Abstract
Food-borne salmonellosis is a major manifestation of gastrointestinal disease in humans across the globe. Accurate and rapid identification methods could positively impact the identification of isolates, enhance outbreak investigation, and aid infection control. The SNaPshot multiplex system is a primer extension-based method that enables multiplexing of single nucleotide polymorphisms (SNPs). Here the method has been developed for the identification of five Salmonella serotypes, commonly detected in the United Kingdom, based on serotype-specific SNPs identified in the multilocus sequence typing (MLST) database of Salmonella enterica. The SNPs, in genes hemD, thrA, purE, and sucA, acted as surrogate markers for S. enterica serovars Typhimurium, Enteritidis, Virchow, Infantis, and Braenderup. The multiplex primer extension assay (MPEA) was conducted in two separate panels and evaluated using 152 Salmonella enterica isolates that were characterized by MLST. The MPEA was shown to be 100% specific and sensitive, within this collection of isolates. The MPEA is a sensitive and specific method for the identification and detection of Salmonella serotypes based upon SNPs seen in MLST data. The method can be applied in less than 6 h and has the potential to improve patient care and source tracing. The utility of the assay for identification of Salmonella serotypes directly from clinical specimens and food samples warrants further investigation.
Food-borne salmonellosis is an important public health problem, causing substantial morbidity and mortality worldwide. Transmission of salmonella to humans has been linked to multiple sources, including contaminated or undercooked food. Therefore, development of rapid and sensitive methods for detection of Salmonella serovars directly from clinical specimens and environmental samples may have a significant impact on the disease burden caused by this pathogen. Tools developed for such purposes could help both in preventing the spread of outbreaks and in clinical diagnosis. Traditional methods for diagnosis of Salmonella strains, including culture on selective media and biochemical and serological identification methods, have been successful in clinical diagnostic laboratories and in epidemiological surveillance (1, 10, 13, 26). However, these methods may require up to 3 days to provide results and are not as amenable to high-throughput analyses as some more-recently proposed molecular typing methods. Many molecular methods have been described for detection of Salmonella serovars (4, 6, 24, 25, 31, 33); however, these methods are not always rapid assays and generally do not permit serovar identification. In addition, these approaches cannot be applied directly to clinical and environmental samples and require a bacterial culture, which increases analysis time and can lead to culture-based biases.
Multilocus sequence typing (MLST) is a high-resolution genotyping technique that has emerged as a powerful tool for determining the global epidemiology and population structure of many bacterial pathogens, including Salmonella enterica (18, 32). MLST detects sequence variability within particular genes to determine the genetic relatedness of organisms (12). The method is reproducible, and the data generated are easy to interpret and directly comparable among laboratories (9). However, MLST remains expensive and impractical for routine examination of bacterial genotype. Therefore, the use of informative single nucleotide polymorphisms (SNPs) has been recently suggested as a cost-effective alternative to full MLST characterization (2, 3). SNP typing, using various genetic targets, has been reported to be a robust method for analysis of relationships of S. enterica serovar Typhi (20, 22, 23). An advantage of the SNPs observed across MLST loci is that they represent neutral genetic variation in the bacterial genome (12) and thus could be used for developing accurate, rapid, economical, and phylogenetically meaningful typing methods that may lead to significant improvements in diagnosis and surveillance for Salmonella serotypes.
The SNaPshot multiplex system (Applied Biosystems) can be used to genotype multiple known SNPs simultaneously using a multiplex primer extension assay (MPEA) approach. These assays monitor the single-base extension of unlabeled oligonucleotide primers, which bind to a cDNA template adjacent to target SNPs. DNA polymerase extends the primer at the polymorphic site, incorporating a fluorescently labeled dideoxynucleoside triphosphate (ddNTP), and the fluorescently labeled extension products are separated and visualized using capillary electrophoresis. The SNaPshot approach has been demonstrated to be efficient in various fields of research, such as forensic investigation (5, 21) and clinical diagnosis (11). The approach has been applied in microbiological investigations, though until recently, this has been for analysis of mammalian systems when interacting with bacteria (8, 14-16, 19, 28).
