Abstract
Rapid and sensitive detection of Aspergillus from clinical samples may facilitate the early diagnosis of invasive pulmonary aspergillosis (IPA). A real-time nucleic acid sequence-based amplification (NASBA) method was investigated by use of an inhalational rat model of IPA. Immunosuppressed male Sprague-Dawley rats were exposed to Aspergillus fumigatus spores for an hour in an aerosol chamber. Bronchoalveolar lavage (BAL) fluid, lung tissues, and whole blood were collected from five infected rats at 1, 24, 48, 72, and 96 h postinfection and five uninfected rats at the end of the experiment. Total nucleic acid (TNA) was extracted on an easyMAG instrument. A primer-molecular beacon set targeting 28S rRNA was designed to detect Aspergillus spp. The results were compared to those of quantitative PCR (qPCR) (18S rDNA) and quantitative culture. The analytical sensitivity of the real-time NASBA assay was <1 CFU/assay. A linear range of detection was demonstrated over 5 log units of conidia (10 to 105 spores). Both NASBA and qPCR showed a progressive increase in lung tissue burdens, while the CFU counts were stable over time. The fungal burdens in BAL fluid were more variable and not indicative of a progressive infection. The results of both real-time assays correlated well for both sample types (r = 0.869 and P < 0.0001 for lung tissue, r = 0.887 and P < 0.0001 for BAL fluid). For all whole-blood specimens, NASBA identified Aspergillus-positive samples in the group from which samples were collected at 72 h postinfection (three of five samples) and the group from which samples were collected at 96 h postinfection (five of five samples), but no positive results were obtained by culture or PCR. Real-time NASBA is highly sensitive and useful for the detection of Aspergillus in an experimental model of IPA.
Invasive pulmonary aspergillosis (IPA) is a major cause of morbidity and mortality in immunocompromised patients. Therapeutic success depends on an early diagnosis and the early initiation of antifungal therapy (5). The rapid and sensitive detection of Aspergillus from clinical samples may facilitate the early diagnosis of IPA. Molecular diagnostics have emerged as promising methods for the diagnosis of a variety of infectious diseases, including fungal infections (22). Given the advantages of detection sensitivity and speed, real-time quantitative PCR (qPCR) has been extensively studied and explored as a tool for use for the detection and identification of Aspergillus and other pathogenic fungi in clinical samples (9-12, 31). However, a lack of standardization often makes it difficult to compare results between laboratories, which is critical for the ultimate approval of a test as a standard diagnostic test for IPA (2).
Nucleic acid sequence-based amplification (NASBA) is an isothermal amplification process that specifically amplifies RNA even in the presence of genomic DNA (30). The amplification is highly robust, yielding >1012 amplicons in 30 min (4). The amplified single-stranded RNA can easily be detected in real time by the use of molecular beacon (MB) probes, which are self-reporting, hairpin-structured, single-stranded nucleic acid (NA) probes that brightly fluoresce when they are bound to their targets (33). NASBA-based assays for the detection of Aspergillus have been described by a few study groups (14, 36). However, to our knowledge, real-time NASBA has not been applied to the detection of Aspergillus fumigatus from an animal model of IPA.
The object of the study described here was to evaluate a highly sensitive real-time NASBA assay for its ability to detect Aspergillus burdens in an inhalational rat model of IPA at selected time points in the course of infection and to compare the diagnostic yield with the yields obtained by culture and qPCR.
MATERIALS AND METHODS
Animal model.
Thirty male Sprague-Dawley rats (Charles River, Margate, United Kingdom) weighing 200 to 250 g were used in the experiment, after being allowed 7 days to acclimatize. The rats were housed in cages provided with HEPA filters and were allowed free access to food and water. The rats were immunosuppressed with cyclophosphamide (75 mg/kg of body weight intraperitoneally twice) plus prednisolone (Depo-medrone; 16 mg/kg intramuscularly once) 2 days before infection. The animals received eurofloxacin (Baytril; 10 mg/kg subcutaneously daily) from day 2 preinfection to prevent secondary bacterial infections. A frozen glycerol stock of A. fumigatus A1163 was cultured into Sabouraud dextrose agar (SAB) for 10 days. The spores were harvested by the use of phosphate-buffered saline (PBS) plus 0.05% Tween 80 and counted with a hemocytometer. The rats were infected with 4 × 108/ml spores in an inhalation acrylic chamber in a class II hood for an hour (28). The rats (n = 5) were checked for symptoms such as body weight loss, dyspnea, and lethargy and were euthanized at 1 h, 24 h, 48 h, 72 h, and 96 h postinfection. Five uninfected rats were euthanized at 96 h after the inoculation. Bronchoalveolar lavage (BAL) fluid and lung tissue were collected from the euthanized rats. Whole-blood samples were also collected from the rats at 48 h, 72 h, and 96 h postinfection as well as from uninfected rats.
Aspergillus fumigatus burden assessment.
