Abstract
This study compared the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) real-time PCR assay to culture by the use of BBL CHROMagar MRSA for the detection of MRSA in 627 nasal surveillance specimens collected from intensive care unit (ICU) patients. The PCR assay had a sensitivity, specificity, positive predictive value, and negative predictive value of 100%, 96.7%, 70.3%, and 100%, respectively. Nine of 19 false-positive PCR specimens grew methicillin-susceptible S. aureus (MSSA) from broth enrichment culture, of which two demonstrated evidence of mecA gene dropout. Compared to culture by the use of BBL CHROMagar MRSA, the BD GeneOhm MRSA PCR assay demonstrated sensitivity and specificity above 95% for the detection of MRSA nasal colonization and provided shorter turnaround time in generating positive and negative final results.
Methicillin-resistant Staphylococcus aureus (MRSA) continues to be a leading and important health care-associated (HC-A) pathogen, and its prevalence has reached epidemic proportions in the U.S. health care system with rates being reported as high as 64% (www.cdc.gov/ncidod/dhqp/ar_mrsa_surveillanceFS.html). An estimate from data from 1999 to 2000 determined that 126,000 (43.2%) of 292,000 hospitalizations with a discharge diagnosis of S. aureus infection were due to MRSA (19). Of greater importance is that patients with MRSA bacteremia have higher rates of morbidity and mortality than patients with infection caused by methicillin-sensitive strains (3, 6, 13). Complicating the situation is the financial impact that MRSA infections have on the health care system, including increased length of stay and additional days of antibiotic therapy (1, 15, 18). Hospital-acquired infections account for a total of approximately $30 billion per year, of which $2.5 billion is attributed to MRSA. This amounts to nearly $20,000 per case of added costs for the treatment of MRSA infections.
These statistics are concerning, yet within the health care setting, it is accepted that MRSA transmission, infection, and colonization are preventable through strict adherence to recommended infection control measures, such as hand hygiene, cleaning of equipment and rooms, and most recently, the implementation of active surveillance measures to screen and identify patients who are carriers of MRSA. Active surveillance for MRSA involves culturing the nares of patients at the time of admission or during hospitalization. This practice is recommended by the Society of Healthcare Epidemiology of America (22). The goal of these intervention measures is to limit the transmission of MRSA and prevent outbreaks in critical care units as well as in other patient settings.
In 2005, we compared a chromogenic medium with the BD GeneOhm MRSA real-time PCR system (BDG PCR) (BD Diagnostics, San Diego, CA) and determined that it was superior to, and faster than, culture. PCR-based diagnostic testing has been successfully implemented in and reported by other health care institutions (2, 7, 12, 31, 33, 35). A recent study conducted in a population of inmates at a correctional facility reported that chromogenic agar combined with an enrichment broth was more sensitive and specific than is PCR (14). These disparate conclusions led us to reinitiate a comparison study between PCR-based MRSA screening methodology and chromogenic culture to determine which diagnostic tool would be best applied in our hospital.
MATERIALS AND METHODS
This study was conducted at the University of Louisville Hospital, a 350-bed tertiary care facility that serves as the primary teaching hospital for the University of Louisville School of Medicine. The current MRSA surveillance screening is restricted to patients admitted to the respective intensive care units (medical, surgical, neurological, and cardiac). Patients found to be positive for MRSA with PCR-based technology are placed into contact precautions. The use of additional infection control measures, such as the administration of nasal mupirocin and chlorhexidine baths, is restricted to those patients scheduled for cardiac surgery. Patients who are negative for MRSA at admission are retested at discharge or transfer to determine if MRSA colonization was acquired during their stay in the intensive care unit.