In the present study, we have developed an MPEA for rapid “molecular serotyping” of Salmonella isolates by targeting 15 SNPs identified by interrogation of the MLST database for salmonella.
(Some of this work was presented as a poster at the Health Protection Agency Conference, Warwick, United Kingdom, September 2009.)
MATERIALS AND METHODS
Bacterial isolates and direct DNA extraction.
A total of 152 Salmonella enterica strains belonging to 33 serotypes were used in this study. The isolates were collected by the Laboratory for Gastrointestinal, Emerging and Zoonotic Infections, Centre for Infections, Health Protection Agency (HPA), Colindale, United Kingdom (Table 1).
TABLE 1.
STs of Salmonella isolates used in this study
| Serovar (no. of isolates) | ST |
|---|---|
| Agona (1) | 13 |
| Albany (1) | 292 |
| Anatum (1) | 64 |
| Arizonae (1) | 195 |
| Bareilly (1) | 203 |
| Bilthoven (1) | 10 |
| Blockly (1) | 52 |
| Braenderup (1) | 22 |
| Brandenburg (1) | 20 |
| Chameleon (1) | 596 |
| Dar-Es-Salaam (1) | 675 |
| Degania (1) | 598 |
| Derby (3) | 40 |
| Enteritidis (33) | 11 |
| Enteritidis (1) | 136 |
| Enteritidis (1) | 183 |
| Florida (1) | 357 |
| Goldcoast (5) | 358 |
| Hadar (7) | 33 |
| Haifa (3) | 49 |
| Heidelberg (3) | 15 |
| Indiana (1) | 17 |
| Infantis (6) | 32 |
| Infantis (1) | 361 |
| Java (1) | 28 |
| Java (1) | 307 |
| Kentucky (3) | 198 |
| London (1) | 155 |
| Mbandaka (1) | 315 |
| Montevideo (6) | 316 |
| Newport (3) | 45 |
| Newport (2) | 118 |
| Newport (1) | 166 |
| Newport (1) | 156 |
| Newport (1) | 360 |
| Poona (1) | 317 |
| Poona (1) | 402 |
| Seftenberg (5) | 14 |
| Seftenberg (1) | 210 |
| Sofia (1) | 597 |
| Stanley (4) | 29 |
| Stanley (4) | 51 |
| Typhimurium (25) | 19 |
| Typhimurium (1) | 34 |
| Typhimurium (1) | 328 |
| Typhimurium (1) | 313 |
| Virchow (6) | 16 |
| Virchow (1) | 197 |
| Virchow (1) | 359 |
| Virchow (1) | 181 |
DNA extraction and MLST analysis.
The 152 isolates were cultured on nutrient and MacConkey agars at 37°C overnight, and chromosomal DNA was extracted using PrepMan Ultra sample preparation reagent, according to the manufacturer's instructions (PrepMan Ultra; Applied Biosystems).
MLST was performed, as described previously (17), on DNA extracted from all cultured isolates. Sequenced products were separated and detected using a CEQ 8000 Beckman Coulter DNA analysis sequencer. The sequence of each DNA strand was determined and assembled from the chromatograms by using Sequencher software (Sequencher 4.7; Gene Codes Corporation). Allele numbers were assigned and converted to sequence type (ST) after the sequences were submitted, via the Internet, to the Salmonella MLST database (http://mlst.ucc.ie/mlst/dbs/Senterica).
Identification of diagnostic alleles and SNPs.
Key alleles for each Salmonella serovar are specific for certain serovars or STs, ideally having a positive predicted value close to 1 (2). Key alleles were identified in the current study for the different STs seen in each of the serovars under examination (Table 2). The predictive alleles identified for each ST/serovar were verified by matching them against all MLST alleles in the Salmonella MLST database (http://mlst.ucc.ie/mlst/dbs/Senterica). Sequences were downloaded and aligned using the BioEdit Sequence Alignment Editor, version 7.0.0 (Tom Hall, North Carolina State University). A visual screen of alignments was carried out to allow identification of SNPs diagnostic for each allele, and a multiplex primer extension, or SNaPshot, assay was designed, including each of the diagnostic SNPs.
TABLE 2.