One gram of lung tissue was homogenized with 2 ml of sterile PBS. Tenfold dilutions of lung tissue homogenates were plated onto SAB plates, and the plates were incubated at 37°C for 24 to 36 h, after which the A. fumigatus burdens (CFU per gram of lung tissue) were determined. The primary lung tissue homogenates were spun down, and the pellets were resuspended in 1 ml of RNAlater RNA stabilization reagent and stored at −20°C until NA extraction for real-time detection. The fungal burdens in the BAL fluid samples were also determined by CFU counting. One milliliter of BAL fluid was mixed with 2 ml of the RNAlater reagent and stored at −20°C until NA extraction. qPCR and real-time NASBA of lung tissue homogenates, BAL fluid, and blood samples were performed; and the fungal burden in each sample was calculated on the basis of the standard curve included in each run.
TNA extraction.
Frozen lung tissue homogenates (1 g lung tissue per sample) in RNAlater reagent were thawed at room temperature before extraction was performed. Following centrifugation at 16,000 × g for 10 min, the supernatant was discarded and the pellet was resuspended in 1 ml of NucliSENS lysis buffer (bioMérieux, Durham, NC) with proteinase K (100 μg/ml). The entire solution was transferred to a tube with lysing matrix D (containing 1.4 mm ceramic spheres) (MP, Biomedicals, Inc., Solon, OH) and processed in a FastPrep instrument (MP, Biomedicals, Inc.) at speed level 6 twice for 45 s each time. After 1 h of incubation at 55°C, the cell lysates were centrifuged at 16,000 × g for 10 min. The supernatant was transferred and mixed with 2 ml of NucliSENS lysis buffer in the NucliSENS easyMAG sample cartridge (bioMérieux) for the automatic total nucleic acid (TNA) extraction. Magnetic silica solution was added, and the extraction was performed according to the easyMAG user manual. The final elution volume was 110 μl. Sterile PBS was processed in parallel as the negative extraction control.
Frozen BAL fluid samples (1 ml BAL fluid plus 2 ml RNAlater reagent) were thawed at room temperature and centrifuged at 16,000 × g for 10 min. A 1-ml mixture of NucliSENS lysis buffer with proteinase K was added to resuspend the pellet, and then the entire resuspension was transferred into the lysing matrix tube and the tube was vigorously vortexed in the FastPrep instrument, as described above. The following TNA extraction process was the same as that for lung tissue samples, and the final elution volume was 60 μl.
One milliliter whole blood was immediately treated with an equal volume of in-house nucleus lysis buffer (containing 1% Triton X-100, 0.01 M Tris, pH 7.6, 0.005 M MgCl2, and 0.32 M sucrose) and centrifuged at 16,000 × g for 10 min. The supernatant was discarded and another 1 ml of nucleus lysis buffer was added without disturbing the pellet. After the second centrifugation at 16,000 × g for 10 min, the supernatant was discarded and the pellet was resuspended in 1 ml of NucliSENS lysis buffer with proteinase K (100 μg/ml). The specimen was transferred to a lysing matrix tube for mechanical disruption, and automatic TNA extraction was performed as stated above. The extracts were eluted in 50 μl of elution buffer.
For external standardization, TNAs from titrated A. fumigatus conidia (5 × 106 to 5 × 100 spores per ml) were extracted in parallel. A 200-μl aliquot of each dilution was mixed with 800 μl of lysis buffer (with 100 μg/ml of proteinase K) for the extraction, and the final elution volume was 50 μl.
Quantitative real-time NASBA assay.
A real-time NASBA assay targeting 28S rRNA, the so-called pan-Aspergillus NASBA, was developed for the detection of Aspergillus spp. The primers and MB probe were designed as described in a previously published report (37). The P1 primer was 5′-AATTCTAATACGACTCACTATAGGGGAGAATCCACATCCAGGTGC-3′, and the P2 primer was 5′-CAGCAGTTGGACATGGGTTA-3′. The MB probe was 5′-CGCGAGTGCGCCGTGTGCCGAAATCGCG-3′ and was labeled with 5-carbofluorescein (FAM) fluorophores at the 5′ end and 4-(4-dimethylaminophenyl)diazenylbenzoic acid (DABCYL) quencher at the 3′ end.
The NASBA reactions were performed with a NucliSENS basic kit (version 2; bioMérieux). Each reaction mixture consisted of 0.2 μM each primer, 0.1 μM MB probe, 5 μl of TNA template, and 5 μl of NASBA enzyme mixture (T7 RNA polymerase, avian myeloblastosis virus reverse transcriptase, RNase H, bovine serum albumin) in a total volume of 20 μl. The enzyme mixture was added after the rest of the reaction mixture was incubated at 65°C for 2 min and 41°C for another 2 min, and then the reaction was run at 41°C for 90 min on a NucliSENS EasyQ analyzer (bioMérieux). The fluorescence signal was measured at time intervals of 30 s for each independent reaction at two wavelengths by using the accompanying NucliSENS EasyQ Director software (version 2).
