Abstract
The molecular epidemiology of multidrug-resistant Acinetobacter baumannii was investigated in two intensive care units of the V. Monaldi university hospital in Naples, Italy, from May 2006 to December 2007. Genotype analysis by pulsed-field gel electrophoresis (PFGE), trilocus sequence-based typing (3LST), and multilocus sequence typing (MLST) of A. baumannii isolates from 71 patients identified two distinct genotypes, one assigned to PFGE group A, 3LST group 1, and ST2 in 14 patients and the other to PFGE group B, 3LST group 6, and ST78 in 71 patients, that we named ST2/A and ST78/B, respectively. Of these, ST2/A corresponded to European clone II identified in the same hospital during 2003 and 2004; ST78/B was a novel genotype that was isolated for the first time in May 2006 but became prevalent during 2007. The ST78/B profile was also identified in five patients from two additional hospitals in Naples during 2007. The ST2/A and ST78/B isolates were resistant to all antimicrobials tested, including carbapenems, but were susceptible to colistin. Both ST2/A and ST78/B isolates possessed a plasmid-borne carbapenem-hydrolyzing oxacillinase gene, blaOXA-58, flanked by ISAba2 and ISAba3 elements at the 5′ and 3′ ends, respectively. The selection of the novel ST78/B A. baumannii clone might have been favored by the acquisition of the blaOXA-58 gene.
Acinetobacter baumannii is an emerging opportunistic nosocomial pathogen, with increasing prevalence worldwide, responsible for a variety of nosocomial infections, especially in intensive care unit (ICU) patients (10, 15). Several hospital outbreaks caused by the selection of multiresistant A. baumannii clones have been described in Europe and worldwide (1, 10, 15, 27). Genotypic characterization of epidemic A. baumannii isolates through amplified fragment length polymorphism analysis has identified clusters of highly similar strains, which were assumed to represent distinct clonal lineages and were defined as European clones I, II, and III (9, 24). Similarly, three distinct groups were recently identified among A. baumannii isolates from five different countries by sequence-based typing (ST): group 1, corresponding to European clone II; group 2, corresponding to European clone I; and group 3, corresponding to European clone III (22). Moreover, epidemics caused by A. baumannii genotypes assigned to novel ST groups 4 and 5 have been recently described in different Greek and Turkish cities (11). The majority of the outbreaks occurring in Europe were caused by carbapenem-resistant strains that carried the blaOXA-58 gene or a distinct carbapenem-hydrolyzing oxacillinase (CHDL) gene (4, 8, 11-13, 16, 20, 27, 28).
We have previously reported the occurrence of two sequential outbreaks from August 1999 to February 2001 and from January 2002 to December 2002 along with the emergence of carbapenem-resistant A. baumannii in the ICU of the Federico II university hospitals in Naples, Italy, during 2002 (26). More recently, we have shown that the same epidemic A. baumannii clone isolated during 2002 was responsible for a large and sustained outbreak in the V. Monaldi tertiary care teaching hospital of Naples between June 2003 and June 2004 (25). An increase in the number of cases of A. baumannii infection was observed after 2 years in the V. Monaldi hospital. The objectives of the present study were (i) to investigate the molecular epidemiology of A. baumannii in the V. Monaldi hospital, (ii) to study the genetic characteristics of A. baumannii isolates responsible for the epidemic, and (iii) to analyze the antimicrobial susceptibilities of the A. baumannii isolates and their mechanisms of resistance.
MATERIALS AND METHODS
Setting and study period.
The V. Monaldi hospital is a 600-bed tertiary care teaching hospital serving approximately 20,000 admissions per year. The hospital is provided with five ICUs: a neonatal ICU, a coronary ICU, a cardiac surgery ICU, a general and specialist surgery ICU (namely, a postoperative ICU [PO-ICU]), and a cardiorespiratory ICU (CR-ICU). The PO-ICU and CR-ICU are located in a recently renovated area of the hospital and are connected by a short internal corridor, and each has eight beds and an isolation box. Although spatially very close, the two wards have distinct staffs and medical equipment. Patients admitted to the PO-ICU are inpatients undergoing major elective surgery, while the CR-ICU admits both inpatients requiring intensive care and outpatients from other municipal or regional ICUs. The present study analyzed all 71 available A. baumannii isolates from 71 patients in the CR-ICU and PO-ICU wards between May 2006 and December 2007.
Microbiological surveillance and epidemiological data.
Patient microbiological screening is routinely performed at admission to the CR-ICU and PO-ICU; further specimens are collected during patients' stays upon clinical judgment. Moreover, starting from January 2007 monthly reporting to the local infection control team of all microbiological isolations in high-risk areas was implemented. Analysis of data for the first part of the study period (May 2006 to December 2006) was performed retrospectively. Epidemiological data for the 76 A. baumannii-positive patients in the V. Monaldi, Cotugno, and A. Cardarelli hospitals (age, gender, primary diagnosis, infectious comorbidities, and outcome) were retrospectively collected from hospital discharge cards. A. baumannii-associated mortality was defined as death occurring during A. baumannii infection. Epidemiological and microbiological data were analyzed using SPSS v.11.0 (SPSS Inc., Chicago, IL) by means of Student's t test or Pearson's chi-square test as appropriate. Results were considered to be statistically significant at P < 0.05.
Bacterial strains and microbiological methods.
A. baumannii isolates were obtained from clinical specimens by standard methods, followed by isolation in pure culture on MacConkey agar plates, and were stored at −80°C in nutrient broth containing 20% (vol/vol) glycerol. Strains were originally identified as Acinetobacter baumannii-A. calcoaceticus complex by using the Vitek 2 automatic system with the ID-GNB card for identification of Gram-negative bacilli (bioMerieux, Marcy-l'Etoile, France). A. baumannii species identification was confirmed by amplification of the blaOXA-51-like gene and PCR amplification and sequence analysis of the 16S-23S rRNA intergenic spacer region (6, 22).
Antimicrobial susceptibilities.
MICs were determined by a microdilution method according to Clinical and Laboratory Standards Institute document M7-A6 (7). Breakpoint values were those recommended by the CLSI (7). Breakpoints for colistin were those from the British Society for Antimicrobial Chemotherapy (BSAC) (5). Etest MBL strips (AB Biodisk, Solna, Sweden) were used to evaluate the presence of metallo-beta-lactamase (MBL) activity according to the manufacturer's procedure. The role of oxacillinase production in carbapenem resistance was assessed by determining carbapenem MICs by microdilution in the presence and absence of 200 mM NaCl, as described previously (28).
PFGE and dendrogram analysis.
ApaI DNA macrorestriction and pulsed-field gel electrophoresis (PFGE) and dendrogram analysis of A. baumannii isolates were performed as previously reported (25). Because A. baumannii isolates were epidemiologically related, interpretation of genomic relatedness was performed using the criteria of Tenover et al. (19).
Identification of PCR-based sequence groups and ST.
Multiplex PCRs and sequence-based typing (ST) were performed as previously described (22). Assignment of novel alleles and ST types was performed using the bioinformatic tools at the Health Protection Agency website on A. baumannii sequence typing that has been developed and maintained by J. F. Turton and R. Meyers (http://www.hpa-bioinformatics.org.uk/AB/home.php).