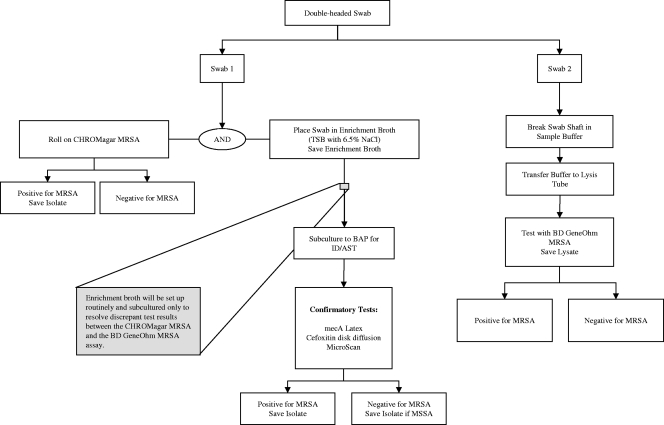
Anterior nasal swabs were collected with double-headed rayon-tipped swabs (BBL CultureSwab liquid Stuart double swab; BD Diagnostics, Sparks, MD). Both swabs were inserted simultaneously and rotated inside the anterior nares, first within one nostril and then within the other nostril, resulting in the acquisition of a paired swab specimen. All samples were collected by nursing personnel trained by microbiology and infection control personnel. Collected specimens were transported to the laboratory; cultures were processed within 1 h, and PCR testing was performed once daily. The first swab of the paired specimen was inoculated into BBL CHROMagar MRSA medium (C-MRSA). The swab was then inoculated onto enrichment broth, BBL tryptic soy broth (TSB) with 6.5% NaCl (BD Diagnostics, Sparks, MD). The C-MRSA medium plates and enrichment broth were incubated at 35°C overnight. C-MRSA medium plates were examined for the presence of mauve-colored colonies. Mauve-colored colonies were confirmed for S. aureus by Gram staining, a positive Staphaurex latex agglutination (Remel Europe Ltd., Dartford, Kent, United Kingdom) or tube coagulase. C-MRSA medium plates that were absent of any mauve colonies in the first 24 h were reincubated for an additional 24 h and reinspected for the presence of mauve-colored colonies. No additional positive cultures were detected following extended incubation. Discrepant results were either (i) C-MRSA culture negative and BDG PCR positive or (ii) C-MRSA culture positive and BDG PCR negative. When a discrepant result was encountered, the enrichment broth was subcultured to BBL Trypticase soy agar with 5% sheep blood (BAP) (BBL; BD Diagnostics, Sparks, MD) and incubated overnight at 35°C. Various selected colonies having the phenotypic characteristics of S. aureus were tested to confirm the presence of MRSA with mecA latex agglutination (Basing Stoke, Hant, United Kingdom), cefoxitin (30 μg) disc diffusion (5), and the MicroScan Walkaway (Siemens Healthcare Diagnostics, Inc., Sacramento, CA). The second swab of the paired swab specimen was processed and tested using the BD GeneOhm MRSA assay according to the manufacturer's instructions. A summary of the processing protocol is depicted in Fig. 1.
FIG. 1.
BDG PCR and C-MRSA specimen processing protocol.
We compared the turnaround times (TATs) for PCR and C-MRSA in reporting positive and negative results for MRSA because PCR testing is performed once daily in our laboratory. Time intervals (minimum, maximum, and average) were determined for the following: (i) specimen collection to receipt in the laboratory; (ii) receipt in the laboratory and test initiation; and (iii) time to reporting of results.
RESULTS
A total of 639 nasal samples were collected and tested as described above. Twenty-four nasal specimens were inhibited in the PCR assay, 12 of which were resolved following freeze-thawing of the lysate and repeat PCR testing. The remaining 12 unresolved samples were excluded in the data analysis, resulting in 627 total nasal specimens. There was 97% concordance between the two methods, with 45 positive specimens and 563 negative specimens by both the C-MRSA and the BDG PCR assay (Table 1). Sixty-four (10.2%) nasal specimens were positive for MRSA by PCR. However, only 44 (7%) of these specimens were MRSA culture positive on C-MRSA agar. No additional positive cultures were detected following incubation extended to 48 h. One additional specimen was determined to be MRSA culture positive following recovery from enrichment broth. This was regarded as a resolved sample and a true positive by the BDG PCR assay. Thus, the sensitivity and specificity for the BDG PCR were 100% and 97.8%, respectively. Performance comparison for both methods, including that for positive predictive values (PPVs) and negative predictive values (NPVs) for PCR, is summarized in Table 1.
TABLE 1.
BDG PCR assay compared to culturec
| PCR result | No. of specimens with indicated culture result |
Total | |
|---|---|---|---|
| POS | NEG | ||
| POS | 45a | 19b | 64 |
| NEG | 0 | 563 | 563 |
| Total | 45 | 582 | 627 |
Includes 44 specimens recovered by direct inoculation into C-MRSA and one specimen recovered from inoculation of an enrichment broth culture onto BAP.
Includes nine specimens that grew MSSA from the subculture of the enrichment broth onto BAP and 10 specimens that were “no growth” after the same process.