The specificity of the real-time NASBA assay was tested as described before (37). Briefly, total nucleic acids from Aspergillus spp. (A. fumigatus, A. flavus, A. terreus, and A. niger) as well as other fungal species (Candida albicans, Candida glabrata, Candida krusei, Candida tropicalis, Candida parapsilosis, Candida guilliermondii, Candida dubliniensis, Cryptococcus neoformans, Saccharomyces cerevisiae, Fusarium spp.) and bacteria (Escherichia coli, Staphylococcus aureus) were tested in parallel for cross-reactivity by the pan-Aspergillus NASBA assay. Nuclease-free water was also included as the no-template control (NTC). The extraction of TNA from the culture was performed as described previously (37).
The analytical sensitivity of the pan-Aspergillus NASBA assay was assessed by using triplicate TNA samples extracted from 10-fold serially titrated A. fumigatus conidia (106 to 100 spores). The time to positivity (TTP), which is the time at which the fluorescence signal rises above the cutoff value (20% higher than the end-point signal from NTC), was determined, as well as linearity between TTP and the log input of conidia. These TNA samples were also used as external standards in each run. The conidial equivalents (CEs) in the rat samples were calculated by comparing the TTP of the sample with that of the external standards.
Real-time PCR.
An Aspergillus genus-specific real-time PCR assay was performed in this study so that the results could be compared with those of the NASBA assay. The primers and probe were modified from those used in a previous study (14) and targeted the 18S rRNA gene. The sequences were as follows: forward primer, 5′-GCCGCGGTAATTCCAGCTCCAATA-3′; reverse primer, 5′-GAGCAAAGGCCTGCTTTGAACA-3′; MB probe, 5′-CGCACGGGTCCGCCTCACCGCGAGTACTGCGTGCG-3′. The PCR master mixture contained 0.2 μM each primer, 0.2 μM MB probe, 2 μl of TNA, and 12.5 μl of 2× Brilliant II qPCR master mixture (Stratagene) in a total reaction volume of 25 μl. The cycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The same standard TNAs used in the NASBA assay were run in conjunction with each set of samples to quantify the Aspergillus burden.
Statistical analysis.
The Aspergillus burdens quantified by the three different methods were analyzed by the use of SPSS (version 11.0) software (SPSS Inc., Chicago, IL). The multiple comparison among samples collected at different times postinfection was performed by two-way analysis of variance on the basis of the sample types. The relationship between the results of qPCR and those of the real-time NASBA assay were analyzed by using Spearman's correlation coefficient. The level of statistical significance was set at a P value of <0.05.
RESULTS
Specificity, sensitivity, and linear range of real-time NASBA assay.
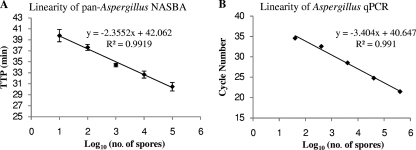
The pan-Aspergillus real-time NASBA assay was designed to detect Aspergillus spp. and showed a high degree of specificity (>95%). However, because Penicillium chrysogenum is 99% homologous to Aspergillus spp. in the amplification region, the cross-reaction from this species would be expected. Other species, such as the AIDS pathogen Penicillium marneffei (35), have the potential to cross-react. The NASBA assay did not cross-react with the other fungal species or the bacterial species tested in this study. The lowest detection limit of 0.1 CFU per reaction, corresponding to 1 CFU/ml of conidial suspension, was observed in 6 of 10 runs, and the level of 1 CFU per reaction was consistently detected by the pan-Aspergillus NASBA assay. The linear range of detection was shown to be from 10 to 105 Aspergillus conidia (Fig. 1A). Given that only 5 μl of TNAs was used for the NASBA assay, the detection limits were 22 CFU per assay for lung tissue, 12 CFU for BAL fluid, and 10 CFU for whole blood. Similarly, the lower quantification limits were 220 CFU for lung tissue, 120 CFU for BAL fluid, and 100 CFU for whole blood.
FIG. 1.
(A) Linearity of the pan-Aspergillus NASBA assay determined by triplicate amplification of total nucleic acids extracted from serially diluted A. fumigatus conidia with 105 to 101 spores. (B) Linearity of Aspergillus qPCR assay. The same TNA series used for the experiment whose results are presented in panel A were used to generate the standard curve.
Reproducibility of real-time NASBA assay.
To assess the reproducibility of the pan-Aspergillus NASBA assay, 5 log units of TNAs (10 to 105 conidia) were tested in triplicate. The coefficients of variation (CVs) of the TTP values for 105 to 10 conidia were 2.54, 1.96, 0.90, 1.43, and 2.80%, respectively.
Linearity of qPCR assay.
Because the qPCR assay was modified from the Aspergillus NASBA detection assay reported by Loeffler et al. (14), the analytical sensitivity and the linear detection range were reassessed by using TNA extracts from 1 to 106 spores of A. fumigatus conidia. The lower detection limit of 4 CFU was achieved in 5 of 10 repeated runs, while 40 CFU was consistently achievable in all runs. The linear range was from 40 to 4 × 105 CFU (Fig. 1B). The calculated PCR efficiency was 1.96. Again, considering that the portion of TNAs taken for the test varied, we calculated the lower limit of quantification to be 2,200 CFU for lung tissue, 1,200 CFU for BAL fluid, and 1,000 CFU for whole blood.