MLST.
Multilocus sequence typing (MLST) analysis was performed using the Institut Pasteur's MLST scheme, publicly available from the MLST website at http://www.pasteur.fr/mlst. This MLST scheme is based on sequencing of an internal portion of the seven genes encoding 60-kDa chaperonin (cpn60), protein elongation factor EF-G (fusA), citrate synthase (gltA), CTP synthase (pyrG), homologous recombination factor (recA), 50S ribosomal protein L2 (rpIB), and RNA polymerase subunit B (rpoB). Primer pairs for three of these genes (cpn60, gltA, and recA) were previously designed by Bartual et al. (3). Primer pairs for three other genes (fusA, pyrG, and rplB) are derived from primers initially proposed by Santos and Ochman (18). Finally, primers for gene rpoB were designed previously (17). PCR conditions were 35 cycles (denaturation at 94°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 1 min) preceded by a 3-min denaturation at 94°C and followed by a 5-min extension at 72°C. Further details on this MLST scheme can be found at www.pasteur.fr/mlst.
PCR analysis of carbapenemase genes and ISs.
PCR analysis for carbapenemase-encoding genes in Acinetobacter spp. (blaIMP, blaVIM, blaSIM, blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58) was performed as previously described (28). PCR characterization of the blaOXA-58-surrounding insertion sequence (IS) was performed as described previously (16). The colinearity between IS elements and the blaOXA-66 or blaOXA-90 gene was analyzed using primers for IS elements described previously by Poirel and Nordmann (16) and for the blaOXA-51-like gene described previously by Turton et al. (23).
Plasmid analysis.
Plasmid DNA preparations were performed by using the QIAfilter plasmid purification maxikit adapted for low-copy-number plasmids (Qiagen Corporation, Milan, Italy) according to the manufacturer's procedure. HindIII-generated fragments were separated on 1% agarose gels, transferred onto nylon membranes, and hybridized with PCR-generated probes specific for blaOXA-58.
Mating experiments.
Filter mating was performed using A. baumannii isolates of PFGE type A or B, resistant to imipenem and susceptible to rifampin, and Acinetobacter genomic species 3 strain 4442 (13), susceptible to imipenem but resistant to rifampin, as donor and recipient cells, respectively. Transconjugants were selected on brain heart infusion (BHI) agar plates containing imipenem (16 mg/liter) plus rifampin (100 mg/liter). The frequency of transfer was calculated as the number of transconjugants divided by the number of surviving recipients.
DNA sequencing and computer analysis of sequencing data.
DNA sequences of plasmid pABNA1 and pABNA2 were amplified using primers 5′-GTCACGCCAGTATTAACCAA-3′ and 5′-TCGTTTACCCCAAACATAAGC-3′, spanning plasmid oriV and ISAba3, respectively. DNA sequencing of PCR products was performed using the ABI Prism BigDye Terminator v3.1 ready reaction cycle sequencing kit and the 3730 DNA analyzer (Applied Biosystems, Foster City, CA). DNA sequences were assembled using the program Autoassembler version 1.4 (Applied Biosystems, Foster City, CA) and annotated using the BLAST program (2) and the sequence annotation tools integrated into the Sequin program version 7.9 available at http://www.ncbi.nlm.nih.gov/Sequin/index.html.
Nucleotide sequence accession numbers.
The nucleotide sequences of the novel alleles identified for A. baumannii isolates of ST group 6 have been deposited in the GenBank nucleotide database under accession numbers EU433384 (ompA allele 7), EU433383 (csuE allele 10), and EU433382 (blaOXA-51-like allele 9 corresponding to blaOXA-90). The nucleotide plasmid sequences pABNA1and pABNA2, amplified from isolates 3979 (PFGE type A) and 3957 (PFGE type B), have been assigned accession numbers GQ338082 and GQ338083, respectively, in the GenBank nucleotide database. Allele sequences of ST2 and ST78 are available from the Institut Pasteur's A. baumannii MLST website at www.pasteur.fr/mlst.
RESULTS
Molecular epidemiology of A. baumannii in the hospital.
We have recently described an A. baumannii outbreak between June 2003 and June 2004 in the V. Monaldi hospital of Naples, Italy (25). During the two subsequent years, only a few sporadic cases of A. baumannii infection were detected in the hospital. The epidemiology of A. baumannii was studied in the CR-ICU and PO-ICU of the hospital between May 2006 and December 2007, when a further increase of the number of A. baumannii isolates was observed in the two wards, while no isolates were obtained from other wards. During the study period, a total of 1,760 patients were admitted to the two wards (514 and 1,240 to the CR-ICU and PO-ICU, respectively). A. baumannii was isolated from 101 patients, with an overall A. baumannii isolation rate of 5.7% (14.6% and 2.1% for the CR-ICU and PO-ICU, respectively; P < 0.05). A. baumannii was isolated in 75 CR-ICU patients who showed a mean number of positive specimens of 2.17 ± 1.58. In the PO-ICU 26 patients proved to be positive for A. baumannii, with a mean number of positive specimens of 2.08 ± 1.35 (P > 0.05). Mean length of stay in the two wards was calculated and proved to be 10.98 ± 22.32 and 2.22 ± 2.94 days for the CR-ICU and PO-ICU, respectively (P < 0.05).