POS, positive; NEG, negative. Sensitivity, 100%; specificity, 96.7%; PPV, 70.3%; NPV, 100%.
Nineteen samples (3%) were classified as false positive by PCR. Of the 19 discrepant specimens, nine grew methicillin-susceptible S. aureus (MSSA) from the enrichment broth, and there was no growth detectable for the remaining 10 specimens. Further investigation of these 19 specimens was undertaken to characterize these “false positives.” Computer-generated cycle threshold (Ct) data for all 19 specimens were analyzed. The nine MSSA isolates were confirmed to be MSSA. Two of the nine specimens were confirmed as S. aureus isolates with evidence of an excised mecA gene (referred to as mecA dropout), as these two strains were positive when the strains were directly tested with the BDG PCR but had no evidence of the mecA gene by the use of PCR testing. An analysis of the computer-generated data output of the remaining 17 specimens demonstrated amplification curves consistent with the presence of the molecular targets for MRSA, 14 with late Ct values and three with mid-range Ct values.
The average time to report positive MRSA results by culture was 28.1 h versus 17.4 h for PCR. In contrast, the average time for reporting negative culture results was 51.3 h versus 14.4 h for PCR (Table 2). Positive and negative results generated by PCR were reported 11 and 36 h sooner than culture, respectively.
TABLE 2.
Time to reporting of PCR- and chromogenic agar-generated results from specimen collection to final results
| Result | Time to report | Time interval (h) from: |
Total TAT (h) | ||
|---|---|---|---|---|---|
| Collection to receipta | Receipt to beginning of testingb | Beginning to endc | |||
| MRSA-positive: | |||||
| C-MRSA | Avg | 0.8 | 0.5 | 26.8 | 28.1 |
| Minimum | 0.02 | 0.2 | 13.9 | 13.9 | |
| Maximum | 3.1 | 0.7 | 45.8 | 49.6 | |
| BDG PCR | Avg | 0.8 | 15.1 | 1.5 | 17.4 |
| Minimum | 0.02 | 2.6 | 1.5 | 4.12 | |
| Maximum | 3.1 | 26.5 | 1.5 | 31.1 | |
| MRSA-negative: | |||||
| C-MRSA | Avg | 0.7 | 1.5 | 49.1 | 51.3 |
| Minimum | 0.02 | 0.3 | 34 | 34.32 | |
| Maximum | 4.3 | 0.75 | 60.9 | 65.95 | |
| BDG PCR | Avg | 0.7 | 12.2 | 1.5 | 14.4 |
| Minimum | 0.02 | 1.6 | 1.5 | 3.12 | |
| Maximum | 4.3 | 28 | 1.5 | 33.8 | |
Time from collection of the specimen until it was received and accessioned in the laboratory.
Time from receipt in the laboratory until actual testing began.
Time from when testing began until testing was completed and results were reported.
DISCUSSION
Previous studies have shown the sensitivity of PCR to be superior to that of culture, but a definitive comparison of the PCR results obtained in the present study to other studies involving the BDG PCR assay is difficult, since we employed C-MRSA while previous studies utilized other chromogenic media or nonchromogenic selective media (2, 9, 16, 22, 29, 30, 31). The BDG PCR sensitivity (100%), although slightly higher than that reported in other studies, is similar to that reported by Boyce and Havill, who used C-MRSA (4). Boyce and Havill suggested that the sensitivity reported in their study may have been overestimated as a result of samples having been inoculated only on selective agar medium with the omission of enrichment broth (4). The addition of an enrichment broth culture has been reported to increase sensitivity, ranging from 2% to 23% (2, 26, 30). The purpose of using enrichment broth in this study was to resolve discrepant results between PCR and C-MRSA. Broth enhancement resulted in only one additional positive culture. Furthermore, C-MRSA served as the “gold standard,” and the manufacturer does not have a claim that supports the use of enrichment broth for such purposes. The exclusion of enrichment broth also avoids the expenditure of technologist time and eliminates an additional 2 to 3 days to obtain final results (9, 10). One plausible explanation for the higher sensitivity is that a separate swab was used for the PCR assay apart from the C-MRSA and broth enrichment method. Although this could be considered a limitation of the present study, every effort was made to ensure equivalent specimen distributions between the paired swabs and random assignment to either PCR or culture. Therefore, our experience and results suggest that the need for enrichment broth is questionable and remains controversial. Broth enrichment is frequently mentioned as an adjunct to plated medium and molecular methods to enhance the detection of MRSA in patient specimens. Many publications have reported increases in the recovery of MRSA and calculated statistical performance achieved at the expense of marginally increased recoveries of MRSA, increased work load, increased effort, longer time to detection and the impediment to expedient enforcement of hospital infection control practices (17, 20, 24, 25, 34), in addition to the associated increased costs. Broth enrichment methods need to be standardized as to optimal growth medium, incubation time and temperature, the influence of sampling schedules, single versus multiple body sites, and the correlation of MRSA bio-burden levels from body sites other than nares, and the correlation of such MRSA bio-burden loads in broth enrichment only to the actual transmission rates of MRSA to others. Broth enrichment would seemingly negate the benefits of an active surveillance program. Furthermore, MRSA carrier status results can and should be determined as soon as possible following admission of patients to the hospital. An informal survey of clinical microbiologists revealed that a significant minority of laboratories embraced the use of broth enrichment techniques (25).