Detection of A. fumigatus in lung tissue.
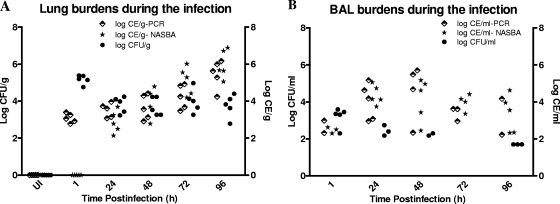
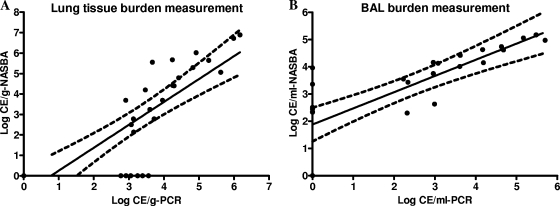
At 1 h postinfection, the pulmonary fungal burden counts were very high by quantitative culture (5.17 ± 0.25 log10 CFU/g), reflecting the presence of conidia, whereas the burden obtained by qPCR was lower (3.07 ± 0.25 log10 CE/g) and no positive signal was obtained by the NASBA assay (Table 1). At 24 h postinfection, the qPCR-based Aspergillus burden counts were close to those obtained by quantitative culture (3.50 ± 0.39 log10 CE/g versus 3.80 ± 0.44 log10 CFU/g), while the NASBA-based burden counts (2.87 ± 0.61 log10 CE/g) were still lower than the measurements obtained by the other two assays. After 24 h postinfection, the CFU counts stayed relatively stable throughout the experiment. In contrast, both qPCR and the real-time NASBA assays demonstrated progressive increases in the Aspergillus burden over the time of infection (by linear regression analysis, F = 40.66 and P < 0.0001 for qPCR measurements and F = 141.10 and P < 0.0001 for real-time NASBA measurements) (Fig. 2), and the results from both real-time platforms correlated well (r = 0.869, Spearman's correlation; P < 0.0001) (Fig. 3A). All lung tissue samples from the uninfected control group were negative by the qPCR and NASBA assay and by CFU counting.
TABLE 1.
Aspergillus burden in lung tissue and BAL fluid measured by CFU, qPCR, and real-time NASBA assays
| Sample type | Time (h) postinfection (no. of rats) |
Aspergillus burden determined bya: |
|||||
|---|---|---|---|---|---|---|---|
| Culture |
qPCR |
NASBA |
|||||
| No. of positive samples | Mean log10 CFU/g (ml)b | No. of positive samples | Mean log10 CE/g (ml)b | No. of positive samples | Mean log10 CE/g (ml)b | ||
| Lung tissue | 1 (5) | 5 | 5.17 ± 0.25 | 5 | 3.07 ± 0.25 | 0 | |
| 24 (5) | 5 | 3.80 ± 0.44 | 5 | 3.50 ± 0.39 | 5 | 2.87 ± 0.61 | |
| 48 (5) | 5 | 3.70 ± 0.50 | 5 | 3.66 ± 0.67 | 4 | 3.92 ± 0.88 | |
| 72 (5) | 5 | 4.00 ± 0.64 | 5 | 4.23 ± 0.66 | 5 | 5.10 ± 0.77c | |
| 96 (5) | 5 | 3.75 ± 0.62 | 5 | 5.46 ± 0.76c | 5 | 6.00 ± 0.78c | |
| BAL fluid | 1 (5) | 5 | 3.20 ± 0.51 | 2 | 2.66 ± 0.47 | 3 | 2.48 ± 0.17 |
| 24 (5) | 3 | 2.44 ± 0.28 | 5 | 4.01 ± 0.97c | 5 | 4.37 ± 0.53c | |
| 48 (5) | 2 | 2.24 ± 0.09 | 4 | 4.55 ± 1.54c | 5 | 4.13 ± 1.16c | |
| 72 (5) | 0 | 3 | 3.40 ± 0.38 | 4 | 3.99 ± 0.45c | ||
| 96 (5) | 3 | 1.70 ± 0.00 | 2 | 3.20 ± 1.37 | 5 | 3.37 ± 1.01c | |
Mean log10 ± standard deviation was used to describe the Aspergillus burden.
Mean log10 CFU/g and mean log10 CE/g are units for lung tissue burden measurement, and mean log10 CFU/ml and mean log10 CE/ml are units for BAL fluid burden assessment.
Statistically significant difference (P < 0.05) compared to culture CFU counts.
FIG. 2.
Aspergillus burdens in lung tissues (A) and BAL fluids (B) in terms of CFU counting, qPCR, and real-time NASBA measurements. UI, uninfected group. A linear increase in the Aspergillus burden in lung tissues was observed by PCR (F = 40.66, P < 0.0001) and NASBA (F = 141.10, P < 0.0001).
FIG. 3.
The correlation between the results of the real-time NASBA and qPCR assays for the detection of the Aspergillus burden in lung tissue (A) and BAL fluids (B) from rats with IPA was statistically significant (P < 0.0001). Spearman's correlation coefficients (r) were 0.869 (A) and 0.887 (B). The black lines represent the linear regression, and the dotted lines represent the 95% confidence intervals.
