslya
| Phenotypic characteristic or relevant factor |
G. sanguinis |
G. sulfidifaciens |
|||||
|---|---|---|---|---|---|---|---|
| Isolate(s) from previous study by: |
HDP50220b | HDP60616b | Isolates from previous study by Vandamme et al. (11) | HDP60617b | |||
| Collins et al. (2) | Shewmaker et al. (8) | Seegmuller et al. (6) | |||||
| Hippurate hydrolysis | + | + | + | + | + | − | ± |
| H2S production | − | ND | − | − | − | + | − |
| Production of: | |||||||
| Pyrrolidonyl-arylamidase | + | V | + | + | + | − | + |
| β-Glucuronidase | − | − | + | − | − | + | − |
| β-Galactosidase | + | + | − | + | + | − | + |
| N-Acetyl-β-glucosaminidase | + | − | + | − | − | − | − |
| Acidification of: | |||||||
| Inulin | − | + | ND | + | + | + | + |
| Ribose | + | V | + | + | + | − | − |
| Mannitol | + | + | + | + | + | − | + |
| No. of strains examined | Type strain and 8 others | 28c | 1 | 1 | 1 | Type strain and 3 others | 1 |
| Host(s) | Humans | Humans | Human | Human | Human | Animals | Human |
| Clinical context | Bacteremia | Meningitis and bacteremia | Meningitis | Meningitis | Carriage | Bacteremia | Carriage |
| Specimen(s) of origin | Blood | CSF, blood | CSF | CSF | Groin specimen | Blood | Rectal specimen |
Isolates were compared to those described in previous reports (2, 6, 8, 11). +, 85% or more of the strains tested positive for the indicated characteristic; −, 15% or less of the strains tested positive; V, 16 to 84% of the strains tested positive; ±, strain tested positive with the API 20 Strep system and negative with the rapid ID 32 Strep system; ND, not determined.
HDP50220, HDP60616, and HDP60617 strains presented the common characteristics of Globicatella spp. They grew in 6.5% NaCl brain heart infusion broth. They hydrolyzed esculin and produced pyrrolidonyl-arylamidase; they gave negative results in the leucine aminopeptidase, bile-esculin, urease, and Voges-Proskauer tests. No reaction of the Lancefield extracts with group A, B, C, D, F, or G antisera (from the Prolex streptococcal grouping latex kit; Pro-Lab, Strasbourg, France) was observed.
Including the 9 strains from the study by Collins et al. (2).
The antibiotic susceptibility of the isolate was investigated by the disk diffusion method, and MICs were determined with the Etest (AES, Combourg, France). The criteria of the Antibiogram Committee of the French Society for Microbiology for Streptococcus spp. were used for the interpretation of antimicrobial drug susceptibility results (http://www.sfm.asso.fr). The strain was susceptible to benzylpenicillin (MIC = 0.125 μg/ml), amoxicillin (MIC ≤ 0.016 μg/ml), chloramphenicol, tetracycline, pristinamycin, rifampin, fosfomycin, vancomycin, and teicoplanin and resistant to cefotaxime (MIC ≥ 32 μg/ml), erythromycin (MIC = 32 μg/ml), and clindamycin (MIC ≥ 256 μg/ml). The isolate was tested for the presence of the mefA, ermB, and ermA genes by multiplex PCR as described previously (7, 9). An amplification product from ermA, but not from the other genes, was obtained.
Swab samples from the nasal fossae, throat, armpit, groin, and rectum were plated onto colistin-nalidixic acid-blood agar plates (Oxoid) for the investigation of colonization. Cefotaxime-resistant alpha-hemolytic colonies growing around disks loaded with 30 μg of cefotaxime (Bio-Rad, Marnes-La-Coquette, France) placed on the agar surface were selected for further identification. Two Globicatella strains, HDP60616 and HDP60617, obtained from the groin and the rectum, respectively, were identified. They were both resistant to cefotaxime (MICs ≥ 32 μg/ml).