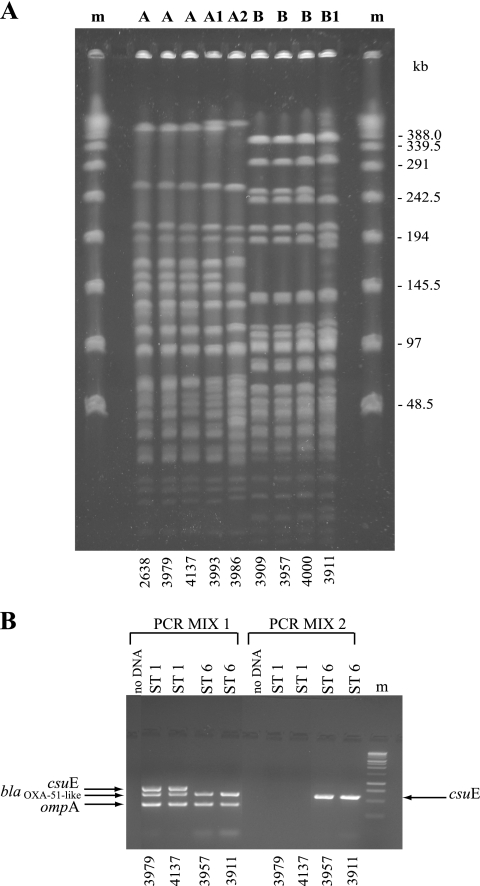
To investigate whether the increase in the frequency of isolation of A. baumannii during the study period was caused by the spread of epidemic clones, all 71 available, nonrepetitive A. baumannii isolates from 71 patients between May 2006 and December 2007 were genotyped: 54 were from CR-ICU patients (73.3% of patients) and 17 were from PO-ICU patients (61.5% of patients). One Acinetobacter isolate was not included in the study because its species identification as A. baumannii was not confirmed by molecular methods. Molecular typing by PFGE identified two major PFGE groups, which differed in the migration of more than six bands, in 14 and 57 isolates, which we named A and B, respectively. Of the 14 isolates of PFGE group A, 12 showed an identical macrorestriction pattern (type A) whereas two differed in the migration of at least two and three DNA fragments and were classified into subtypes A1 and A2, respectively. Fifty-six isolates of PFGE group B showed an identical macrorestriction pattern (type B),whereas one differed in the migration of up to six bands and was designated subtype B1 (Fig. 1A and Table 1). The epidemic PFGE profile A was identical to that of the epidemic A. baumannii strain 2638 isolated in the V. Monaldi hospital in 2004 (25) (Fig. 1A). Genotype analysis using the multiplex PCRs and trilocus sequence-based typing (3LST) approach described by Turton et al. (22) assigned all 14 isolates of PFGE group A to previously defined three-locus sequence type group 1 (Fig. 1B and Table 1). The multiplex PCR approach identified a distinct PCR pattern in the other 57 isolates of PFGE group B with the amplification of blaOXA-51-like and ompA but not csuE alleles in PCR mix 1 and amplification of csuE but not blaOXA-51-like and ompA alleles in PCR mix 2 (Fig. 1B). Trilocus sequence-based typing identified an identical allele profile, 7/10/9, at ompA/csuE/blaOXA-51-like loci in the 57 isolates of PFGE group B that were assigned to novel 3LST group 6 (Table 1). The differences in group-specific PCR patterns facilitated rapid identification of the A. baumannii isolates belonging to different ST groups, 1 and 6, in the hospital and were used as a preliminary typing approach for the isolates. MLST based on the conserved regions of cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB housekeeping genes identified ST2 for isolates of PFGE type A and subtypes A1 and A2 (group A), allelic profile 25/3/6/2/28/1/29, which corresponds to a novel ST assigned as ST78 for isolates of PFGE type B and subtype B1 (group B), respectively (Table 1). Based on all the above data, two distinct genotypes were identified, one assigned to PFGE group A (including PFGE type A and subtypes A1 and A2), 3LST group 1, and ST2, and the other assigned to PFGE group B (which included PFGE type B and subtype B1), 3LST group 6, and ST78, which we named ST2/A and ST78/B, respectively. The differences in PFGE patterns observed between subtypes A1 and A2 and type A and between subtype B1 and type B were probably due to the higher discriminatory power of PFGE than of MLST and were not further considered.
FIG. 1.
(A) ApaI PFGE profiles of representative A. baumannii strains included in the study. Capital letters above the lanes indicate PFGE types identified; m, phage lambda DNA molecular mass markers. The isolate number is shown below each lane. Sizes of lambda DNA molecular mass markers are shown on the right. (B) Multiplex PCR to selectively amplify ompA, csuE, and blaOXA-51-like alleles. ST groups identified are indicated above the lanes; m, 1-kb DNA ladder molecular mass markers (Promega, Milan, Italy). The isolate number is shown below each lane.
TABLE 1.
Epidemiological, phenotypic and genotypic features of A. baumannii strains included in the studya
| Hospital | Strain | Culture date (day/mo/yr) | Ward | Age (yr)/ gender | Primary diagnosis | Infectious comorbidity | Outcome | Isolate source | PFGE type | 3LST group | MLST ST | MIC (mg/liter) of antibiotic: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | SAM | AK | GM | RIF | COL | ||||||||||||
| Monaldi | 3978 | 15/05/2006 | CR-ICU | 61/M | Respiratory failure | Pneumonia | E | BA | B | 6 | 78 | 32 | 16 | 125 | 125 | 32 | 4 | ≤0.5 |
| 3979 | 29/05/2006 | CR-ICU | 85/F | Respiratory failure | Pneumonia | VD | BA | A | 1 | 2 | 32 | 8 | 32 | 125 | 8 | 4 | ≤0.5 | |
| 3980 | 05/06/2006 | CR-ICU | 67/F | Respiratory failure | Pneumonia* | E | BA | B | 6 | 78 | 16 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | |
| 3982 | 09/06/2006 | PO-ICU | 81/F | Acute myocardial infarction | E | BA | A | 1 | 2 | 16 | 16 | 32 | 125 | 8 | 2 | ≤0.5 | ||
| 3993 | 19/06/2006 | CR-ICU | 83/F | Pneumonia | E | BA | A1 | 1 | 2 | 16 | 4 | 32 | 125 | 8 | 2 | ≤0.5 | ||
| 3983 | 28/06/2006 | CR-ICU | 67/M | Lung cancer | Sepsis | E | BC | A | 1 | 2 | 16 | 4 | 125 | 4 | 8 | >500 | ≤0.5 | |
| 3991 | 17/07/2006 | CR-ICU | 79/F | Pneumonia* | T | PS | A | 1 | 2 | 16 | 4 | 32 | 4 | 8 | 4 | ≤0.5 | ||
| 3994 | 17/07/2006 | CR-ICU | 75/M | Aortic dissection | E | BA | B | 6 | 78 | 8 | 4 | 125 | 125 | 32 | 2 | ≤0.5 | ||
| 3984 | 17/07/2006 | CR-ICU | 81/F | Respiratory failure | D | BA | A | 1 | 2 | 16 | 8 | 32 | 32 | 8 | 4 | ≤0.5 | ||
| 4137 | 31/07/2006 | CR-ICU | 67/F | Respiratory failure | Pneumonia | E | BA | A | 1 | 2 | 8 | 4 | 125 | 125 | 256 | >500 | ≤0.5 | |
| 3987 | 03/08/2006 | CR-ICU | 47/F | Respiratory failure | Pneumonia | E | BA | B | 6 | 78 | 16 | 4 | 64 | 125 | 32 | 4 | ≤0.5 | |
