Abstract
Four flagellin allelic types (I to IV) of Burkholderia pseudomallei were identified based on their sequence variation and restriction fragment length polymorphism (RFLP) analysis of the amplified flagellin gene. Flagellin allelic type I was the most predominantly (75.0%) found among the 100 clinical isolates of B. pseudomallei investigated in this study.
Melioidosis is a serious infection caused by the soil saprophyte Burkholderia pseudomallei, and it is capable of becoming a dangerous pathogen particularly in individuals with one or more preexisting conditions associated with an altered immune response. This infection, commonly reported from southeast Asia and northern Australia, is being recognized increasingly around the world (6, 8, 13). The flagellin gene has been used to differentiate or subtype many bacterial strains (15). PCR amplification combined with restriction fragment length polymorphism (RFLP) or sequence analysis has been used to investigate the variation of the B. pseudomallei flagellin gene (fliC) (9, 10, 12, 14). However, no sequence variation has been reported so far, probably due to the small number of isolates investigated in these studies. Recently, B. pseudomallei flagellin protein has been recognized as a protective immunogen and a potential vaccine target for melioidosis (2, 4, 7, 16). The flagellin protein also has been found to be an efficient antigen for serodiagnosis (5). As antigenic variation may be generated by different flagellin types, knowledge of a particular flagellin type of B. pseudomallei in the regions in which it is endemic may be useful for designing effective diagnostic reagents and vaccine.
This study was undertaken first to determine the sequence polymorphism of the fliC gene of a collection of genetically diverse B. pseudomallei isolates (as indicated by different randomly amplified polymorphism DNA [RAPD] profiles). A PCR-RFLP method then was developed to facilitate the recognition of B. pseudomallei flagellin allelic types. The distribution of the flagellin allelic types of 100 Malaysian clinical isolates then was determined.
The PCR primers used for fliC amplification were BC6E (5′-ACCAACAGCCTGCAGCGTATC-3′) and BCR14 (5′-TTATTGCAGGAGCTTCAGCAC-3′) (14). The amplification mixture (total volume of 50 μl) contained 5 μl of 10× Taq buffer with KCl; 2 mM MgCl2; 0.2 mM (each) dATP, dCTP, dGTP, and dTTP; 0.5 μM primers; 2.5 U Taq DNA polymerase (MBI Fermentas); and 2.5 μl of purified bacterial DNA. Amplification was performed with denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1.5 min, with a final extension at 72°C for 10 min. The amplicons were purified and sequenced using the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, CA) using primers BC6E and BCR14. B. pseudomallei ATCC 23343 and Burkholderia thailandensis ATCC 700388 were included in this investigation.
A total of 730 nucleotides (corresponding to bp 342 to 1072 of the variable region of fliC of B. pseudomallei ATCC 23343) were determined for 17 RAPD types of B. pseudomallei. Sequence analysis demonstrated four flagellin allelic types (I to IV) (Fig. 1). The fliC gene sequence of B. thailandensis greatly varied from that of B. pseudomallei, with 76 nucleotide differences and a deletion of 15 nucleotides. The nucleotide sequences of flagellin allelic type I were 100% identical to the fliC of B. pseudomallei reference strain ATCC 23343 as well as of strains 1106a, 668, 1710b, and E503 in GenBank. The flagellin allelic type II showed only one nucleotide difference (C instead of G at position 456) compared to the sequences of flagellin allelic type I, but it matched exactly with the fliC gene of B. pseudomallei K9623 and B. mallei NCTC 10247, 10229, SAVP1, and ATCC 23344.
FIG. 1.
Sequence alignment of the variable region of the flagellin (fliC) genes of four allelic types of B. pseudomallei. The reference strains of B. pseudomallei ATCC 23343 and E503, B. mallei NCTC 10247, and B. thailandensis ATCC 700388 were included for comparison. Identical nucleotides are indicated by a dot, whereas deletions are indicated by a minus.