The specificity and PPV of the BD GeneOhm MRSA assay (96.7% and 70.3%, respectively) in the current study are similar to results reported by others. Several studies have reported PPVs ranging from 63% to 95.8% which most likely reflects the differences in MRSA prevalence in the respective study populations (4, 14). The specificity reported in the present study is higher than that reported by Farley et al. (14) but similar to that reported by Boyce and Havill (4). The NPV reported in the present study is similar to what has been reported in other studies and may influence how laboratories utilize a PCR assay in support of a surveillance program. For example, as suggested by Farley et al., culture verification would be limited to PCR-positive samples, although it may not be justified due to the additional costs and added complexity to testing protocols (14, 26).
There are many possible explanations for PCR-positive, culture-negative results. In this study, two categories of false-positive results were observed: (i) evidence of excision of the mecA gene from the SCCmec cassette (0.3% of specimens in this study) and (ii) evidence of amplification of MRSA but no growth of MRSA in culture. Deletions of the mecA gene from the SCCmec cassette have been reported (11). Researchers have shown that the BDG PCR assay can amplify the SCCmec insertion sequence with the respective strains lacking the mecA gene (14, 32). However, the low frequency of mecA deletion in this study is consistent with another report (29) and does not compromise the utilization of the assay in support of a surveillance program where high NPV is extremely important. Possible factors that could account for the inability to establish viability of MRSA in a nasal specimen include the bioburden of MRSA being below the limit of detection of culture and the presence of substances in the specimen, such as antibiotics, which could inhibit the growth of MRSA but not interfere with PCR amplification of the organism, or the presence of nonviable organisms. It is unclear why the MRSA could not be recovered from the three specimens with mid-range Ct values; however, the aforementioned factors are applicable as well. Fifteen of the 19 patients with PCR-positive and culture-negative results received antibiotics within 1 week prior to or at the time of nasal swab collection; the remaining four patients had no evidence of having received antibiotics. The majority of these patients had received a combination of antibiotics including tobramycin, cefazolin, piperacillin-tazobactam, ciprofloxacin, clindamycin, gentamicin, imipenem, levofloxacin, ceftriaxone, and moxifloxacin; three patients received only vancomycin. We did not determine if any of these patients had a past or current history of MRSA infection or colonization.