Detection of A. fumigatus in BAL fluid.
Real-time NASBA was more sensitive than culture in detecting the Aspergillus burden in BAL fluid. It detected A. fumigatus in 22 of 25 (88%) BAL fluid specimens from rats with experimental IPA, which was significantly more than the number detected by culture (13 of 25 [52%]) (Fisher's exact test, P = 0.0121). The PCR assay detected Aspergillus in 16 of 25 (64%) positive BAL fluid samples, but there was no significant difference in the rate of detection compared to the rates by the other two methods, possibly due to the small sample size. At 1 h postinfection, quantitative culture found all five samples to be positive and to have mean counts of 3.20 ± 0.51 log10 CFU/ml; in the meantime, only two and three of five samples were positive for Aspergillus by PCR (2.66 ± 0.47 log10 CE/ml) and NASBA (2.48 ± 0.17 log10 CE/ml), respectively. With the progression of the infection, the real-time assays became more sensitive than culture (Table 1), especially at 72 h postinfection, when all five BAL fluid samples were culture negative, while three of five specimens were PCR positive and four of five specimens were NASBA positive, with the mean counts by the two assays being 3.40 ± 0.38 and 3.99 ± 0.45 log10 CE/ml, respectively. Although the burdens in BAL fluid varied during the infection and no obvious trend was observed by any measurement method (Fig. 2B), the PCR measurements correlated (r = 0.887, P < 0.0001) well with the NASBA assay results (Fig. 3B), consistent with the results observed for lung tissue.
Detection of A. fumigatus in whole blood.
Apart from lung tissue and BAL fluid, whole-blood samples from rats with experimental IPA failed to demonstrate Aspergillus infection by culture or PCR, although at 72 h postinfection, three of five samples elicited positive signals by the real-time NASBA assay, with the TTPs ranging from 42 min to 47 min. At 96 h postinfection, all samples were NASBA positive, with the average TTP being 40 min, which is on the borderline of the lowest linear quantification range of the pan-Aspergillus NASBA assay. The estimated fungal burdens were about or below 100 CFU per ml of blood, although the extrapolation was not applied to obtain the exact load in blood because the TTPs were out of the lowest linear quantification limit. Meanwhile, the result of the PCR assay and culture remained negative throughout the experiment.
DISCUSSION
The early diagnosis of IPA continues to be problematic for immunocompromised patients due to the lack of rapid, sensitive, and specific diagnostic methods. Use of the combination of galactomannan (GM) detection and standard clinical, radiological, and histological examinations has been an important advance in the detection of IPA. However, the fact that testing for GM frequently has false-positive results and decreased sensitivity for patients receiving antifungal prophylaxis (17, 23, 32) highlights the critical need for more sensitive, specific, and reliable diagnostic techniques. Nucleic acid-based detection is a viable option, although the PCR technique still has not been approved for use for defining IA because of a lack of standardization and the absence of a commercially available system (1, 2).
As an alternative to PCR for molecular detection, real-time NASBA has been widely used for the detection of RNA viruses, such as enterovirus and HIV (13, 18, 25); certain microbial pathogens, including Legionella species and Vibrio cholerae (7, 16); and pathogens from food and environmental samples (3, 20). However, only studies of small scope have been reported on the use of NASBA to detect Aspergillus species (14, 36); nevertheless, the good diagnostic value of NASBA was demonstrated by those studies. To extend the applicability of the real-time NASBA assay to the quantification of the fungal load in order to monitor the progression of Aspergillus infection, we established a pan-Aspergillus real-time NASBA assay and used it in tests with an experimented model of IPA. We chose to develop a pan-Aspergillus assay to gain the complete capture of Aspergillus species, even though A. fumigatus remains the most abundant invasive species. The results were compared to those obtained by quantitative culture and qPCR. To our knowledge, this is the first report of a study that has used the real-time NASBA assay to assess the Aspergillus burden in an animal model of IPA.
A rapid real-time NASBA assay aimed at the detection of medically important Aspergillus spp. was developed by using the multicopy 28S rRNA target. The pan-Aspergillus NASBA assay demonstrated a high sensitivity, with the detection limit being 1 CFU of conidia or even lower (66% of the runs detected 0.1 CFU), which is comparable to the detection limit of the qPCR used in this and other studies (6, 27, 31). Good linearity was also observed by this assay when TNAs extracted from serially diluted conidial suspensions were used. This good linearity provides a reasonable basis for quantification of the Aspergillus burden by NASBA.