Positive and negative predictive values of each of the key alleles chosen as being diagnostic for Salmonella isolates of the five major serovars in the United Kingdom
| Serovar | No. of STs per MLST database | Key allele (%)a | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|
| Typhimurium | 29 | hemD12 (86.2) | 0.925 (92.5) | 0.993 (99.3) |
| Enteritidis | 15 | thrA11 (53.3) | 0.615 (61.5) | 0.988 (98.8) |
| Virchow | 12 | thrA14 (81.8) | 0.692 (69.2) | 0.995 (99.5) |
| Infantis | 8 | sucA21 (75) | 1.0 (100) | 0.996 (99.6) |
| Braenderup | 3 | purE11 (100) | 1.0 (100) | 1.0 (100) |
The values shown in parentheses indicate the percentage of STs for each serovar that are represented by the chosen key allele in the MLST database.
SNaPshot primer design.
In this study, an MPEA was performed to detect 15 SNPs in the following two panels. Panel I targeted 7 SNPs among two key alleles (hemD and thrA) to identify and differentiate S. enterica serovar Typhimurium and S. enterica serovar Enteritidis, which are the most common serovars causing infection in the United Kingdom. Panel II included 8 SNPs within three alleles (thrA, sucA, and purE) for identification of S. enterica serovar Virchow, S. enterica serovar Infantis, and S. enterica serovar Braenderup (Table 3). The primers (forward and reverse) were tested individually to assess performance and validate migration size before being tested in multiplex reactions.
TABLE 3.
Details of primers, SNPs, and observed migration of extension products for the Salmonella serovars under study
| Panel | Serovar (key allele) | PCR primer | Amplified product length (bp) | SNP position | Extension base variation | Extension primer sequence (5′ to 3′) | Probe size (bp) | Observed migration of extension products (LIZ-120 parameter)a |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | T | G | ||||||||
| I | Typhimurium | F-GAA GCG TTA GTG AGC CGT CTG CG | 643 | 10 | T/C | GCAAAAACCAGATCGTTTTCCG | 22 | — | 27.3 | 29.6 | — |
| (hemD12) | R-ATC AGC GAC CTT AAT ATC TTG CCA | 48 | T/C | CTGGTTTTTGCCCTTTCACAGCACGC | 26 | — | 28.4 | 31.1 | — | ||
| 100 | G/T | CCAGCTCCAGCGGGATGGTCGAAACTGGCCTGTG | 34 | — | — | 39.3 | 35.8 | ||||
| 166 | A/G | CGCACCACGGCGCTCGCCCTTCATACCGTTAGCGGGT TCGAT | 42 | 45.7 | — | — | 44.4 | ||||
| Enteritidis (thrA11) | F-GTC ACG GTG ATC GAT CCG GT | 852 | 192 | A/G/C | GCGTAATGGTGCGAGGGTGAAG | 22 | 33.2 | NA | — | 30.6 | |
| R-CAC GAT ATT GAT ATT AGC CCG | 309 | G/A/C | GTTAAGGTTAGAGATCCCTTTAACCGGCAG | 30 | 35.4 | NA | — | 25.8 | |||
| 369 | C/A/T | AAAACACGCGCCGCCATCCCAATCATCCCTTTCATTCC | 38 | 40.2 | 40.1 | 41.60 | — | ||||
| II | Virchow (thrA14) | As above for thrA11 | As above for thrA11 | 291 | C/T/A/G | TAAGGTTAGAGATCCCTTTAACCGGCAGGTTATCATCGTCGCTGG A | 46 | NA | 48.7 | 50.78 | NA |
| 339 | A/G | CCAATCATCCCTTTCATTCCAGGGCCGGAGACGCTAAACATCGCCATGTT | 50 | 53.6 | — | — | 52.3 | ||||
| Infantis (sucA21) | F-AGC ACC GAA GAG AAA CGC TGC | 666 | 69 | C/T/G/A | CGAACTGACTGCCGCTGAAGGGCTGGAACGTTATCTGGGGGCCAAATTCCCCGG | 54 | NA | 57.52 | 60.2 | 57.5 | |
| R-GGT TGT TGA TAA CGA TAC GTA | 137 | C/T | TTCTCGCTGGAAGGGGGAGATGCTCTGGTACCCATGCTGAAAGAGATGGTTCGCCATG | 58 | — | 61.3 | NA | — | |||
| 438 | C/T | GTGAGCCCGGTGGTGATGGGCTCCGTGCGCGCCCGTCTGGACAGACTGGACGAACCGAGCAG | 62 | — | 65.5 | NA | — | ||||