The clinical implications for a patient having a positive PCR result in the absence of a positive culture are unknown. In some cases in which a positive PCR result from the nares is obtained in the absence of a positive culture, the MRSA could be present at other anatomical sites, namely, the groin, axilla, or rectum (23). When using culture to assess colonization with S. aureus or MRSA, it has been demonstrated that 13 to 26% of colonized patients may not be positive in the nares (8, 21). It is an accepted axiom in the clinical testing arena that no test is 100% sensitive and 100% specific. In our comparative study with 19 “false-positive” specimens, the BDG PCR method exhibited excellent specificity compared to culture. However, if one accepts the fact that only two of the isolates were false positive by the demonstration of mecA dropout, the specificity increases to 99.7%. The significance of the remaining seven MSSA isolates recovered from broth for which mecA dropout was not detected is unclear. This may have been due either to the coexistence of a small population of MRSA that was overgrown by a larger population of MSSA or to the MRSA, if present, being nonviable. This further highlights the occasional occurrence of false positives that occur with molecular assays. Overall, our false-positive rate was low (<4%) and would result in those patients being placed in contact precautions and add to the cost of care. Although statistical differences were noted to exist between the performance of the two methods, the BDG PCR assay achieved better specificity and provided shorter TAT in generating positive (17.4 h versus 28.1 h for PCR and culture, respectively) and negative (14.4 h versus 51.3 h, respectively) results. The recommendation for extended incubation for C-MRSA cultures that are negative at 24 h, which did not result in the detection of additional positive cultures, accounted for the additional time for reporting final results. The use of PCR testing would promote the placement of MRSA-colonized patients into contact precautions in a shorter period of time and potentially influence a reduction in the rate of transmission of MRSA to noncolonized patients. Laboratories similar to ours may experience staffing constraints, thereby limiting the performance of the BDG PCR assay to a single shift, usually the day shift, which normally has the highest concentration of staff that are trained and experienced in molecular techniques. However, to compensate for the lack of staffing to support a 24/7 PCR testing service, inoculating nasal swabs onto C-MRSA agar at designated periods of the day, with the confirmation of MRSA within 20 to 24 h, or reserving PCR testing for negative cultures and those swabs received in the early morning may serve as suitable alternatives for a laboratory to provide continuous support to the institution's surveillance program. However, based on our data, the decision was made to schedule PCR testing once per day between 10:00 a.m. and 12:00 p.m., which is more beneficial than culture and provides shorter TATs for reporting positive and negative MRSA results. Following the implementation of PCR testing in our setting, we have noted a 15% overall decline in MRSA infections in the intensive care setting. However, additional studies such as those reported by Peterson et al. and Robicsk et al. that focus on the impact that PCR assays have on patient outcome and reduction in transmission and infection rates due to S. aureus, especially MRSA, are necessary to justify the utilization of such assays along with their associated costs and current level of reimbursement (27, 28).
In conclusion, the BDG PCR assay is a rapid and accurate method for determining the MRSA carrier status of intensive care unit patients, in support of an active surveillance program. The assay displayed sensitivity and specificity above 95% for the detection of MRSA nasal colonization compared to chromogenic culture. The specificity was similar to that reported by others, and the number of false positives was due in part to mecA dropout and possibly the use of antistaphylococcal antibiotics. This requires further investigation, and even though the false-positive rate was low (<4%), this results in patients being placed in isolation, which adds to the cost of hospitalization. Furthermore, the lower PPV is dependent on the institutional prevalence of MRSA. A significant and important advantage of this assay, even when testing is limited to once per day, is the time to report positive and negative results being shorter than the 18 to 48 h required for chromogenic agar.
Acknowledgments