The flagellin allelic types III and IV identified in this study are considered new sequence variants for B. pseudomallei, as identical sequences have not been reported in the public sequence databases. Flagellin allelic type III demonstrated two nucleotide differences (T instead of C at position 377 and C instead of G at position 456) from the sequence of flagellin allelic type I. Flagellin allelic type IV showed 12 nucleotide differences from allelic type I (Fig. 1). The flagellin allelic type IV in this study show 98% sequence similarity to the flagellin sequences of B. pseudomallei and B. mallei but 89% to that of B. thailandensis. Four sequence entries (AJ617736, AJ617737, AJ617738, and AJ617739) in the GenBank database had partial sequence identities (only 58% sequence coverage) to the flagellin allelic type IV identified in this study. These isolates had been detected with different melting temperatures by real-time PCR assays (11). While the single-nucleotide substitution for flagellin allelic type II did not result in an amino acid change, one nonsynonymous change (valine instead of alanine) was noted for flagellin allelic type III, whereas five nonsynonymous changes (two alanines instead of serines, one serine instead of threonine, one alanine instead of threonine, and one threonine instead of serine) were observed for flagellin allelic type IV.
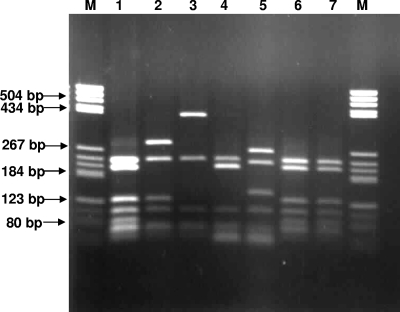
Restriction analyses of amplified flagellin PCR products (10 μl) were performed using 5 U each of MspI (5′CCGG3′) and AsuI (5′GGNCC3′) (MBI Fermentas) incubated overnight at 37°C. Four restriction profiles (Fig. 2) were generated for B. pseudomallei corresponding to the flagellin sequence analysis. The PCR-RFLP approach also was able to differentiate B. thailandensis from B. pseudomallei (Fig. 2). Of the 100 clinical isolates of B. pseudomallei investigated in this study, flagellin allelic type I was the most predominant (75.0%), followed by allelic type II (20.0%). Flagellin allelic types III and IV were identified in three (3.0%) and two (2.0%) clinical isolates, respectively (Table 1). No major difference was found in the distribution of flagellin allelic types for isolates obtained from blood cultures or other clinical specimens (Table 1).
FIG. 2.
Restriction profiles of four flagellin allelic types of B. pseudomallei isolates. Lanes: M, pBR322/HaeIII DNA marker; 1, flagellin allelic type I; 2, flagellin allelic type II; 3, flagellin allelic type III; 4, flagellin allelic type IV; 5, B. thailandensis ATCC 700388; 6, B. pseudomallei NCTC 13178; 7, B. pseudomallei ATCC 23343.
TABLE 1.
Distribution of flagellin allelic types in clinical isolates of B. pseudomallei by PCR-RFLP analysis
| Flagellin allelic type | No. (%) of isolates |
Total | |
|---|---|---|---|
| Blood culture | Other clinical specimen | ||
| I | 35 (70.0) | 40 (80.0) | 75 (75.0) |
| II | 11 (22.0) | 9 (18.0) | 20 (20.0) |
| III | 2 (4.0) | 1 (2.0) | 3 (3.0) |
| IV | 2 (4.0) | 0 (0.0) | 2 (2.0) |
| Total | 50 (100) | 50 (100) | 100 (100.0) |
Due to the lack of comprehensive patient follow-up data, we are not able to comment on the clinical presentation and outcome of the patients infected by B. pseudomallei of different flagellin allelic types. Our preliminary study showed that the motility of the isolates was not affected by the allelic types. It has yet to be investigated whether minor amino acid changes observed for flagellin types III and IV of B. pseudomallei will have some effect on the expression and antigenicity of flagellin. In some bacteria, for example, the switch from alanine to serine or alanine to threonine in the central region of flagellin may cause posttranslational modifications that are responsible for the changes in the antigenicity of flagellin protein (1, 3).