| Braenderup (purE11) | F-ATG TCT TCC CGC AAT AAT CC | 510 | 81 | A/G | TCACTTGATATCCGTTCTCTTCCGCCGTTTCGGCGAAGCTGAACAGTTTATCGGGGGTGCGATG | 64 | 67.6 | — | — | 66.7 | |
| R-TCA TAG CGT CCC CCG CGG ATC | 171 | C/T | GCAGCGCTTTGTACCGGCACGCCGAGTACCGGGACCAGCGTTTTTGCCGCAATCATTCCCGGCAGGTG | 68 | — | 68.8 | 69.90 | — | |||
| 360 | C/G | CATTCCGGTGGGTACGCTGGCGATCGGTAAAGCCGGTGCCGCTAACGCCGCCCTGCTCGCCGCGCAGATTCT | 72 | — | 75.17 | — | 74.3 | ||||
NA, extension products corresponding to these loci were not included among the study collection; —, nucleotides not covered by extension primers.
Template preparation.
Templates for the MPEA were amplified using the primers shown in Table 3. PCR amplifications were carried out in a total volume of 50 μl and included the following: 1 μl of genomic DNA; 5 μl of mix primer (10 pmol/μl); 5 μl of 10× PCR buffer (Qiagen, Crawley, West Sussex, United Kingdom); 10 μl of 1 mM deoxynucleotide triphosphates (dNTPs; Roche); 2 μl of 25 mM magnesium chloride (MgCl2); and 0.25 units of Hot Taq DNA polymerase (Qiagen). The targets were amplified under the following conditions: initial denaturation of 95°C for 15 min; 35 cycles of denaturation at 94°C for 40 s, annealing at 55°C for 40 s, and extension at 72°C for 1 min; and a final extension step of 72°C for 2 min and a hold at 4°C. The samples were placed on an Eppendorf Mastercycler thermal cycler (Helena Biosciences, Gateshead, United Kingdom). Amplification products were checked and quantified by electrophoresis on a 2% agarose gel.
PCR products were purified to remove unincorporated primers and dNTPs by preparing 7 μl of mix with the following: 1.0 μl (1.0 unit) of shrimp alkaline phosphatase (SAP; USB Corporation), 0.05 μl (0.5 units) of exonuclease I (ExoI; New England BioLabs, Inc.), and 2.0 μl of PCR product. The mix was incubated at 37°C for 1 h, followed by 72°C for 15 min to inactivate enzymes.
Primer extension reactions and postextension treatment.
The multiplex extension reactions were performed by following the manufacturer's recommendations with a 10-μl mix (2 μl treated PCR mix, 5 μl SNaPshot multiplex ready reaction mix from the SNaPshot multiplex kit [Applied Biosystems], and 1 μl of mix extension primer [8 pmol]). The reaction mix was incubated in an Eppendorf Mastercycler thermal cycler (Helena Biosciences) for 25 cycles of 95°C for 10 s, 50°C for 15 s, and 60°C for 30 s. Primer extension products were treated using 1.5 μl of SAP at 37°C for 1 h, followed by 72°C for 15 min to prevent migration of unincorporated fluorescent ddNTPs during electrophoresis.
Analysis of primer extension products.
Primer extension product analysis was carried out on an ABI Prism 3100 genetic analyzer (Applied Biosystems), using dye set E and the SNP36_POP-4 polymer default module (Applied Biosystems). A total of 1.2 μl of the purified primer extension products was mixed with 9.5 μl of Hi-Di formamide (Applied Biosystems) and 0.5 μl of LIZ-120 internal sizing standard (Applied Biosystems). The data were analyzed using GeneScan version 3.5, GeneScan-120 LIZ standard analysis parameter files, and Genotyper software version 3.7 (Applied Biosystems). All resulting data were exported to and analyzed using Microsoft Excel.