| 3985 | 14/08/2006 | CR-ICU | 67/M | Pneumonia | E | BA | B | 6 | 78 | 16 | 16 | 125 | 16 | 4 | 2 | 1 | ||
| 3992 | 21/08/2006 | CR-ICU | 50/F | Respiratory failure | E | BA | A | 1 | 2 | 32 | 8 | 32 | 125 | 8 | 4 | ≤0.5 | ||
| 3990 | 23/08/2006 | CR-ICU | 76/M | Pneumonia | T | CVC | A | 1 | 2 | 32 | 16 | 64 | 16 | 4 | >500 | ≤0.5 | ||
| 3989 | 28/08/2006 | CR-ICU | 59/M | Esophageal cancer | Pneumonia* | E | BA | B | 6 | 78 | 16 | 8 | 125 | 125 | 256 | 2 | 1 | |
| 4138 | 12/09/2006 | PO-ICU | 68/M | Lung cancer | E | BA | A | 1 | 2 | 8 | 8 | 32 | 16 | 4 | >500 | ≤0.5 | ||
| 3986 | 06/11/2006 | CR-ICU | 19/M | Respiratory failure | E | CT | A2 | 1 | 2 | 16 | 4 | 32 | >250 | 8 | 4 | ≤0.5 | ||
| 3988 | 06/11/2006 | CR-ICU | 76/F | Respiratory failure | D | BA | A | 1 | 2 | 32 | 4 | 32 | 125 | 8 | 4 | ≤0.5 | ||
| 3956 | 05/01/2007 | PO-ICU | 26/M | Respiratory failure | Sepsis | E | BC | B | 6 | 78 | 32 | 8 | 64 | 125 | 32 | 4 | ≤0.5 | |
| 4155 | 08/01/2007 | PO-ICU | 81/M | Abdominal aortic aneurysm | Vascular prosthesis infection* | E | BA | A | 1 | 2 | 16 | 16 | 125 | 16 | 64 | >500 | ≤0.5 | |
| 4156 | 08/01/2007 | PO-ICU | 68/M | Larynx cancer | D | BA | B | 6 | 78 | 8 | 8 | 125 | >250 | 8 | 6 | ≤0.5 | ||
| 3958 | 15/01/2007 | CR-ICU | 81/M | Respiratory failure | E | BA | B | 6 | 78 | 32 | 8 | 125 | 32 | 32 | 16 | ≤0.5 | ||
| 4157 | 15/01/2007 | PO-ICU | 66/M | Colorectal cancer | VD | BA | B | 6 | 78 | 16 | 8 | 125 | >250 | 32 | 4 | ≤0.5 | ||
| 3957 | 18/01/2007 | PO-ICU | 44/M | Bronchiectasis | Lung abscess | D | BA | B | 6 | 78 | 16 | 8 | 125 | 32 | 32 | 4 | ≤0.5 | |
| 3942 | 24/01/2007 | PO-ICU | 81/F | Respiratory failure | VD | BA | B | 6 | 78 | 16 | 8 | 125 | 125 | 32 | 16 | ≤0.5 | ||
| 3997 | 29/01/2007 | CR-ICU | 85/F | Respiratory failure | E | BA | B | 6 | 78 | 16 | 8 | 64 | 16 | 32 | 2 | ≤0.5 | ||
| 4158 | 03/02/2007 | PO-ICU | 56/M | Lung cancer | E | CVC | A | 1 | 2 | 16 | 16 | 64 | >250 | 8 | >500 | ≤0.5 | ||
| 3944 | 12/02/2007 | PO-ICU | 65/F | Intestinal occlusion | E | BA | B | 6 | 78 | 16 | 8 | 125 | >250 | 32 | 4 | ≤0.5 | ||
| 3945 | 16/02/2007 | PO-ICU | 39/M | Aortic dissection | T | BA | B | 6 | 78 | 8 | 8 | 64 | 32 | 64 | 4 | ≤0.5 | ||
| 3960 | 23/02/2007 | CR-ICU | 45/M | Pneumocystosis* | Sepsis | E | BC | B | 6 | 78 | 16 | 8 | 125 | 32 | 32 | 2 | ≤0.5 | |
| 3998 | 03/03/2007 | CR-ICU | 52/F | Respiratory failure | Sepsis | E | BC | B | 6 | 78 | 16 | 8 | 125 | >250 | 32 | 4 | ≤0.5 | |
| 3946 | 13/03/2007 | PO-ICU | 75/M | Unspecified hemorrhage | E | WS | B | 6 | 78 | 8 | 8 | 125 | >250 | 32 | 4 | ≤0.5 | ||
| 3999 | 19/03/2007 | CR-ICU | 66/F | Mediastinitis | E | BA | B | 6 | 78 | 8 | 4 | 64 | 125 | 32 | 4 | ≤0.5 | ||
| 3961 | 21/03/2007 | CR-ICU | 77/M | Aortic dissection | E | BA | B | 6 | 78 | 8 | 8 | 125 | 125 | 64 | 2 | ≤0.5 | ||
| 4000 | 21/03/2007 | CR-ICU | 62/M | Respiratory failure | E | BA | B | 6 | 78 | 16 | 16 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 3962 | 10/04/2007 | CR-ICU | 71/F | Respiratory failure | E | BA | B | 6 | 78 | 32 | 4 | 125 | 32 | 32 | 2 | ≤0.5 | ||
| 4001 | 30/04/2007 | CR-ICU | 78/M | Respiratory failure | D | BA | B | 6 | 78 | 32 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 4002 | 30/04/2007 | CR-ICU | 75/F | Pneumonia | E | BA | B | 6 | 78 | 8 | 4 | 125 | 32 | 64 | 4 | ≤0.5 | ||
| 3963 | 04/05/2007 | CR-ICU | 76/F | Respiratory failure | E | BA | B | 6 | 78 | 16 | 4 | 125 | 32 | 32 | 4 | ≤0.5 | ||
| 3947 | 08/05/2007 | PO-ICU | 75/M | Bladder cancer | D | UC | B | 6 | 78 | 16 | 4 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 4139 | 14/05/2007 | CR-ICU | 57/F | Respiratory failure | E | CT | B | 6 | 78 | 16 | 8 | 64 | 125 | 32 | 4 | ≤0.5 | ||
| 3948 | 26/05/2007 | PO-ICU | 64/M | Colorectal cancer | Sepsis | E | BC | B | 6 | 78 | 8 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | |
| 3969 | 04/06/2007 | CR-ICU | 74/M | Wegener's granulomatosis | E | BA | B | 6 | 78 | 8 | 8 | 64 | 16 | 32 | 4 | ≤0.5 | ||
| 4140 | 05/06/2007 | CR-ICU | 79/F | Respiratory failure | E | BA | B | 6 | 78 | 8 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 3970 | 19/06/2007 | CR-ICU | 52/F | Acute myocardial infarction | Pneumonia | E | BA | B | 6 | 78 | 16 | 8 | 64 | 16 | 64 | 4 | ≤0.5 | |
| 4141 | 21/06/2007 | CR-ICU | 49/M | Respiratory failure | T | BA | B | 6 | 78 | 16 | 4 | 64 | 16 | 32 | 4 | ≤0.5 | ||
| 3971 | 25/06/2007 | CR-ICU | 68/F | Aortic dissection | Sepsis | VD | BC | B | 6 | 78 | 32 | 8 | 125 | 125 | 64 | 4 | ≤0.5 | |
| 4142 | 28/06/2007 | CR-ICU | 76/M | Respiratory failure | E | BA | B | 6 | 78 | 8 | 4 | 125 | 16 | 32 | 4 | ≤0.5 | ||
| 4143 | 02/07/2007 | CR-ICU | 54/M | Amyotrophic lateral sclerosis | Pneumonia | D | BA | B | 6 | 78 | 8 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | |
| 3972 | 16/07/2007 | CR-ICU | 85/M | Respiratory failure | Pneumonia | T | BA | B | 6 | 78 | 8 | 8 | 125 | >250 | 32 | 4 | ≤0.5 | |
| 4145 | 23/07/2007 | CR-ICU | 90/M | Acute myocardial infarction | E | BA | B | 6 | 78 | 16 | 8 | 125 | 125 | 32 | 8 | ≤0.5 | ||
| 4144 | 24/07/2007 | CR-ICU | 70/F | Respiratory failure | E | BA | B | 6 | 78 | 8 | 8 | 64 | 16 | 32 | 4 | ≤0.5 | ||
| 4136 | 30/07/2007 | CR-ICU | 84/F | Respiratory failure | E | BA | B | 6 | 78 | 16 | 4 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 3966 | 01/08/2007 | CR-ICU | 51/M | Renal ptosis | Sepsis | T | BC | B | 6 | 78 | 32 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | |
| 4146 | 02/08/2007 | CR-ICU | 53/F | Respiratory failure | E | BA | B | 6 | 78 | 8 | 4 | 125 | 16 | 32 | 4 | ≤0.5 | ||
| 4147 | 04/08/2007 | CR-ICU | 70/M | Respiratory failure | E | BA | B | 6 | 78 | 32 | 16 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 3967 | 03/09/2007 | CR-ICU | 77/F | Respiratory failure | T | BA | B | 6 | 78 | 8 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 4148 | 03/09/2007 | CR-ICU | 70/M | Respiratory failure | E | BA | B | 6 | 78 | 16 | 8 | 125 | >250 | 32 | 2 | ≤0.5 | ||
| 4149 | 05/09/2007 | CR-ICU | 58/F | Respiratory failure | Pneumonia | E | BA | B | 6 | 78 | 8 | 4 | 125 | 125 | 32 | 4 | ≤0.5 | |
| 4150 | 10/09/2007 | CR-ICU | 76/F | Respiratory failure | E | BA | B | 6 | 78 | 16 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 3974 | 12/09/2007 | CR-ICU | 72/M | Respiratory failure | Sepsis | E | BC | B | 6 | 78 | 8 | 4 | 125 | 16 | 32 | 4 | ≤0.5 | |
| 4151 | 01/10/2007 | CR-ICU | 77/M | Respiratory failure | E | BA | B | 6 | 78 | 16 | 16 | 64 | 125 | 32 | 4 | ≤0.5 | ||
| 4152 | 12/10/2007 | CR-ICU | 63/M | Respiratory failure | E | BA | B | 6 | 78 | 8 | 4 | 125 | >250 | 8 | 4 | ≤0.5 | ||
| 3950 | 12/10/2007 | PO-ICU | 72/M | Respiratory failure | E | BA | B | 6 | 78 | 8 | 4 | 64 | 16 | 32 | 4 | ≤0.5 | ||
| 3911 | 24/10/2007 | CR-ICU | 74/M | Respiratory failure | Sepsis | VD | BC | B1 | 6 | 78 | 32 | 8 | 125 | >250 | 64 | 4 | ≤0.5 | |
| 4153 | 03/11/2007 | CR-ICU | 69/F | Respiratory failure | E | BA | B | 6 | 78 | 8 | 8 | 125 | 125 | 64 | 4 | ≤0.5 | ||
| 3909 | 05/11/2007 | CR-ICU | 44/F | Respiratory failure | Pneumonia* | E | BA | B | 6 | 78 | 32 | 8 | 125 | 2 | 32 | 4 | ≤0.5 | |
| 3951 | 14/11/2007 | PO-ICU | 49/M | Pancreatic cancer | E | BA | B | 6 | 78 | 16 | 8 | 125 | 2 | 256 | 2 | ≤0.5 | ||
| 4154 | 19/11/2007 | CR-ICU | 62/M | Respiratory failure | Pneumonia | E | BAL | B | 6 | 78 | 16 | 8 | 125 | 2 | 32 | 4 | ≤0.5 | |
| 3952 | 24/11/2007 | PO-ICU | 79/M | Abdominal aortic aneurysm | Sepsis | E | BC | B | 6 | 78 | 16 | 8 | 125 | 2 | 32 | 4 | ≤0.5 | |
| 3912 | 22/10/2007 | CR-ICU | 45/M | Acute myocardial infarction | E | BA | B | 6 | 78 | 8 | 8 | 125 | >250 | 32 | 4 | ≤0.5 | ||
| Cotugno | 3678 | 07/04/2007 | ICU | 60/M | Respiratory failure | Pneumonia | D | BA | B | 6 | 78 | 32 | 16 | 125 | 125 | 4 | 4 | ≤0.5 |
| 3679 | 24/05/2007 | ICU | 75/M | Cholangiocarcinoma | Sepsis | D | BC | B | 6 | 78 | 16 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | |
| 3701 | 06/06/2007 | ICU | 60/F | Sepsis | E | BC | B | 6 | 78 | 16 | 8 | 125 | 125 | 32 | 4 | ≤0.5 | ||
| 3696 | 03/10/2007 | ICU | 60/F | Tetanus* | D | CVC | B | 6 | 78 | 16 | 8 | 125 | >250 | 64 | 4 | ≤0.5 | ||
| Cardarelli | 3933 | 04/02/2007 | ICU | 19/M | Polytrauma | Sepsis | D | BC | B | 6 | 78 | 1 | 0.5 | 125 | 32 | 32 | 4 | ≤0.5 |
Abbreviations: CR-ICU, cardiorespiratory intensive care unit; PO-ICU, postoperative intensive care unit; ICU, intensive care unit; F, female; M, male; PFGE, pulsed-field gel electrophoresis; 3LST, trilocus sequence-based typing; MLST, multilocus sequence typing; ST, sequence type; E, exitus; VD, voluntary discharge; T, transferred; D, discharge; BA, bronchial aspirate; BAL, bronchoalveolar lavage; BC, blood culture; CVC, central venous catheter; CT, catheter tip; UC, urine culture; PS, pharyngeal swab; WS, wound swab; IPM, imipenem; MEM, meropenem; SAM, sulbactam-ampicillin; AK, amikacin; GM, gentamicin; RIF, rifampin; COL, colistin. Infections not attributable to A. baumannii are indicated by an asterisk.
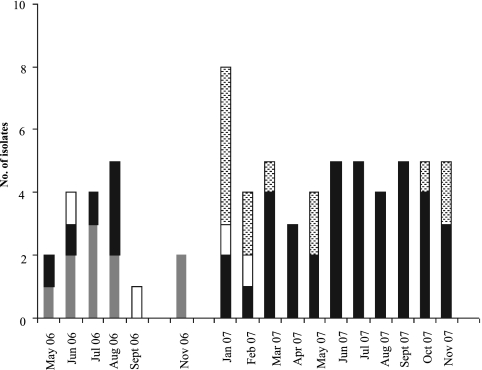
Molecular epidemiology of A. baumannii isolates showed that the outbreak in the two wards of the V. Monaldi hospital was caused by the spread of ST2/A and ST78/B genotypes, which were isolated in two consecutive temporal clusters, one clone predominating over the other. In fact, ST2/A was identified in 10 CR-ICU patients and in 4 PO-ICU patients, while ST78/B was found in 44 CR-ICU patients and in 13 PO-ICU patients. Moreover, ST2/A isolates occurred between May 2006 and February 2007 in both wards, while ST78/B isolates were first identified in the CR-ICU ward in May 2006 and predominated in both wards from March 2007 onward (Fig. 2). The lower respiratory tract was the most frequent site of isolation (9 of 14 and 45 of 57 patients for ST2/A and ST78/B, respectively) (P > 0.05) and was associated with clinical infection as the primary diagnosis or infectious comorbidity in 3 of 10 and 11 of 44 patients for ST2/A and ST78/B, respectively (P > 0.05). Nine A. baumannii isolates from blood were assigned to ST78/B, and one was assigned to ST2/A (P > 0.05), and they were always associated with clinical infection. ST78/B was also isolated from one wound swab, the urinary tract of one patient, and one catheter tip, while ST2/A was also isolated from the upper respiratory tract of one patient and the central venous catheter and the catheter tip of two patients and one patient, respectively. The mean ages of patients infected by ST2/A and ST78/B were 75.5 ± 9.84 and 59.79 ± 14.42 years, respectively (P > 0.05). The female/male ratios for patients infected by ST2/A and ST78/B were 3:1 and 0.46:1, respectively (P > 0.05). Crude mortality and A. baumannii-associated mortality were 64% (9/14) and 21% (3/14), respectively, for patients with isolation of ST2/A A. baumannii and 75% (43/57) and 25% (14/57), respectively, for patients with isolation of ST78/B A. baumannii (Table 1).