We thank BD Diagnostics, Inc., for providing the media in support of this study.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Abramson, M. A., and D. J. Sexton. 1999. Nosocomial methicillin-resistant and methicillin-susceptible Staphylococcus aureus primary bacteremia: at what costs? Infect. Control Hosp. Epidemiol. 20:408-411. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, E. J., E. A. Grabsch, S. A. Ballard, B. Mayall, S. Xie, R. Martin, and M. L. Grayson. 2006. Concurrent analysis of nose and groin swab specimens by the IDI-MRSA PCR assay is comparable to analysis by individual-specimen PCR and routine culture assays for detection of colonization by methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2904-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blot, S. I., K. H. Vandewoude, E. A. Hoste, and F. A. Colardyn. 2002. Outcome and attributable mortality in critically ill patients with bacteremia involving methicillin-susceptible and methicillin-resistatn Staphylococcus aureus. Arch. Intern. Med. 162:2229-2235. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, J. M., and N. L. Havill. 2008. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR versus the CHROMagar assay for screening patients for the presence of MRSA strains. J. Clin. Microbiol. 46:350-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement, 8th ed. M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, R., P. Jenks, J. Northwood, M. Wallis, S. Ferguson, and S. Hunt. 2007. Effect on MRSA transmission of rapid PCR testing of patients admitted to critical care. J. Hosp. Infect. 65:24-28. [DOI] [PubMed] [Google Scholar]
- 8.Currie, A., L. Davis, E. Odrobina, S. Waldman, D. White, J. Tomassi, and K. C. Katz. 2008. Sensitivities of nasal and rectal swabs for detection of methicillin-resistant Staphylococcus aureus colonization in an active surveillance program. J. Clin. Microbiol. 46:3101-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de San, N., O. Denis, M. F. Gasria, R. De Mendonca. C. Nonhoff, and J. J. Struelens. 2007. Controlled evaluation of the IDI-MRSA assay for detection of colonization by methicillin-resistant Staphylococcus aureus in diverse mucocutaneous specimens. J. Clin. Microbiol. 45:1098-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desjardins, M., C. Guibord, B. Lalonde, B. Toye, and K. Ramotar. 2006. Evaluation of the IDI-MRSA assay for detection of methicilliln-resistant Staphylococcus aureus from nasal and rectal specimens pooled in a selective broth. J. Clin. Microbiol. 44:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnio, P. Y., F. Février, P. Bifnai, M. Dehem, C. Kervégant, N. Wilhelm, A. L. Gautier-Lerestif, N. Lafforgue, M. Cormeir, the MR-MSSA Study Group of the Collège de Bactériologie-Virologie-Hygiène des Hôpitaux de France, and A. Le Coustumier. 2007. Molecular and epidemiological evidence for spread of multiresistant methicillin-resistant Staphylococcus aureus strains in hospitals. Antimicrob. Agents Chemother. 51:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drews, S. J., B. M. Willey, N. Kreiswirth, M. Wang, T. Ianes, J. Mitchell, M. Latchford, A. J. McGreer, and K. C. Katz. 2006. Verification of the IDI-MRSA assay for detecting methicillin-resistant Staphylococcus aureus in diverse specimen types in a core clinical laboratory. J. Clin. Microbiol. 44:3794-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engemann, J. J., Y. Carmeli, S. E. Cosgrove, V. G. Fowler, M. Z. Bronstein, S. L. Trivette, J. P. Briggs, D. J. Sexton, and K. S. Kaye. 2003. Adverse clinical and economic outcomes attributable to methicillin-resistance among patients with Staphylococcus aureus surgical site infection. Clin. Infect. Dis. 36:592-598. [DOI] [PubMed] [Google Scholar]
- 14.Farley, J. E., P. D. Stamper, T. Ross, M. Cai, S. Speser, and K. C. Carroll. 2008. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of the BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from an at-risk community population. J. Clin. Microbiol. 46:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farr, B. M. 2004. Prevention and control of methicillin-resistant Staphylococcus aureus infections. Curr. Opin. Infect. Dis. 17:317-322. [DOI] [PubMed] [Google Scholar]
- 16.Flayhart, D., J. F. Hindler, D. A. Bruckner, G. Hall, R. K. Shrestha, S. A. Vogel, S. S. Richter, W. Howard, R. Walther, and K. C. Carroll. 2005. Multicenter evaluation of BBL CHROMagar MRSA medium for direct detection of methicillin-resistant Staphylococcus aureus from surveillance cultures of the anterior nares. J. Clin. Microbiol. 43:5536-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley, P. G., E. A. Grabsch, B. P. Howden, W. Gao, and M. L. Grayson. 2009. Comparison of the XPert Methicillin-Resistant Staphylococcus aureus (MRSA) assay, BD GeneOhm MRSA assay, and culture for detection of nasal and cutaneous groin colonization by MRSA. J. Clin. Microbiol. 47:3769-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopp, B. J., E. Nix, and E. P. Armstrong. 2004. Clinical and economic analysis of methicillin-susceptible and -resistant Staphylococcus aureus infections. Ann. Pharmacother. 38:1377-1382. [DOI] [PubMed] [Google Scholar]