RESULTS
MLST analysis of salmonella isolates.
MLST resolved the 152 isolates into 50 STs (Table 1). Good correlation was seen between ST and serovar for all isolates, and some serovars were subgrouped by MLST, generally showing a predominance of one ST. For example, the 28 S. Typhimurium isolates were resolved into STs 19 (n = 25), 34 (n = 1), 313 (n = 1), and 328 (n = 1). In contrast, isolates of S. enterica serovar Newport were more genetically diverse, being represented by STs 45 (n = 3), 118 (n = 2), 156 (n = 1), 166 (n = 1), and 360 (n = 1).
Identification of diagnostic alleles and corresponding SNPs specific for differentiation of salmonella serovars.
Using the approach adopted by Best and colleagues (2, 3), key alleles were identified for differentiation among five Salmonella serovars. The strategy used in our MPEA involved the identification of discriminatory key allele combinations that were predictive for each ST of interest. Where possible, STs were selected that had high positive and negative predictive values and that were the most common in the MLST database (Table 2).
Up to four SNPs were required for precise identification of some serovars, leading to a large collection of oligonucleotides covering the SNPs contained in all five serovars under investigation (Table 3). As a consequence, the MPEA was run with the following two panels of probes: one panel covering S. Typhimurium and S. Enteritidis and the second covering S. Virchow, S. Infantis, and S. Braenderup (Table 3).
The fluorophore attached to the oligonucleotides will affect the migration pattern of the individual sample, in that the observed fragment size may not be the expected fragment size. Therefore, samples were initially tested individually to validate the migration pattern of the individual SNPs prior to multiplexing.
Specificity of the MPEA.
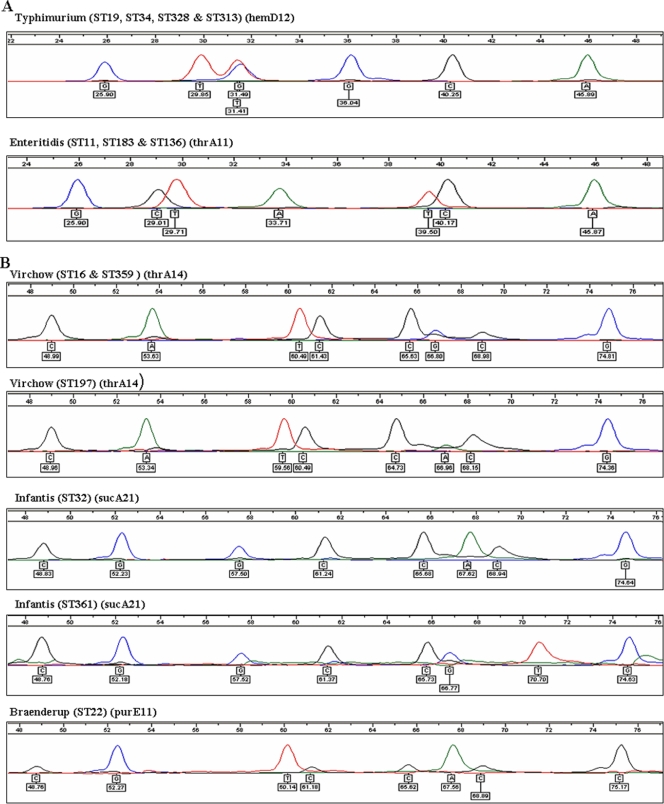
The MPEA was demonstrated to be a reliable and accurate method for identification of the five target Salmonella serovars (Fig. 1). All isolates (n = 152) were tested with both panels of extension primers in order to determine the specificity and sensitivity. The assay was shown to be specific and data obtained were consistent with the corresponding MLST database profiles. The specificity of the assay was 100% (P < 0.0001), following evaluation using isolates representing 33 Kauffmann-White serovars and 50 different STs, with a sensitivity of 100%.
FIG. 1.
Electropherograms of isolates of five Salmonella serovars targeted in the multiplex primer extension assay. (A) Panel I (hemD12 and thrA11); (B) panel II (thrA14, sucA21, and purE11). Colored peaks indicate the nature of the base incorporated at each respective extension site.
