Abstract
Since the 1970s, influenza B viruses have diverged into two antigenically distinct virus lineages called the Yamagata and Victoria lineages. We present the first real-time PCR assay for virus lineage differentiation to supplement classical antigenic analyses. The assay was successfully applied to 310 primary samples collected in Germany from 2007 to 2009.
Influenza viruses are members of the family Orthomyxoviridae and are divided into three genera, A, B, and C (8). Influenza A and B viruses are most relevant clinically, since they cause severe respiratory infections in humans (2). While influenza A viruses comprise a large group of different subtypes (8), influenza B viruses formed a homogenous group and started to diverge into two antigenically distinguishable lineages only in the 1970s (3, 4, 6). These virus lineages were named after their first representatives, B/Victoria/2/87 and B/Yamagata/16/88, as the Victoria and Yamagata lineages (6). Today, the antigenic differences between the lineages allow their differentiation by hemagglutination inhibition testing (HIT) by using specific immune sera raised against contemporary strains of either lineage. However, HIT is a time-consuming and tedious process and needs virus isolation as a prerequisite. In contrast, PCR is well known to be a fast, specific, and sensitive diagnostic method, and furthermore, real-time PCR reduces the risk of carryover contamination and allows large-scale diagnostics (5). However, to date, there has been no real-time PCR assay described that enables the differentiation of influenza B viruses, which would greatly speed up and thus improve influenza virus surveillance. We therefore present an assay that not only amplifies viruses of both lineages but also discriminates between them by the application of two differently labeled minor-groove binder (MGB) probes, with either one being specific for one lineage.
The target region of the assay was chosen from an alignment with recent influenza B virus hemagglutinin (HA) database sequences (from the years 2000 to 2008). The 81-bp amplicon comprises a 13-bp stretch that differs in 6 positions between the two lineages. The stability of the characteristic nucleotide changes was confirmed by an alignment comprising all available influenza B virus hemagglutinin database sequences (1,622 sequences, from the years 1954 to 2008). The distinctive nucleotides have been stable from the late 1990s until today, so nucleotide changes are not impossible but are unlikely to occur in the near future. Thus, an MGB probe was designed for either lineage targeting this 13-bp stretch. By the application of both probes with different color labels (6-carboxyfluorescein [FAM] and VIC) in a single PCR, both virus lineages can be detected and discriminated simultaneously, as only one of the two probes will give a fluorescence signal.
Reaction conditions were established for the LightCycler 480 system in a total reaction mixture volume of 25 μl containing 1× PCR buffer, 5 mM MgCl2, 1.25 μM deoxynucleoside triphosphate (dNTP) (Invitrogen) with dUTP (GE Healthcare, Great Britain), 0.5 U Platinum Taq polymerase (Invitrogen), 900 nM forward primer F432 (5′-ACCCTACARAMTTGGAACYTCAGG-3′), 600 nM reverse primer R479 (5′-ACAGCCCAAGCCATTGTTG-3′), 150 nM Yamagata probe MGB437 (5′-FAM-AATCCGMTYTTACTGGTAG-MGB-3′), 100 nM Victoria probe MGB470 (5′-VIC-ATCCGTTTCCATTGGTAA-MGB-3′), and 3 μl of template cDNA. Cycling conditions were 5 min at 95°C, followed by 45 cycles of 15 s at 95°C and 30 s at 60°C.
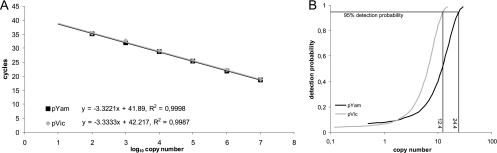
The assay was evaluated by using two plasmids that were cloned according to routine procedures (1) and contained 610 and 613 bp of the hemagglutinin genes of B/Bayern/7/08 (plasmid pYam) and B/Berlin/38/08 (plasmid pVic), two contemporary German isolates representing the Yamagata and Victoria lineages, respectively. Thus, the complete primer- and probe-binding regions represent the original sequences of these two isolates. Amplification of 10-fold serial dilutions of each plasmid in λ DNA (1 ng/μl) revealed a linear detection range from 107 to 102 genome equivalents per reaction with a correlation (R2) of >0.998 and slopes of −3.32 (pYam) and −3.33 (pVic) (Fig. 1A), resembling a PCR efficiency of 1 (E = 10−1/slope − 1). We performed a probit analysis as a model of nonlinear regression that indicated a 95% detection probability of 24.4 genome equivalents per reaction for plasmid pYam and 12.4 genome equivalents per reaction for pVic (Fig. 1B). Additionally, from virus culture material of the corresponding virus isolates B/Bayern/7/08 (Yamagata) and B/Berlin/38/08 (Victoria), the 95% detection probabilities were determined to be 1.3 × 10−5 and 3.8 × 10−5 HA units per reaction, respectively. The overall variability was assessed by the repeated examination of three different plasmid copy numbers as well as virus culture material with a high, medium, or low virus load. The standard deviations of threshold cycle (CT) values were found to be very low and were comparable for Yamagata and Victoria viruses and plasmids (Table 1). We found no cross-reactivity with DNA/cDNA of isolates from seasonal influenza A virus subtypes H1N1 and H3N2; pandemic influenza A/H1N1 virus; respiratory syncytial viruses A and B; adenovirus serotypes 2, 3, and 4; human metapneumovirus; parainfluenza viruses 1, 2, and 3; coxsackievirus; and rhinovirus as well as human DNA from swab samples.