FIG. 2.
Molecular epidemiology of A. baumannii in the V. Monaldi hospital, Naples, Italy, during 2006 and 2007. Gray and white bars represent ST2/A isolates from the cardiorespiratory ICU (CR-ICU) and postoperative ICU (PO-ICU), respectively; black and dotted bars represent ST78/B isolates from the CR-ICU and PO-ICU, respectively.
Five additional multidrug-resistant (MDR) A. baumannii strains with the ST78/B profile were isolated in the ICU wards of the Cotugno and A. Cardarelli hospitals in Naples (4 isolates and 1 isolate, respectively) during 2007 (Table 1).
Antimicrobial susceptibility patterns of A. baumannii isolates.
Both ST2/A and ST78/B A. baumannii isolates from the V. Monaldi hospital showed a multidrug-resistant antibiotype. In particular, they were resistant to ampicillin-sulbactam, piperacillin-tazobactam, broad-spectrum cephalosporins, fluoroquinolones, and aminoglycosides; intermediate or resistant to imipenem and rifampin; and intermediate to meropenem but were susceptible to colistin sulfate (Table 2). Interestingly, 5 isolates with the ST2/A profile showed high-level resistance to rifampin (Tables 1 and 2). All four A. baumannii isolates with the ST78/B profile from the Cotugno hospital in Naples showed antimicrobial susceptibility profiles identical to those of ST78/B isolates from the V. Monaldi hospital; the single A. baumannii ST78/B isolate from the A. Cardarelli hospital was susceptible to imipenem (MIC, 1.0 mg/liter) and meropenem (MIC, 0.5 mg/liter) (Table 1). Tests with Etest MBL strips showed that all carbapenem-resistant ST2/A and ST78/B isolates were intermediate or resistant to imipenem (MICs, 8 to 16 mg/liter) but were negative for MBL production (imipenem-EDTA MICs, 4 to 8 mg/liter). To study the contribution of oxacillinases to imipenem resistance, imipenem MICs were analyzed in the presence of 200 mM NaCl for carbapenem-resistant ST2/A and ST78/B isolates by a microdilution method. These experiments showed that imipenem MICs (16 mg/liter) were inhibited by up to 8-fold in the presence of NaCl (2 mg/liter).
TABLE 2.
Antibiotic susceptibility profiles of A. baumannii isolates from the V. Monaldi hospitala
| Antibiotic | MIC (mg/liter) |
|||||
|---|---|---|---|---|---|---|
| ST2/A (14 total strains) |
ST78/B (57 total strains) |
|||||
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |
| Sulbactam-ampicillin | 32 | 125 | 32-125 | 125 | 125 | 64-125 |
| Piperacillin-tazobactam | 125 | 250 | 125->250 | 250 | 250 | 32->250 |
| Ceftazidime | 250 | 250 | 125->250 | 250 | 250 | 125->250 |
| Cefepime | 125 | 250 | 16-250 | 16 | 32 | 16-250 |
| Imipenem | 16 | 32 | 8-32 | 16 | 32 | 8-32 |
| Meropenem | 8 | 16 | 4-16 | 8 | 16 | 4-16 |
| Amikacin | 125 | >250 | 4->250 | 125 | >250 | 2->250 |
| Gentamicin | 8 | 64 | 4->250 | 32 | 64 | 4->250 |
| Ciprofloxacin | 64 | 250 | 32->250 | 64 | 250 | 32->250 |
| Rifampin | 4 | 500 | 2-500 | 4 | 4 | 2-16 |
| Colistin | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5-1 |
A. baumannii isolates were analyzed by a microdilution method for MIC determination according to CLSI guidelines.
Molecular analysis of carbapenem resistance in A. baumannii isolates.
PCR and sequence analysis identified a blaOXA-58 gene flanked by ISAba2 and ISAba3 elements at the 5′ and 3′ ends, respectively, in plasmid DNA from all carbapenem-resistant A. baumannii ST2/A and ST78/B isolates but not from the single carbapenem-susceptible A. baumannii strain of PFGE type B isolated in the A. Cardarelli hospital. No amplification products were obtained from chromosomal or plasmid DNA of A. baumannii ST2/A and ST78/B isolates using primers for blaIMP-type, blaVIM-type, or blaSIM-type MBLs or blaOXA-23 or blaOXA-24/40 CHDLs. Also, PCR experiments failed to identify any IS element upstream of the naturally occurring blaOXA-66 or blaOXA-90 genes in A. baumannii ST2/A and ST78/B isolates, respectively, thus excluding the notion that IS-mediated overexpression of these oxacillinases may account for the resistance to imipenem (23).
Genetic location and characterization of the genetic structures surrounding the blaOXA-58 gene.
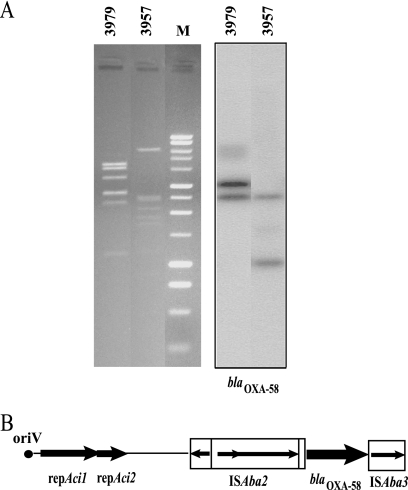
Digestion of plasmid DNA from A. baumannii ST2/A and ST78/B isolates with HindIII enzyme revealed different restriction patterns that generated two different positive bands of approximately 3.0 and 2.7 kb and 2.7 and 1.0 kb, respectively, when hybridized with a blaOXA-58-specific probe (Fig. 3A). The direct sequence of amplicons generated from plasmid DNA preparation of A. baumannii ST2/A and ST78/B isolates using primers spanning the 5′ end of A. baumannii origin of plasmid replication (oriV) and the 3′ end of the ISAba3 element identified two similar fragments of 6,095 and 6,073 bp that were designated pABNA1 and pABNA2, respectively.