DISCUSSION
This paper provides the first report of the detection and identification of five Salmonella serovars by using a multiplex primer extension, or SNaPshot assay, which targets 15 SNPs in four key genes (hemD, thrA, sucA, and purE). SNP typing based on the data generated from MLST analyses has been described for other organisms, including campylobacter, Neisseria meningitidis, and Staphylococcus aureus (27, 29, 30). Such assays represent powerful tools in that the assays give phylogenetically representative information.
We have adopted an approach used previously for phylotyping Brucella species (28) and Escherichia coli (16). We have studied 152 isolates, representing 50 STs and 33 Kauffmann-White Salmonella serovars, and have selected key alleles from the Salmonella MLST database. At the time of writing (July 2009), the Salmonella MLST database had more than 670 STs, representing various Salmonella serovars, with some serovars being represented by more than 1 ST.
The MPEA was separated into panel I (seven SNPs diagnostic for S. Typhimurium and S. Enteritidis) and panel II (eight SNPs specific for S. Virchow, S. Infantis, and S. Braenderup). The extension primers were designed to be of different lengths and could be distinguished by migration times during capillary electrophoresis. The MPEA had 100% specificity and sensitivity for detecting the target Salmonella serovars, and we suggest that the MPEA is useful for molecular serotyping S. enterica isolates.
Our strategy was based on the identification of key allele combinations for the five most prevalent serovars in the United Kingdom. The approach was complicated by the diversity seen in Salmonella MLST alleles, and more than one target was required for identification of each serovar. The key alleles were selected, where possible, to have high positive and negative predictive values and be present with a high frequency within the serovar of interest. For example, with S. Typhimurium and S. Infantis, the key alleles hemD12 and sucA21 had high positive (0.92 and 1.0, respectively) and negative (0.99 and 0.99, respectively) predictive values, which would allow identification of 25 of 29 STs observed for S. Typhimurium in the MLST database and 6 of 8 STs for S. Infantis. Isolates of the above-mentioned serovars that were not recognized by the assay are present in low numbers in the MLST database and were genetically unrelated to the correctly identified isolates, having allelic profiles that placed them in different clonal complexes.
For S. Enteritidis and S. Virchow, the key alleles thrA11 and thrA14 had low positive predictive values (0.61 and 0.69, respectively) and high negative predictive values (0.98 and 0.99, respectively). In this case, the MPEA scheme is predicted to correctly identify the majority of S. Enteritidis (53%) and S. Virchow (82%) STs in the MLST database. However, local epidemiological variations would probably lead to much higher accuracy, with a more select range of STs circulating in any given region (7). In the current study, all S. Enteritidis isolates were correctly identified using the MPEA.
For identification of all S. Enteritidis isolates using the MPEA, additional key alleles targeting STs 460, 77, 335, 334, 6, 172, and 180 will need to be identified, followed by creation of a new primer panel. However, as alluded to above, this would be necessary only if these STs were thought to be present in the locality in which the assay was being performed and if it is feasible given the flexibility and adaptability of the assay.
In summary, the MPEA developed here represents a novel diagnostic tool with utility for detection and identification of the five most prevalent Salmonella serovars in the United Kingdom. The data produced are clear and objective and reflect the phylogeny of salmonella, though further studies with application of the assay to more diverse isolates and more recently discovered STs are necessary. The assay is currently separated into two panels, but it is extremely flexible and could be developed to target any serovar/ST of interest. The MPEA could be used to type (and in some instances, predict the serovars of) organisms that are not able to be identified using conventional serotyping approaches. We also suggest that the preliminary work we have carried out using DNA extracted directly from a small number of clinical and food samples (data not shown) warrants further investigation of what may be a powerful tool for identification of salmonella isolates.
The MPEA technique could be semiautomated, has a high-throughput capability, and can easily and cheaply be performed in a single day. As such, it could have an impact on the care of individual patients or be applied in the wider context of detecting the source of infection or food contamination.
Acknowledgments
E.B.-D. was supported by a studentship from the government of the Great Socialist People's Libyan Arab Jamahiriya.