Nucleotide sequence accession numbers. The sequences for B. pseudomallei flagellin allelic types I to IV were deposited in GenBank under accession numbers HM003221 to HM003224.
Acknowledgments
This study was supported by a research grant (FS167/2007C) provided by the University of Malaya, Kuala Lumpur, Malaysia.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Alm, R. A., P. Guerry, M. E. Power, and T. J. Trust. 1992. Variation in antigenicity and molecular weight of Campylobacter coli VC167 flagellin in different genetic backgrounds. J. Bacteriol. 174:4230-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brett, P. J., D. C. Mah, and D. E. Woods. 1994. Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect. Immun. 62:1914-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brimer, C. D., and T. C. Montie. 1998. Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J. Bacteriol. 180:3209-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y. S., Y. S. Hsiao, H. H. Lin, Y. Liu, and Y. L. Chen. 2006. CpG-modified plasmid DNA encoding flagellin improves immunogenicity and provides protection against Burkholderia pseudomallei infection in BALB/c mice. Infect. Immun. 74:1699-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y. S., D. Shiuan, S. C. Chen, S. M. Chye, and Y. L. Chen. 2003. Recombinant truncated flagellin of Burkholderia pseudomallei as a molecular probe for diagnosis of melioidosis. Clin. Diagn. Lab. Immunol. 10:423-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puthucheary, S. D., N. Parasakthi, and M. K. Lee. 1992. Septicaemic melioidosis: a review of 50 cases from Malaysia. Trans. R. Soc. Trop. Med. Hyg. 86:683-685. [DOI] [PubMed] [Google Scholar]
- 9.Sonthayanon, P., P. Krasao, V. Wuthiekanun, S. Panyim, and S. Tungpradabkul. 2002. A simple method to detect and differentiate Burkholderia pseudomallei and Burkholderia thailandensis using specific flagellin gene primers. Mol. Cell Probes 16:217-222. [DOI] [PubMed] [Google Scholar]
- 10.Tanpiboonsak, S., A. Paemanee, S. Bunyarataphan, and S. Tungpradabkul. 2004. PCR-RFLP based differentiation of Burkholderia mallei and Burkholderia pseudomallei. Mol. Cell Probes 18:97-101. [DOI] [PubMed] [Google Scholar]
- 11.Tomaso, H., T. L. Pitt, O. Landt, S. AlDahouk, H. C. Scholz, E. C. Reisinger, L. D. Sprague, I. Rathmann, and H. Neubauer. 2005. Rapid presumptive identification of Burkholderia pseudomallei with real-time PCR assays using fluorescent hybridization probes. Mol. Cell Probes 19:9-20. [DOI] [PubMed] [Google Scholar]
- 12.Tungpradabkul, S., S. Wajanarogana, S. Tunpiboonsak, and S. Panyim. 1999. PCR-RFLP analysis of the flagellin sequences for identification of Burkholderia pseudomallei and Burkholderia cepacia from clinical isolates. Mol. Cell Probes 13:99-105. [DOI] [PubMed] [Google Scholar]
- 13.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 14.Winstanley, C., B. A. Hales, J. E. Corkill, M. J. Gallagher, and C. A. Hart. 1998. Flagellin gene variation between clinical and environmental isolates of Burkholderia pseudomallei contrasts with the invariance among clinical isolates. J. Med. Microbiol. 47:689-694. [DOI] [PubMed] [Google Scholar]
- 15.Winstanley, C., and J. A. W. Morgan. 1997. The bacterial flagellin gene as a biomarker for detection, population genetics, and epidemiological analysis. Microbiology 143:3071-3084. [DOI] [PubMed] [Google Scholar]
- 16.Ye, Z., C. M. Lee, G. W. Sun, and Y. H. Gan. 2008. Burkholderia pseudomallei infection of T cells leads to T-cell costimulation partially provided by flagellin. Infect. Immun. 76:2541-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]