We measured the lung tissue burden by three different methods. As expected, throughout the course of the infection, the lung tissue burden assessed by NASBA or qPCR showed distinctly similar trends of consistent increases. The quantitative culture, instead, showed an initial drop, followed by a relatively stable lung tissue burden. This is in line with the findings of studies described in previous reports (29, 34). The mean value of the log10 CE per gram of lung tissue obtained by NASBA was almost 1 log10 unit lower than those assessed by quantitative culture at 24 h postinfection. However, the NASBA-based burdens increased more than 1 log10 unit 1 day later and climbed to 6.00 log10 CE per gram of lung tissue at 96 h postinfection, which was 2.25 log10 units more than the burdens obtained by CFU counting. In addition, we noticed that at 1 h postinfection, the NASBA assay failed to detect lung tissue infection and the qPCR obtained lung tissue burden measurements with mean values of 3.07 ± 0.25 log10 CE per gram of lung tissue, which is just between the limit of detection and the lower linear quantification range of the PCR assay (220 to 2,200 log10 CE/gram lung tissue). In contrast, the CFU counts for the rats at this time point yielded an average of 5.17 log10 CFU per gram of lung tissue. This paradox could partly be explained by the low efficiency of extraction of conidia from tissue samples, especially for the recovery of RNA. It was estimated that less than 0.1% of the fungal DNA was yielded when the A. fumigatus conidia in lung tissue and BAL fluid samples from the rabbits were subjected to the extraction (21). Similar difficulties were also seen in other studies (8, 19). This result may be accounted for by the fact that RNA is less stable than DNA and is easily digested by the endogenous nuclease released from the tissue during homogenization.
The fungal burden in BAL fluid specimens did not show clear trends over time by either measurement method. Although the burden counts varied from time to time, the counts obtained by the two real-time methods still correlated very well (r = 0.887) and both were more sensitive than CFU counting from 24 h postinfection throughout the experiment. It is interesting to point out that a similar paradox was encountered in assessment of the BAL fluid burden at 1 h postinfection. Quantitative culture found that at this time point all five rats were infected with A. fumigatus with a mean value of 3.20 log10 CFU per ml of BAL fluid, while both nucleic acid-based methods obtained positive results for only two or three of the five rats and the mean log10 CE/ml counts for positive samples were also lower than the values obtained by CFU counting. However, NASBA detected A. fumigatus in almost all the samples at later time points, and the burden counts were also higher than those obtained by culture. The lack of sensitivity of using BAL fluid to detect A. fumigatus in IPA cases has been well documented (26); thus, the need to improve the methodology of detection of A. fumigatus in BAL fluid was proposed (21). Our study demonstrated that the real-time NASBA assay can be an option for the detection of A. fumigatus in BAL fluid, because the sensitivity of the NASBA assay is comparable to or even greater than that of qPCR. NASBA consistently detected the infection throughout the study and provided higher estimates of the fungal burden.
Although PCR was unable to detect Aspergillus in whole blood (and the culture results for all whole-blood samples were negative), NASBA showed positive results when the infection was getting severe and rats started showing some early clinical symptoms, such as slight weight loss but no breathing problems at 72 h postinfection. Although PCR has been proved to be able to detect fungal DNA in whole-blood specimens (15, 24), there are a couple of possible reasons for the negative PCR results in this study. First, a relatively small volume of blood (1 ml) was used for detection, whereas previous studies used 3 to 5 ml (15, 24) and the use of a small volume decreased the sensitivity. Second, according to Loeffler and colleagues, only 25% of the blood samples were PCR positive even after 6 days of infection with A. fumigatus and the fungal load in blood was low (10 to 100 CFU/ml) (15). Thus, the relatively small sample size (25 infected rats) of our study and the relatively short postinfection time (4 days) could also have led to the negative PCR results. Nevertheless, the positive NASBA results with whole blood were encouraging because they demonstrated that it is possible that the NASBA assay may be used to detect fungal RNA in whole blood. Exact measurement of the fungal burden in blood was difficult, because the TTPs of positive samples were about 40 min or later, which could be translated as being the time when the burden was close to or below the lower quantification limit (100 CFU). This is consistent with the findings of a study described in a previous report (15).
There are several limitations of our study. First, the nucleic acid extraction method remains inefficient, especially for blood samples and when conidia are the majority of fungal cells in the samples. Our blood spiking experiment showed that the rate of recovery of RNA was below 0.03% when less than 1 ng of A. fumigatus RNA was spiked in 2.5 ml of sheep blood (data not shown). More efficient extraction methods are urgently needed, and such studies are ongoing. Second, for exact measurements, the TNAs extracted from the titrated conidia spiked in uninfected lung tissues or BAL fluids should be used as the quantification standards.
In conclusion, the real-time NASBA assay developed in the present study is highly sensitive and useful for the detection of Aspergillus from lung tissue and BAL fluid in an experimental model of IPA.
Acknowledgments
This work was supported by NIH grant AI066561 to D.S.P., an NIH IAAM contract, and in-kind support from bioMérieux.
Footnotes
Published ahead of print on 3 February 2010.