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Bale, J. A., E. de Pinna, E. J. Threlfall, and L. R. Ward. 2007. Kauffmann-White scheme 2007: Salmonella identification; serotypes and antigenic formulae. Centre for Infections, Health Protection Agency, London, United Kingdom.
- 2.Best, E. L., A. J. Fox, J. A. Frost, and F. J. Bolton. 2004. Identification of Campylobacter jejuni multilocus sequence type ST-21 clonal complex by single-nucleotide polymorphism analysis. J. Clin. Microbiol. 42:2836-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best, E. L., A. J. Fox, J. A. Frost, and F. J. Bolton. 2005. Real-time single-nucleotide polymorphism profiling using Taqman technology for rapid recognition of Campylobacter jejuni clonal complexes. J. Med. Microbiol. 54:919-925. [DOI] [PubMed] [Google Scholar]
- 4.Best, E. L., M. D. Hampton, S. Ethelberg, E. Liebana, F. A. Clifton-Hadley, and E. J. Threlfall. 2009. Drug-resistant Salmonella Typhimurium DT 120: use of PFGE and MLVA in a putative international outbreak. Invest. Microb. Drug Resist. 15:133-138. [DOI] [PubMed] [Google Scholar]
- 5.Brandstatter, A., H. Niederstatter, M. Pavlic, P. Grubwieser, and W. Parson. 2007. Generating population data for the EMPOP database—an overview of the mtDNA sequencing and data evaluation processes considering 273 Austrian control region sequences as example. Forensic Sci. Int. 166:164-175. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, C. H., and J. T. Ou. 1996. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 34:2619-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooke, F. J., D. J. Brown, M. Fookes, D. Pickard, A. Ivens, J. Wain, M. Roberts, R. A. Kingsley, N. R. Thomson, and G. Dougan. 2008. Characterization of the genomes of a diverse collection of Salmonella enterica serovar Typhimurium definitive phage type 104. J. Bacteriol. 190:8155-8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalmasso, A., T. Civera, and M. T. Bottero. 2009. Multiplex primer-extension assay for identification of six pathogenic vibrios. Int. J. Food Microbiol. 129:21-25. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 10.Feder, I., J. C. Nietfeld, J. Galland, T. Yeary, J. M. Sargeant, R. Oberst, M. L. Tamplin, and J. B. Luchansky. 2001. Comparison of cultivation and PCR-hybridization for detection of Salmonella in porcine fecal and water samples. J. Clin. Microbiol. 39:2477-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippini, S., A. Blanco, A. Fernandez-Marmiesse, V. Alvarez-Iglesias, C. Ruiz-Ponte, A. Carracedo, and A. Vega. 2007. Multiplex SNaPshot for detection of BRCA1/2 common mutations in Spanish and Spanish related breast/ovarian cancer families. BMC Med. Genet. 8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foley, S. L., A. M. Lynne, and R. Nayak. 2009. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect. Genet. Evol. 9:430-440. [DOI] [PubMed] [Google Scholar]
- 13.Gerner-Smidt, P., K. Hise, J. Kincaid, S. Hunter, S. Rolando, E. Hyytia-Trees, E. M. Ribot, and B. Swaminathan. 2006. PulseNet USA: a five-year update. Foodborne Pathog. Dis. 3:9-19. [DOI] [PubMed] [Google Scholar]
- 14.Ghebremicael, S. B., J. R. Hasenstein, and S. J. Lamont. 2008. Association of interleukin-10 cluster genes and Salmonella response in the chicken. Poult. Sci. 87:22-26. [DOI] [PubMed] [Google Scholar]
- 15.Hasenstein, J. R., and S. J. Lamont. 2007. Chicken gallinacin gene cluster associated with Salmonella response in advanced intercross line. Avian Dis. 51:561-567. [DOI] [PubMed] [Google Scholar]