Complete sequencing of the 16S rRNA genes from the three isolates obtained from the patient and the strains G. sanguinis CIP 107044T (ATCC 51173T) and Globicatella sulfidifaciens CIP 107175T was carried out as described previously (3). The two type strains were found to be more than 99% similar. Species differentiation was achieved by sodA sequencing as described by Poyart et al. (5). The sodA sequences of the two type strains were 96.6% identical. The sodA sequences of CSF isolate HDP50220 and groin isolate HDP60616 were 100% identical to each other. They were 99.8 and 96.3% identical to those of G. sanguinis CIP 107044T (GenBank accession no. EU649714) and G. sulfidifaciens CIP 107175T (GenBank accession no. EU649713), respectively. The sodA sequence of the HDP60617 rectal isolate was 99.8% identical to that of G. sulfidifaciens and 96.3% identical to that of G. sanguinis. Pulsed-field gel electrophoresis (PFGE) showed that the HDP50220 and HDP60616 isolates were closely related and clearly differed from the rectal isolate and the two type strains (Fig. 1).
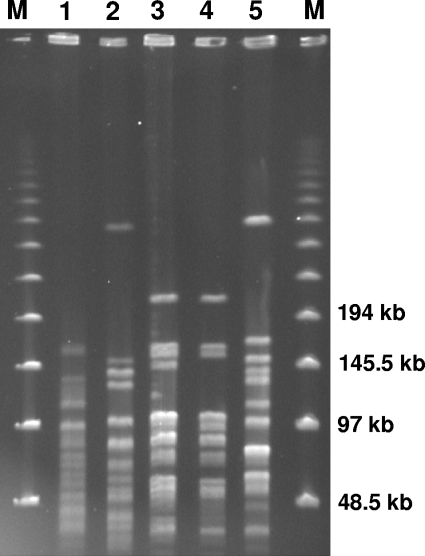
FIG. 1.
PFGE profiles of the three clinical strains of Globicatella from the patient and the type strains of G. sanguinis and G. sulfidifaciens. Patterns were obtained by SmaI macrorestriction, with a method adapted from that of van den Braak et al. (12). Lanes: M, molecular size markers (48.5 to 970 kb [size standard no. 170-3635; Bio-Rad]); 1, G. sanguinis strain CIP 107044T; 2, G. sulfidifaciens strain CIP 107175T; 3, strain HDP50220 (isolated from CSF); 4, strain HDP60616 (isolated from the groin); and 5, strain HDP60617 (isolated from the rectum). According to the criteria of Tenover et al., strain HDP50220 and strain HDP60616 are closely related, as their profiles differ by only three DNA fragments (10). These patterns differed from the pattern of the rectal strain and from the individual patterns of the type strains.
Globicatella was proposed as a new genus in 1992, initially with only one species, G. sanguinis (2). A second species, G. sulfidifaciens, was subsequently discovered in animals (11). The few reports of Globicatella species isolation from humans have concerned only cases of infection with G. sanguinis (1, 4, 6, 8). We report here the first case of human carriage of both species of Globicatella, which was documented several months after the patient had recovered from acute meningitis. A strain of G. sanguinis identical to the meningitis strain was isolated from the groin, while a strain of G. sulfidifaciens was isolated from the rectum. These two bacterial species are difficult to distinguish from related catalase-negative, Gram-positive cocci. Caution is required in interpretation according to the previously described phenotypic identification system, as only four strains were investigated for the description of G. sulfidifaciens (11). Indeed, the various descriptions of Globicatella strains report different phenotypic reactions for the same species (Table 1). Among the phenotypic characteristics, the production of pyrrolidonyl-arylamidase and the absence of production of leucine aminopeptidase may contribute to the presumptive identification of Globicatella spp. but do not distinguish these species from A. viridans. Genotypic study is therefore required for accurate identification. 16S rRNA gene analysis was useful for identifying the three isolates to the genus level but not to the species level due to the high degree of similarity (>99%) of 16S rRNA sequences between the species. In contrast, the sodA gene provided accurate discrimination between the two species.
The pathogenic role of Globicatella spp. remains only partially known because of the difficulties involved in identification and the small number of reports regarding Globicatella. G. sanguinis has been reported to cause meningitis or bacteremia in humans (1, 2, 4), whereas G. sulfidifaciens was thought to be a secondary agent of suppurative processes in animals (11). This report confirms the role of G. sanguinis as a human pathogen and, furthermore, demonstrates that Globicatella spp. are commensal organisms in humans. Indeed, both species were isolated from the patient in the absence of any symptom. The sites from which they were isolated, the rectum and groin, are consistent with an intestinal bacterial reservoir, as suggested previously (4). The close relationship between the CSF and groin isolates was deduced primarily from phenotypic characteristics (Table 1) and sodA sequences. It was confirmed by similarities in PFGE patterns (Fig. 1). As a farmer, the patient had been exposed to the intestinal flora of animals. However, at the time of the carriage investigation, she was no longer keeping cows; therefore, the animals with which she had been in contact previously could not be examined.