REFERENCES
- 1.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Chen, S. C., C. L. Halliday, and W. Meyer. 2002. A review of nucleic acid-based diagnostic tests for systemic mycoses with an emphasis on polymerase chain reaction-based assays. Med. Mycol. 40:333-357. [DOI] [PubMed] [Google Scholar]
- 3.Churruca, E., C. Girbau, I. Martinez, E. Mateo, R. Alonso, and A. Fernandez-Astorga. 2007. Detection of Campylobacter jejuni and Campylobacter coli in chicken meat samples by real-time nucleic acid sequence-based amplification with molecular beacons. Int. J. Food Microbiol. 117:85-90. [DOI] [PubMed] [Google Scholar]
- 4.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 350:91-92. [DOI] [PubMed] [Google Scholar]
- 5.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 6.Francesconi, A., M. Kasai, S. M. Harrington, M. G. Beveridge, R. Petraitiene, V. Petraitis, R. L. Schaufele, and T. J. Walsh. 2008. Automated and manual methods of DNA extraction for Aspergillus fumigatus and Rhizopus oryzae analyzed by quantitative real-time PCR. J. Clin. Microbiol. 46:1978-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fykse, E. M., G. Skogan, W. Davies, J. S. Olsen, and J. M. Blatny. 2007. Detection of Vibrio cholerae by real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 73:1457-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haugland, R. A., J. L. Heckman, and L. J. Wymer. 1999. Evaluation of different methods for the extraction of DNA from fungal conidia by quantitative competitive PCR analysis. J. Microbiol. Methods 37:165-176. [DOI] [PubMed] [Google Scholar]
- 9.Imhof, A., C. Schaer, G. Schoedon, D. J. Schaer, R. B. Walter, A. Schaffner, and M. Schneemann. 2003. Rapid detection of pathogenic fungi from clinical specimens using LightCycler real-time fluorescence PCR. Eur. J. Clin. Microbiol. Infect. Dis. 22:558-560. [DOI] [PubMed] [Google Scholar]
- 10.Jordanides, N. E., E. K. Allan, L. A. McLintock, M. Copland, M. Devaney, K. Stewart, A. N. Parker, P. R. Johnson, T. L. Holyoake, and B. L. Jones. 2005. A prospective study of real-time panfungal PCR for the early diagnosis of invasive fungal infection in haemato-oncology patients. Bone Marrow Transplant. 35:389-395. [DOI] [PubMed] [Google Scholar]
- 11.Kami, M., T. Fukui, S. Ogawa, Y. Kazuyama, U. Machida, Y. Tanaka, Y. Kanda, T. Kashima, Y. Yamazaki, T. Hamaki, S. Mori, H. Akiyama, Y. Mutou, H. Sakamaki, K. Osumi, S. Kimura, and H. Hirai. 2001. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clin. Infect. Dis. 33:1504-1512. [DOI] [PubMed] [Google Scholar]
- 12.Klingspor, L., and S. Jalal. 2006. Molecular detection and identification of Candida and Aspergillus spp. from clinical samples using real-time PCR. Clin. Microbiol. Infect. 12:745-753. [DOI] [PubMed] [Google Scholar]
- 13.Landry, M. L., R. Garner, and D. Ferguson. 2005. Real-time nucleic acid sequence-based amplification using molecular beacons for detection of enterovirus RNA in clinical specimens. J. Clin. Microbiol. 43:3136-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeffler, J., H. Hebart, P. Cox, N. Flues, U. Schumacher, and H. Einsele. 2001. Nucleic acid sequence-based amplification of Aspergillus RNA in blood samples. J. Clin. Microbiol. 39:1626-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeffler, J., K. Kloepfer, H. Hebart, L. Najvar, J. R. Graybill, W. R. Kirkpatrick, T. F. Patterson, K. Dietz, R. Bialek, and H. Einsele. 2002. Polymerase chain reaction detection of aspergillus DNA in experimental models of invasive aspergillosis. J. Infect. Dis. 185:1203-1206. [DOI] [PubMed] [Google Scholar]
- 16.Loens, K., T. Beck, H. Goossens, D. Ursi, M. Overdijk, P. Sillekens, and M. Ieven. 2006. Development of conventional and real-time NASBA for the detection of Legionella species in respiratory specimens. J. Microbiol. Methods 67:408-415. [DOI] [PubMed] [Google Scholar]
- 17.Marr, K. A., M. Laverdiere, A. Gugel, and W. Leisenring. 2005. Antifungal therapy decreases sensitivity of the Aspergillus galactomannan enzyme immunoassay. Clin. Infect. Dis. 40:1762-1769. [DOI] [PubMed] [Google Scholar]
- 18.McClernon, D. R., C. Vavro, and M. St. Clair. 2006. Evaluation of a real-time nucleic acid sequence-based amplification assay using molecular beacons for detection of human immunodeficiency virus type 1. J. Clin. Microbiol. 44:2280-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller, F. M., K. E. Werner, M. Kasai, A. Francesconi, S. J. Chanock, and T. J. Walsh. 1998. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J. Clin. Microbiol. 36:1625-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadal, A., A. Coll, N. Cook, and M. Pla. 2007. A molecular beacon-based real time NASBA assay for detection of Listeria monocytogenes in food products: role of target mRNA secondary structure on NASBA design. J. Microbiol. Methods 68:623-632. [DOI] [PubMed] [Google Scholar]
