Abstract
Rapid laboratory diagnosis is critical for treating, managing, and preventing methicillin-resistant Staphylococcus aureus (MRSA) infections. We evaluated and compared the potential for MRSA detection of five chromogenic media, Brilliance MRSA agar (Oxoid), ChromID (bioMérieux), MRSASelect (Bio-Rad), CHROMagar (CHROMagar Microbiology), and BBL-CHROMagar (BD Diagnostics). Media were tested with log serial dilutions (100 to 106 CFU) of pure isolates of MRSA (n = 60), non-MRSA (n = 27), and defined mixtures thereof simulating clinical samples (n = 84). Further evaluations were done on pre-enriched nasal and groin screening swabs (n = 213) from 165 hospitalized patients. Randomized samples were spiral plated on each medium and independently scored by five investigators for characteristic colonies at 24 and 48 h of incubation. Confirmatory testing of up to five putative MRSA colonies recovered from each medium was done. The cumulative average sensitivity with isolates, mixtures, and clinical samples was the highest for Brilliance MRSA agar (97%) and similar for the other four media (≥92%). The cumulative average specificity was the highest for BBL-CHROMagar (99%), followed by MRSASelect (98%), CHROMagar (97%), ChromID (89%), and Brilliance MRSA agar (86%). All of the media detected MRSA at 10 and 1 CFU, although at these low loads, few MRSA samples harboring SCCmec type III or IV were misinterpreted as non-MRSA by investigators. False-positive results were mainly due to methicillin-resistant S. epidermidis. For an arbitrary MRSA prevalence of 5% and based on patient sample evaluations, the positive predictive values for BBL-CHROMagar and CHROMagar (∼84%) were the highest. The negative predictive values of all of the media were ≥92% for MRSA prevalences ranging from 5% to 30%. In conclusion, BBL-CHROMagar and CHROMagar gave the best overall results for detection of MRSA, irrespective of the sample concentration, investigator, or incubation period.
Methicillin-resistant Staphylococcus aureus (MRSA) has emerged as a major nosocomial pathogen in the last decade. Patients colonized with MRSA serve as reservoirs of self-infection or dissemination to other patients and to the hospital environment (6, 12, 22). Hence, screening for MRSA carriage and contact isolation of MRSA carriers are crucial for effective hospital infection control (9). Employing rapid and sensitive screening assays for MRSA detection could help to further improve infection control, as well as decrease costs (10, 13).
In recent years, the use of chromogenic media has become a key method for the rapid identification of microorganisms in clinical samples (20). These media detect key microbial enzymes as diagnostic markers for pathogens through the use of “chromogenic” substrates incorporated into a solid-agar-based matrix (20). In contrast to conventional culture media, chromogenic media allow direct colony color-based identification of the pathogen from the primary culture. This reduces the need for subculture for further biochemical testing and hence the time until a result is obtained. Currently available chromogenic media for MRSA detection incorporate chromogens to differentiate S. aureus from other pathogens and antibiotics for selective growth of MRSA. These media differ in their chromogenic substrates, antibiotic formulations, and/or concentrations, factors that impact their sensitivity and specificity for MRSA detection (reviewed in reference 13). We compared the potential of five of the most commonly used commercial chromogenic media for MRSA detection using pure MRSA isolates, non-MRSA isolates, and mixtures thereof at defined concentrations simulating clinical samples. Further evaluations of the media were carried out with nasal and groin screening samples from hospitalized patients.
(Part of this work was presented at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 24 to 28 October 2008.)
MATERIALS AND METHODS
Study design.
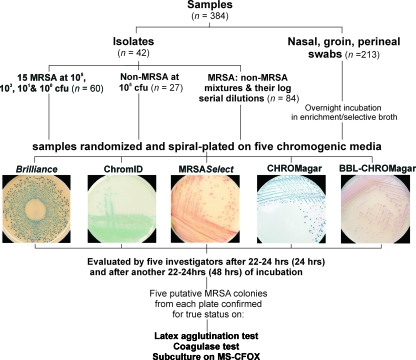
Figure 1 shows the outline of this study. The chromogenic media evaluated were ChromID MRSA (bioMérieux, Marcy l'Etoile, France), MRSASelect (Bio-Rad, Nazareth Eke, Belgium), CHROMagar MRSA (CHROMagar, Paris, France), BBL-CHROMagar MRSA (BD Diagnostics, Erembodegem, Belgium), and the recently introduced Brilliance MRSA agar (Oxoid, Basingstoke, United Kingdom; marketed as Remel Spectra in the United States). As differences between the constituents of BBL-CHROMagar and the original CHROMagar formulation are not disclosed, we tested both products. Each medium was challenged with a total of 384 samples that consisted of well-characterized bacterial isolates and mixtures thereof at various concentrations, as well as patient screening samples (see further). Each sample was assigned an Excel-generated randomization code and inoculated onto all five of the media in random order using a spiral plater (Eddy Jet). All of the media were incubated for 22 to 24 h (referred to as 24 h of incubation) and then again for another 22 to 24 h (referred to as 48 h of incubation) at 37°C under aerobic conditions. The entire set of inoculated chromogenic media (n = 1,920 plates; 384 samples plated on five media) were scored independently by each of the five investigators at both time points, i.e., at 24 and 48 h of incubation. Furthermore, putative MRSA colonies were recovered from each of the five media and confirmed by standard biochemical tests.