- 19.Kuehnert, M. J., H. A. Hill, B. A. Kupronis, J. L. Tokars, S. L. Solomon, and D. B. Jernigan. 2005. Methicillin-resistant Staphylococcus aureus hospitalizations, United States. Emerg. Infect. Dis. 11:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagacé-Wiens, P. R. S., M. J. Alfa, K. Manicham, and G. K. M. Harding. 2008. Reductions in workload and reporting time by use of methicillin-resistant Staphylococcus aureus screening with MRSASelect medium compared to mannitol-salt medium supplemented with oxacillin. J. Clin. Microbiol. 46:1174-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mertz, D., R. Frei, B. Jaussi, A. Tietz, C. Stebler, U. Ruckiger, and A. F. Widmer. 2007. Throat swabs are necessary to reliably detect carriers of Staphylococcus aureus. Clin. Infect. Dis. 45:475-477. [DOI] [PubMed] [Google Scholar]
- 22.Muto, C. A., J. A. Jernigan, B. E. Ostrowsky, H. M. Richet, W. R. Jarvis, J. M. Boyce, and B. M. Farr. 2003. SHEA guideline for preventing noscomial transmission of multidrug-resistant strains of Staphylococcus aureus and Enterococcus. Infect. Control Hosp. Epidemiol. 24:362-386. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen Van, J. C., M. D. Kitzis, A. Ly, A. Chalfine, J. Carlet, A. Ben Ali, and F. Goldstein. 2006. Detection of nasal colonization methicillin-resistant Staphylococcus aureus: a prospective study comparing real-time genic amplification assay vs selective chromogenic media. Pathol. Biol. 54:285-292. (In French.) [DOI] [PubMed] [Google Scholar]
- 24.Pasanen, T., M. Korkeila, S. Mero, E. Tarkka, H. Piiparinen, J. Vuopio-Varkila, M. Vaara, and P. Tissari. 2010. A selective broth enrichment combined with real-time nuc-mecA-PCR in the exclusion of MRSA. APMIS 118:74-80. [DOI] [PubMed] [Google Scholar]
- 25.Paule, S. M., M. Mehta, D. Hacek, T. Gonzaleles, A. Robicsek, and L. Peterson. 2009. Chromogenic media vs real-time PCR for nasal surveillance of methicillin-resistant Staphylococcus aureus. Microbiol. Infect. Dis. 131:532-539. [DOI] [PubMed] [Google Scholar]
- 26.Paule, S. M., D. M. Hacek, B. Kufner, K. Truchon, R. B. Thomson, Jr., K. L. Kaul, A. Robicsek, and L. R. Peterson. 2007. Performance of the BD GeneOhm methicillin-resistant Staphylococcus aureus test before and during high-volume clinical use. J. Clin. Microbiol. 45:2992-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, L. R., D. M. Hacek, and A. Robiesek. 2007. Case study: an MRSA intervention at Evanston Northwestern Healthcare. Jt. Comm. J. Qual. Patient Saf. 33:732-738. [DOI] [PubMed] [Google Scholar]
- 28.Robicsk, A., J. L. Beaumont, S. M. Paule, D. M. Hacek, R. B. Thomson, Jr., K. I. Kaul, P. King, and L. R. Peterson. 2008. Universal surveillance for methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Ann. Intern. Med. 148:409-418. [DOI] [PubMed] [Google Scholar]
- 29.Rossney, S. S., C. M. Herra, M. Fitzgibbon, P. M. Morgan, M. J. Lawrence, and B. O'Connel. 2008. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J. Clin. Microbiol. 46:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossney, S. S., C. M. Herra, M. Fitzgibbon, P. M. Morgan, M. J. Lawrence, and B. O'Connel. 2007. Evaluation of the IDI-MRSA assay on the SmartCycler real-time PCR platform for rapid detection of MRSA from screening specimens. Eur. J. Clin. Microbiol. Infect. Dis. 26:459-466. [DOI] [PubMed] [Google Scholar]
- 31.van Hal, S. J., D. Stark, B. Lockwood, D. Marriott, and J. Harkness. 2007. Methicillin-resistant Staphylococcus aureus (MRSA) detection: comparison of two molecular methods (IDI-MRSA PCR assay and Genotype MRSA Direct PCR assay) with three selective MRSA agars (MRSA ID, MRSASelect, and CHROMagar MRSA) for use with infection control swabs. J. Clin. Microbiol. 45:2486-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vonrentzell, J. E., and P. C. Schreckenberger. 2008. Comparison of direct plating and broth enhanced culture for recovery of MRSA from PCR positive nasal swabs, abstr. D-1137. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 33.Warren, D. K., R. S. Liao, L. R. Merz, M. Eveland, and W. M. J. Dunne. 2004. Detection of methicillin-resistant Staphylococcus aureus directly from nasal swab specimens by a real-time PCR assay. J. Clin. Microbiol. 42:5578-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolk, D. M., J. L. Marx, L. Dominguez, D. Driscoll, and R. B. Schifman. 2009. Comparison of MRSASelect, CHROMagar methicillin-resistant Staphylococcus aureus (MRSA) medium and Xpert MRSA PCR for detection of MRSA in nares: diagnostics accuracy and surveillance samples with various bacterial densities. J. Clin. Microbiol. 47:3933-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wren, M. W. D., C. Carder, P. G. Coen, V. Gant, and A. P. R. Wilson. 2006. Rapid molecular detection of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:1604. [DOI] [PMC free article] [PubMed] [Google Scholar]