FIG. 3.
(A) Plasmid localization of the blaOXA-58 gene in A. baumannii ST2/A and ST78/B isolates. Agarose (1%) gel electrophoresis in 1× Tris-acetate-EDTA buffer of HindIII-digested plasmids from A. baumannii isolates 3979 and 3957, respectively, stained with ethidium bromide and visualized under UV light and Southern blot hybridization with the blaOXA-58 probe are shown. Lane M shows a 1-kb DNA ladder (Promega, Milan, Italy). (B) Schematic map of the genetic structure surrounding the blaOXA-58 gene in A. baumannii ST2/A and ST78/B isolates. The genes and their corresponding transcription orientations are indicated by horizontal arrows. IS elements are represented by open rectangles filled with black arrows indicating the transposase gene and the direction of transcription. Names of relevant features are reported below or above the map.
The two amplicons showed identical origins of replication (oriV); a repeat region composed of five 22-bp-long imperfect direct iterons in pABNA1 and four 22-bp-long iterons in pABNA1 and pABNA2, respectively; identical repAci1 and repAci2 replicase genes; and a single copy of the blaOXA-58 gene that was flanked by ISAbA2 and ISAba3 elements at the 5′ and 3′ ends, respectively (Fig. 3B). Filter mating experiments demonstrated that resistance to imipenem, along with the blaOXA-58 gene, was transferred from A. baumannii ST78/B isolate 3957, but not from A. baumannii ST2/A isolate 3979, to imipenem-susceptible Acinetobacter genomic species 3 isolate 4442 at a frequency of 1 × 10−6. Imipenem MICs for transconjugants were similar (16 mg/liter) to those for donor isolates.
DISCUSSION
In the present report, we studied the molecular epidemiology and the genetic basis of carbapenem resistance in A. baumannii strains isolated between May 2006 and December 2007 during an epidemic occurring in two ICUs of the V. Monaldi hospital in Naples. In accordance with previous data (10, 15), the isolation rate of A. baumannii was significantly associated with length of stay in the ward, being higher in the CR-ICU than in the PO-ICU (10, 15). Based on a previous study of an A. baumannii outbreak occurring in the same institution between June 2003 and June 2004 (25), we could assume that the epidemic described herein was caused by the spread of a single epidemic clone. However, the present report revealed the emergence of two distinct A. baumannii epidemic clones that were isolated in two consecutive temporal clusters in the same wards of the hospital. Indeed, the identity of 3LST, ST, and resistance profiles/genes and the near-identity of PFGE profiles indicate that these two sets of isolates each represent a clone. The first epidemic clone showed identical PFGE profiles of the A. baumannii strains responsible for two epidemics occurring in Naples in the Federico II and V. Monaldi hospitals during 2002 and during 2003 and 2004, respectively (25, 26), and was assigned to 3LST group 1 and ST2, which corresponded to the previously characterized European clone II (9, 10). The second epidemic clone, which was first isolated in the CR-ICU in May 2006 and replaced the previous clone in both wards from March 2007 onward, showed a distinct genotype, assigned to novel 3LST group 6 and ST78, which has never been isolated before and is described for the first time herein. This is consistent with previous studies showing that carbapenem-resistant A. baumannii epidemics in southern Europe are caused by genotypes belonging to 3LST groups 1 and 2, corresponding to the European clones II and I, respectively, but also by additional genotypes of 3LST groups 4 and 5 (10, 11, 15, 22). Our data are also in agreement with a recent report showing that four distinct clones are responsible for a cluster of carbapenem-resistant A. baumannii infections in the ICU of a Greek hospital (21). Also, the isolation of A. baumannii ST78/B strains in the ICUs of two other hospitals in Naples during 2007 suggests that the spread of the novel A. baumannii epidemic clone described herein might have been caused by interhospital transfer of colonized patients in the city. In agreement with previous studies, the respiratory tract was the most frequent site of isolation for both clones (9, 15, 25, 26). However, ST78/B strains caused a higher but not statistically significant proportion of bacteremias than did the other clone in patients from the V. Monaldi hospital, thus suggesting that the novel epidemic clone may possess some inherent properties for developing invasive disease. Moreover, no statistically significant differences in mean age and female/male ratio were observed between patients of the V. Monaldi hospital infected by ST2/A strains and those infected by ST78/B strains of A. baumannii.
Several studies demonstrate that A. baumannii epidemic strains are selected in the hospital setting because of their multiple antimicrobial resistances (1, 9, 13, 15, 21, 27). In particular, the emergence of carbapenem resistance has been reported during hospital outbreaks of multidrug-resistant A. baumannii in Italy and southern Europe (4, 8, 11, 13, 20, 21, 25-28). Accordingly, the two A. baumannii clones described in the present study showed similar antibiotypes, characterized by resistance to all classes of antimicrobials, including carbapenems, and intermediate resistance to rifampin but susceptibility to colistin. Additional epidemiological information was provided by molecular analysis of carbapenem resistance genes. A plasmid-borne blaOXA-58 gene was identified in both A. baumannii clones isolated in the V. Monaldi hospital but not in the single carbapenem-susceptible A. baumannii ST78/B isolate from the A. Cardarelli hospital. Although the plasmids carrying the blaOXA-58 gene from the two epidemic clones showed distinct restriction patterns, two similar amplicons containing an origin of plasmid replication, a repeat region composed of four or five 22-bp imperfect direct iterons, the replicase genes, and a single copy of the blaOXA-58 gene flanked by ISAba2 and ISAba3 sequences at the 5′ and 3′ ends of the gene, respectively, were identified. The above genetic structures were highly homologous with those found in plasmids pOUR and pACICU1 from A. baumannii strains 183 and ACICU, respectively, isolated in Rome, Italy (4, 14). Interestingly, all A. baumannii strains carrying the blaOXA-58 gene isolated in Rome were assigned to ST group 1 and European clone II (4, 8, 14), as were the A. baumannii strains responsible for the outbreak occurring in the V. Monaldi hospital during 2003 and 2004 (11, 25). A blaOXA-58 gene flanked by ISAba2 and ISAba3 sequences has been also found in plasmids isolated in strains from France and Spain showing distinct pulsotypes (16) and in plasmids isolated in strains from Greece assigned to ST groups 1 and 2 (11). The above data all suggest that carbapenem resistance in the two A. baumannii epidemic clones might have been acquired through horizontal gene transfer among distinct clones. Because clone ST2/A carrying a plasmid-borne blaOXA-58 gene was first isolated in the V. Monaldi hospital during 2003 (25) while the first isolation of clone ST78/B carrying a plasmid-borne blaOXA-58 gene occurred during 2006 in the hospital and one carbapenem-susceptible A. baumannii strain with the ST78/B profile was isolated in another hospital of Naples during 2007, we can make the hypothesis that plasmid sequences carrying the blaOXA-58 gene flanked by ISAba2 and ISAba3 elements were transferred from ST2/A strains to ST78/B strains. In further support of this, we demonstrated herein that resistance to imipenem, along with the blaOXA-58 gene, was transferred from ST78/B strains into the imipenem-susceptible Acinetobacter genomic species 3 strain.