MIC determination confirmed the cefotaxime resistance of both invasive and colonizing Globicatella strains. Cefotaxime resistance in 48% of 27 clinical isolates of G. sanguinis was reported previously by Shewmaker et al. (8). This unexpected resistance to cefotaxime may be a useful marker for presumptive identification of this Streptococcus-like bacterium, although half of the strains tested by Shewmaker et al. were susceptible to cefotaxime (8). We found that the invasive strain HDP50220 was also resistant to erythromycin and clindamycin, in relation to the presence of the ermA gene. The presence of the ermA gene in Globicatella spp. has not been reported previously. Indeed, previous reports have implicated only the mefA gene in the erythromycin resistance of Globicatella strains (6, 8, 11).
This case report highlights the difficulties involved in identifying Globicatella, resulting in underestimation of the potential role of this bacterium as a human pathogen, and the need for accurate genomic analysis.
Nucleotide sequence accession numbers.
The sequence of the identical sodA genes from CSF isolate HDP50220 and groin isolate HDP60616 has been deposited in GenBank under accession no. EU649715, and the sequence of the sodA gene from the HDP60617 rectal isolate has been deposited in GenBank under accession no. EU649716.
Acknowledgments
We thank Gislène Collobert and Michèle Virmaux for excellent technical assistance.
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Abdul-Redha, R. J., U. Balslew, J. J. Christensen, and M. Kemp. 2007. Globicatella sanguinis bacteraemia identified by partial 16S rRNA gene sequencing. Scand. J. Infect. Dis. 39:745-748. [DOI] [PubMed] [Google Scholar]
- 2.Collins, M. D., M. Aguirre, R. R. Facklam, J. Shallcross, and A. M. Williams. 1992. Globicatella sanguis gen. nov., sp. nov., a new gram-positive catalase-negative bacterium from human sources. J. Appl. Bacteriol. 73:433-437. [DOI] [PubMed] [Google Scholar]
- 3.Harf-Monteil, C., A. L. Fleche, P. Riegel, G. Prevost, D. Bermond, P. A. Grimont, and H. Monteil. 2004. Aeromonas simiae sp. nov., isolated from monkey faeces. Int. J. Syst. Evol. Microbiol. 54:481-485. [DOI] [PubMed] [Google Scholar]
- 4.Lau, S. K., P. C. Woo, N. K. Li, J. L. Teng, K. W. Leung, K. H. Ng, T. L. Que, and K. Y. Yuen. 2006. Globicatella bacteraemia identified by 16S ribosomal RNA gene sequencing. J. Clin. Pathol. 59:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poyart, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seegmuller, I., M. van der Linden, C. Heeg, and R. R. Reinert. 2007. Globicatella sanguinis is an etiological agent of ventriculoperitoneal shunt-associated meningitis. J. Clin. Microbiol. 45:666-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seppala, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shewmaker, P. L., A. G. Steigerwalt, L. Shealey, R. Weyant, and R. R. Facklam. 2001. DNA relatedness, phenotypic characteristics, and antimicrobial susceptibilities of Globicatella sanguinis strains. J. Clin. Microbiol. 39:4052-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandamme, P., J. Hommez, C. Snauwaert, B. Hoste, I. Cleenwerck, K. Lefebvre, M. Vancanneyt, J. Swings, L. A. Devriese, and F. Haesebrouck. 2001. Globicatella sulfidifaciens sp. nov., isolated from purulent infections in domestic animals. Int. J. Syst. Evol. Microbiol. 51:1745-1749. [DOI] [PubMed] [Google Scholar]
- 12.van den Braak, N., A. van Belkum, M. van Keulen, J. Vliegenthart, H. A. Verbrugh, and H. P. Endtz. 1998. Molecular characterization of vancomycin-resistant enterococci from hospitalized patients and poultry products in The Netherlands. J. Clin. Microbiol. 36:1927-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]