- 16.Hommais, F., S. Pereira, C. Acquaviva, P. Escobar-Paramo, and E. Denamur. 2005. Single-nucleotide polymorphism phylotyping of Escherichia coli. Appl. Environ. Microbiol. 71:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 18.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristensen, R., K. G. Berdal, and A. Holst-Jensen. 2007. Simultaneous detection and identification of trichothecene- and moniliformin-producing Fusarium species based on multiplex SNP analysis. J. Appl. Microbiol. 102:1071-1081. [DOI] [PubMed] [Google Scholar]
- 20.Le, T. A., L. Fabre, P. Roumagnac, P. A. Grimont, M. R. Scavizzi, and F. X. Weill. 2007. Clonal expansion and microevolution of quinolone-resistant Salmonella enterica serotype Typhi in Vietnam from 1996 to 2004. J. Clin. Microbiol. 45:3485-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morley, J. M., J. E. Bark, C. E. Evans, J. G. Perry, C. A. Hewitt, and G. Tully. 1999. Validation of mitochondrial DNA minisequencing for forensic casework. Int. J. Legal Med. 112:241-248. [DOI] [PubMed] [Google Scholar]
- 22.Octavia, S., and R. Lan. 2009. Multiple-locus variable-number tandem-repeat analysis of Salmonella enterica serovar Typhi. J. Clin. Microbiol. 47:2369-2376. [DOI] [PMC free article] [PubMed]
- 23.Octavia, S., and R. Lan. 2007. Single-nucleotide-polymorphism typing and genetic relationships of Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 45:3795-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathmanathan, S. G., N. Cardona-Castro, M. M. Sanchez-Jimenez, M. M. Correa-Ochoa, S. D. Puthucheary, and K. L. Thong. 2003. Simple and rapid detection of Salmonella strains by direct PCR amplification of the hilA gene. J. Med. Microbiol. 52:773-776. [DOI] [PubMed] [Google Scholar]
- 25.Pickard, D., N. R. Thomson, S. Baker, J. Wain, M. Pardo, D. Goulding, N. Hamlin, J. Choudhary, J. Threfall, and G. Dougan. 2008. Molecular characterization of the Salmonella enterica serovar Typhi Vi-typing bacteriophage E1. J. Bacteriol. 190:2580-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popoff, M. Y., J. Bockemuhl, F. W. Brenner, and L. L. Gheesling. 2001. Supplement 2000 (no. 44) to the Kauffmann-White scheme. Res. Microbiol. 152:907-909. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, G. A., V. Thiruvenkataswamy, H. Shilling, E. P. Price, F. Huygens, F. A. Henskens, and P. M. Giffard. 2004. Identification and interrogation of highly informative single nucleotide polymorphism sets defined by bacterial multilocus sequence typing databases. J. Med. Microbiol. 53:35-45. [DOI] [PubMed] [Google Scholar]
- 28.Scott, J. C., M. S. Koylass, M. R. Stubberfield, and A. M. Whatmore. 2007. Multiplex assay based on single-nucleotide polymorphisms for rapid identification of Brucella isolates at the species level. Appl. Environ. Microbiol. 73:7331-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens, A. J., F. Huygens, J. Inman-Bamber, E. P. Price, G. R. Nimmo, J. Schooneveldt, W. Munckhof, and P. M. Giffard. 2006. Methicillin-resistant Staphylococcus aureus genotyping using a small set of polymorphisms. J. Med. Microbiol. 55:43-51. [DOI] [PubMed] [Google Scholar]
- 30.Stone, M., K. Bamford, and J. Wain. 2009. Detection of single nucleotide polymorphisms based on the multilocus sequence typing database of Staphylococcus aureus using locked nucleic acid oligonucleotides. J. Med. Microbiol. 58:693-695. [DOI] [PubMed] [Google Scholar]
- 31.Threlfall, E. J., M. D. Hampton, L. R. Ward, and B. Rowe. 1996. Application of pulsed-field gel electrophoresis to an international outbreak of Salmonella Agona. Emerg. Infect. Dis. 2:130-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torpdahl, M., M. N. Skov, D. Sandvang, and D. L. Baggesen. 2005. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J. Microbiol. Methods 63:173-184. [DOI] [PubMed] [Google Scholar]
- 33.Wise, M. G., G. R. Siragusa, J. Plumblee, M. Healy, P. J. Cray, and B. S. Seal. 2009. Predicting Salmonella enterica serotypes by repetitive sequence-based PCR. J. Microbiol. Methods 76:18-24. [DOI] [PubMed] [Google Scholar]