In conclusion, molecular epidemiology of A. baumannii in the V. Monaldi hospital showed the occurrence of a novel epidemic clone that successfully spread among different wards and was selected because of the presence of a plasmid-borne blaOXA-58 gene. This emphasizes the need to study the global epidemiology of A. baumannii and its associated antimicrobial resistances by using molecular typing methods in order to control the epidemic spread of multidrug-resistant A. baumannii infections in the hospital setting.
Acknowledgments
We thank J. F. Turton, Health Protection Agency, United Kingdom, for help in the identification of the novel alleles and ST types of A. baumannii isolates and D. Vitale, CEINGE Biotecnologie Avanzate, Napoli, Italy, for technical support in DNA sequencing. We also thank J.-W. Chu (Centre for Health Protection, The Government of the Hong Kong SAR, China) for kindly providing Acinetobacter genomic species 3 strain 4442 and Alfonso Baccari, the V. Monaldi hospital, Naples, Italy, for his kind support in epidemiological data collection.
This work was supported in part by a grant from Agenzia Italiana del Farmaco (AIFA2007 contract no. FARM7X9F8K), and from Ministero dell'Istruzione, dell'Universitá e della Ricerca, Italy (PRIN 2008 to R.Z.). Platform Genotyping of Pathogens and Public Health receives financial support from the Institut Pasteur and the Institut de Veille Sanitaire (Saint-Maurice, France).
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Adams, M. D., K. Goglin, N. Molyneaux, K. M. Hujer, H. Lavender, J. J. Jamison, I. J. MacDonald, K. M. Martin, T. Russo, A. A. Campagnari, A. M. Hujer, R. A. Bonomo, and S. R. Gill. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartual, S. G., H. Seifert, C. Hippler, M. A. Luzon, H. Wisplinghoff, and F. Rodrìguez-Valera. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertini, A., L. Poirel, S. Bernabeu, D. Fortini, L. Villa, P. Nordmann, and A. Carattoli. 2007. Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:2324-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.British Society for Antimicrobial Chemotherapy. January 2006. British Society for Antimicrobial Chemotherapy methods for antimicrobial susceptibility testing. Version 6. British Society for Antimicrobial Chemotherapy, Birmingham, United Kingdom.
- 6.Chang, H. C., Y. F. Wei, L. Dijkshoorn, M. Vaneechoutte, C. T. Tang, and T. C. Chang. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing, 18th informational supplement. Approved standards M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.D'Arezzo, S., A. Capone, N. Petrosillo, and P. Visca on behalf of GRAB. 2009. Epidemic multidrug-resistant Acinetobacter baumannii related to European clonal types I and II in Rome (Italy). Clin. Microbiol. Infect. 15:347-357. [DOI] [PubMed] [Google Scholar]
- 9.Dijkshoorn, L., H. Aucken, P. Gerner-Smidt, P. Janssen, M. E. Kaufmann, J. Garaizar, J. Ursing, and T. L. Pitt. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 11.Giannouli, M., F. Tomasone, A. Agodi, H. Vahaboglu, Z. Daoud, M. Triassi, A. Tsakris, and R. Zarrilli. 2009. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii strains in intensive care units of multiple Mediterranean hospitals. J. Antimicrob. Chemother. 63:828-830. [DOI] [PubMed] [Google Scholar]
- 12.Heritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houang, E. T. S., J. W. Chu, W. S. Lo, K. J. Chu, and A. F. B. Cheng. 2003. Epidemiology of rifampin ADP-ribosyltransferase (arr-2) and metallo-β-lactamase (blaIMP-4) gene cassettes in class 1 integrons in Acinetobacter strains isolated from blood cultures in 1997 to 2000. Antimicrob. Agents Chemother. 47:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iacono, M., L. Villa, D. Fortini, R. Bordoni, F. Imperi, R. J. P. Bonnal, T. Sicheritz-Ponten, G. De Bellis, P. Visca, A. Cassone, and A. Carattoli. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peleg, Α. Υ., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel, L., and P. Nordmann. 2006. Genetic structure at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salerno, A., A. Delétoile, M. Lefevre, I. Ciznar, K. Krovacek, P. Grimont, and S. Brisse. 2007. Recombining population structure of Plesiomonas shigelloides (Enterobacteriaceae) revealed by multilocus sequence typing. J. Bacteriol. 189:7808-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos, S. R., and H. Ochman. 2004. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 6:754-759. [DOI] [PubMed] [Google Scholar]
- 19.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towner, K. J., K. Levi, M. Vlassiadi, and the ARPAC Steering Group. 2008. Genetic diversity of carbapenem-resistant isolates of Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 14:161-167. [DOI] [PubMed] [Google Scholar]
- 21.Tsakris, A., A. Ikonomidis, A. Poulou, N. Spanakis, D. Vrizas, M. Diomidous, S. Pournaras, and F. Markou. 2008. Clusters of imipenem-resistant Acinetobacter baumannii clones producing different carbapenemases in an intensive care unit. Clin. Microbiol. Infect. 14:588-594. [DOI] [PubMed] [Google Scholar]
- 22.Turton, J. F., S. N. Gabriel, C. Valderrey, M. E. Kauffmann, and T. L. Pitt. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807-815. [DOI] [PubMed] [Google Scholar]
- 23.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 24.Van Dessel, H., L. Dijkshoorn, T. van der Reijden, N. Bakker, A. Paauw, P. van den Broek, J. Verhoef, and S. Brisse. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105-112. [DOI] [PubMed] [Google Scholar]
- 25.Zarrilli, R., R. Casillo, A. Di Popolo, M.-F. Tripodi, M. Bagattini, S. Cuccurullo, V. Crivaro, E. Ragone, A. Mattei, N. Galdieri, M. Triassi, and R. Utili. 2007. Molecular epidemiology of a clonal outbreak of multidrug-resistant Acinetobacter baumannii in a university hospital in Italy. Clin. Microbiol. Infect. 13:481-489. [DOI] [PubMed] [Google Scholar]
- 26.Zarrilli, R., M. Crispino, M. Bagattini, E. Barretta, A. Di Popolo, M. Triassi, and P. Villari. 2004. Molecular epidemiology of sequential outbreaks of Acinetobacter baumannii in an intensive care unit shows the emergence of carbapenem resistance. J. Clin. Microbiol. 42:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarrilli, R., M. Giannouli, F. Tomasone, M. Triassi, and A. Tsakris. 2009. Carbapenem resistance in Acinetobacter baumannii: the molecular epidemic features of an emerging problem in health care facilities. J. Infect. Dev. Countries 3:335-341. [DOI] [PubMed] [Google Scholar]
- 28.Zarrilli, R., D. Vitale, A. Di Popolo, M. Bagattini, Z. Daoud, A. U. Khan, C. Afif, and M. Triassi. 2008. A plasmid-borne blaOXA-58 gene confers imipenem resistance to Acinetobacter baumannii isolates from a Lebanese hospital. Antimicrob. Agents Chemother. 52:4115-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]