- 21.O'Sullivan, C. E., M. Kasai, A. Francesconi, V. Petraitis, R. Petraitiene, A. M. Kelaher, A. A. Sarafandi, and T. J. Walsh. 2003. Development and validation of a quantitative real-time PCR assay using fluorescence resonance energy transfer technology for detection of Aspergillus fumigatus in experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 41:5676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perlin, D. S., and Y. Zhao. 2009. Molecular diagnostic platforms for detecting Aspergillus. Med. Mycol. 47(Suppl. 1):S223-S232. [DOI] [PubMed] [Google Scholar]
- 23.Pinel, C., H. Fricker-Hidalgo, B. Lebeau, F. Garban, R. Hamidfar, P. Ambroise-Thomas, and R. Grillot. 2003. Detection of circulating Aspergillus fumigatus galactomannan: value and limits of the Platelia test for diagnosing invasive aspergillosis. J. Clin. Microbiol. 41:2184-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez, M., C. Castro, J. C. Palomares, M. J. Torres, A. I. Aller, M. Ruiz, J. Aznar, and E. Martin-Mazuelos. 2009. Molecular detection and identification of Aspergillus spp. from clinical samples using real-time PCR. Mycoses 52:129-134. [DOI] [PubMed] [Google Scholar]
- 25.Rutjes, S. A., R. Italiaander, H. H. van den Berg, W. J. Lodder, and A. M. de Roda Husman. 2005. Isolation and detection of enterovirus RNA from large-volume water samples by using the NucliSens miniMAG system and real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 71:3734-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito, H., E. J. Anaissie, R. C. Morice, R. Dekmezian, and G. P. Bodey. 1988. Bronchoalveolar lavage in the diagnosis of pulmonary infiltrates in patients with acute leukemia. Chest 94:745-749. [DOI] [PubMed] [Google Scholar]
- 27.Scotter, J. M., and S. T. Chambers. 2005. Comparison of galactomannan detection, PCR-enzyme-linked immunosorbent assay, and real-time PCR for diagnosis of invasive aspergillosis in a neutropenic rat model and effect of caspofungin acetate. Clin. Diagn. Lab. Immunol. 12:1322-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheppard, D. C., J. R. Graybill, L. K. Najvar, L. Y. Chiang, T. Doedt, W. R. Kirkpatrick, R. Bocanegra, A. C. Vallor, T. F. Patterson, and S. G. Filler. 2006. Standardization of an experimental murine model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 50:3501-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheppard, D. C., K. A. Marr, D. N. Fredricks, L. Y. Chiang, T. Doedt, and S. G. Filler. 2006. Comparison of three methodologies for the determination of pulmonary fungal burden in experimental murine aspergillosis. Clin. Microbiol. Infect. 12:376-380. [DOI] [PubMed] [Google Scholar]
- 30.Simpkins, S. A., A. B. Chan, J. Hays, B. Popping, and N. Cook. 2000. An RNA transcription-based amplification technique (NASBA) for the detection of viable Salmonella enterica. Lett. Appl. Microbiol. 30:75-79. [DOI] [PubMed] [Google Scholar]
- 31.Spiess, B., D. Buchheidt, C. Baust, H. Skladny, W. Seifarth, U. Zeilfelder, C. Leib-Mosch, H. Morz, and R. Hehlmann. 2003. Development of a LightCycler PCR assay for detection and quantification of Aspergillus fumigatus DNA in clinical samples from neutropenic patients. J. Clin. Microbiol. 41:1811-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulahian, A., F. Boutboul, P. Ribaud, T. Leblanc, C. Lacroix, and F. Derouin. 2001. Value of antigen detection using an enzyme immunoassay in the diagnosis and prediction of invasive aspergillosis in two adult and pediatric hematology units during a 4-year prospective study. Cancer 91:311-318. [DOI] [PubMed] [Google Scholar]
- 33.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 34.Vallor, A. C., W. R. Kirkpatrick, L. K. Najvar, R. Bocanegra, M. C. Kinney, A. W. Fothergill, M. L. Herrera, B. L. Wickes, J. R. Graybill, and T. F. Patterson. 2008. Assessment of Aspergillus fumigatus burden in pulmonary tissue of guinea pigs by quantitative PCR, galactomannan enzyme immunoassay, and quantitative culture. Antimicrob. Agents Chemother. 52:2593-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanittanakom, N., C. R. Cooper, Jr., M. C. Fisher, and T. Sirisanthana. 2006. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin. Microbiol. Rev. 19:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo, J. H., S. M. Choi, D. G. Lee, S. H. Park, J. H. Choi, E. Y. Kwon, and W. S. Shin. 2007. Comparison of the real-time nucleic acid sequence-based amplification (RTi-NASBA) with conventional NASBA, and galactomannan assay for the diagnosis of invasive aspergillosis. J. Korean Med. Sci. 22:672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao, Y., S. Park, B. N. Kreiswirth, C. C. Ginocchio, R. Veyret, A. Laayoun, A. Troesch, and D. S. Perlin. 2009. Rapid real-time nucleic acid sequence-based amplification-molecular beacon platform to detect fungal and bacterial bloodstream infections. J. Clin. Microbiol. 47:2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]