FIG. 1.
PCR assay validation. (A) Mean CT values (double reactions) of plasmid dilutions containing 107 to 102 genome equivalents of pYam and pVic were plotted against the cycle number. The slope and correlation (R2) are indicated. (B) Probit analyses were performed by examination of plasmid dilutions containing 100 to 0.1 genome equivalents of pYam and pVic in 10-fold reactions. Results were analyzed by using SPSS 17.0 statistics software.
TABLE 1.
PCR assay validation: detection variabilitya
| Variability | Material | Virus load (no. of genome equivalents/reaction) | SD of CT value |
|
|---|---|---|---|---|
| Yamagata | Victoria | |||
| Intra-assay | Plasmids | 5 × 105 | 0.03 | 0.05 |
| 5 × 103 | 0.12 | 0.08 | ||
| 5 × 101 | 0.34 | 0.54 | ||
| Cultured virus | High | 0.08 | 0.16 | |
| Medium | 0.21 | 0.10 | ||
| Low | 0.22 | 0.35 | ||
| Interassay | Plasmids | 5 × 105 | 0.04 | 0.07 |
| 5 × 103 | 0.17 | 0.11 | ||
| 5 × 101 | 0.64 | 0.56 | ||
| Cultured virus | High | 0.35 | 0.34 | |
| Medium | 0.27 | 0.40 | ||
| Low | 0.23 | 0.39 | ||
Variability runs were performed by examination of pYam and pVic plasmid dilutions (5×105, 5×103, and 5×101 genome equivalents per reaction) as well as cultured virus material with a high (6.67×108 genome copies/ml), medium (6.67×106 genome copies/ml), or low (6.67×104 genome copies/ml) virus load. Intra-assay variability was tested in sextuplicate reactions. Interassay variability was determined by 2-fold examinations of duplicate reactions with the inclusion of data from the intra-assay variability run (total, 3-fold examination). The standard deviations (SD) of obtained CT values are listed.
Finally, to confirm the applicability of the assay to clinical diagnostics, we examined 310 influenza B virus-positive primary samples from the 2007-2008 and 2008-2009 influenza seasons. All samples were taken from German patients presenting with influenza-like illness and successfully underwent HIT after virus isolation on MDCK2 cells. The nasal and throat swabs were washed in minimal essential medium (MEM) cell culture medium immediately after arrival. RNA was extracted by using either the RTP DNA/RNA virus MiniKit (Invitek) or the MagAttract viral RNA M48 kit (Qiagen) according to the manufacturer's suggestions. cDNA was synthesized from 25 μl of RNA by applying Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) and random hexamer primers as described elsewhere previously (7). Residual RNA was stored at −80°C until further use.
By applying the presented assay, viruses were amplified from all 310 primary samples with CT values between 22 and 37. All samples were genetically identified as Yamagata or Victoria lineage viruses in concordance with HIT results. The 310 primary samples comprised 185 Yamagata and 3 Victoria lineage viruses from the 2007-2008 season as well as 120 Victoria and 2 Yamagata lineage viruses from the 2008-2009 season. Since the assay's introduction into our diagnostic routine in February 2009, it has been run on approximately 5,000 samples, and to our knowledge, no false-positive or false-negative results have been obtained.
In summary, we present the first real-time PCR assay for the differentiation of influenza B viruses. This assay speeds up virus lineage identification in clinical specimens considerably and will therefore help to improve the surveillance of influenza B viruses. Furthermore, it will enable a timely recognition of the circulating B virus lineage during influenza seasons and will thus allow short-term decisions on patient care, e.g., in the case of a nonmatching vaccine, as well as the early onset of on-time epidemiological examinations, including WHO decisions on vaccine composition.
Acknowledgments
We thank Julia Hinzmann and Madlen Sohn for excellent technical assistance.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Chmielewicz, B., A. Nitsche, B. Schweiger, and H. Ellerbrok. 2005. Development of a PCR-based assay for detection, quantification, and genotyping of human adenoviruses. Clin. Chem. 51:1365-1373. [DOI] [PubMed] [Google Scholar]
- 2.Hampson, A. W., and J. S. Mackenzie. 2006. The influenza viruses. Med. J. Aust. 185:S39-S43. [DOI] [PubMed] [Google Scholar]
- 3.Hay, A. J., V. Gregory, A. R. Douglas, and Y. P. Lin. 2001. The evolution of human influenza viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanegae, Y., S. Sugita, A. Endo, M. Ishida, S. Senya, K. Osako, K. Nerome, and A. Oya. 1990. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J. Virol. 64:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackay, I. M. 2004. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 10:190-212. [DOI] [PubMed] [Google Scholar]
- 6.Rota, P. A., T. R. Wallis, M. W. Harmon, J. S. Rota, A. P. Kendal, and K. Nerome. 1990. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 175:59-68. [DOI] [PubMed] [Google Scholar]
- 7.Schweiger, B., I. Zadow, R. Heckler, H. Timm, and G. Pauli. 2000. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J. Clin. Microbiol. 38:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright, P. F., G. Neumann, and Y. Kawaoka. 2007. Orthomyxoviruses, p. 1691-1740. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.