FIG. 1.
Study design. MS-CFOX, mannitol salt agar with 4 μg/ml of cefoxitin.
Samples. (i) Isolates.
Forty-two well-characterized MRSA (n = 15) and non-MRSA (n = 27) strains were inoculated as pure isolates onto the five chromogenic media. The MRSA collection included strains of some of the most prevalent clonal lineages that have disseminated worldwide and harbor various known types of SCCmec (staphylococcal cassette chromosome mec, the mobile genetic element that carries the methicillin resistance gene mecA) (see Table 1). The non-MRSA isolates included methicillin-sensitive S. aureus (MSSA; n = 5), methicillin-resistant coagulase-negative staphylococci (MRCoNS; S. epidermidis, S. warneri, and S. sciuri; n = 5), methicillin-sensitive coagulase-negative staphylococci (MSCoNS; S. capitis, S. epidermidis, and S. lugdunensis; n = 3), Enterococcus spp. (Enterococcus faecalis, E. faecium, and E. casseliflavus; n = 4), and various gram-negative bacteria (Escherichia coli, Klebsiella pneumoniae, K. oxytoca, Acinetobacter sp., and Pseudomonas aeruginosa; n = 10). Staphylococcal isolates were characterized using the coagulase test and identified to the species level by a semiautomated biochemical test (API Staph; bioMérieux, Marcy l'Étoile, France). Nonstaphylococci were identified and identified to the species level either by relevant API tests or by standard biochemical tests (23, 25).
TABLE 1.
Characteristics of the MRSA strains tested in this study
| Strain no. | MRSA phenotypea | Clone | SCCmec type | mecA/pvl | MIC (μg/ml) of: |
|
|---|---|---|---|---|---|---|
| Oxacillin | Cefoxitin | |||||
| 1 | hoMRSA | Iberian | IB | +/− | 256 | >256 |
| 2 | hoMRSA | Iberian | IA | +/− | 256 | >256 |
| 3 | hoMRSA | Sporadic | II | +/− | 8 | >256 |
| 4 | hoMRSA | Brazilian | III | +/− | 256 | >256 |
| 5 | hoMRSA | Hungarian | IIIA | +/− | 256 | >256 |
| 6 | hoMRSA | Hungarian | IIIB | +/− | 256 | >256 |
| 7 | hoMRSA | Hungarian | IIIC | +/− | 256 | >256 |
| 8 | hoMRSA | Hungarian | IIID | +/− | 256 | >256 |
| 9 | hoMRSA | Poland | IIIE | +/− | 16 | >256 |
| 10 | he/hoMRSA | Berlin | IV | +/− | 2 | 32 |
| 11 | he/hoMRSA | Mediterranean | IV | +/+ | 2 | 32 |
| 12 | heMRSA | Mediterranean | IV | +/+ | 16 | 32 |
| 13 | he/hoMRSA | Mediterranean | IV | +/+ | 1.5 | 32 |
| 14 | he/hoMRSA | Mediterranean | IV | +/+ | 1.5 | 64 |
| 15 | hoMRSA | SWb Pacific | -c | +/− | 2 | 64 |
Abbreviations: ho, homogenous resistance; he, heterogenous resistance.
SW, southwest.
-, undefined SCCmec type.
For testing on chromogenic media, all of the isolates were subcultured on Mueller-Hinton (MH) agar and incubated overnight. A colony suspension in brain heart infusion broth (BHI) with a turbidity equivalent to a 0.5 McFarland standard (∼108 CFU/ml) was made from the pure subculture on MH agar. After appropriate dilutions, a 105-CFU colony suspension of each of the 42 isolates was spiral plated onto the five chromogenic media and onto MH agar, which served as a positive control for bacterial growth. The limits of detection of each of the five media were also determined. For this, pure colony suspensions of all 15 MRSA, 3 MRCoNS, and 2 MSCoNS isolates were serially diluted (103, 101, and 100 CFU) and spiral plated onto the five chromogenic media. All of the media were incubated for 24 and 48 h and were evaluated at both time points by all five of the investigators.
(ii) Mixtures of isolates.
To study cross-reactions due to non-MRSA on the chromogenic media, mixtures of MRSA and non-MRSA isolates were prepared at defined concentrations. The strains used were MRSA (n = 2; a hospital-acquired strain with SCCmec type III and a community-acquired strain with SCCmec type IV) (see Table 1), MRCoNS (n = 2; S. epidermidis and S. warneri), enterococci (n = 2, E. faecalis and E. faecium), and one strain each of MSSA, E. coli, K. pneumoniae, Acinetobacter sp., and P. aeruginosa. To prepare various combinations of “parent” mixtures, 105 or 106 CFU each of MRSA, MRCoNS, MSSA, and occasionally nonstaphylococcal species were mixed (n = 20; see Table S1 in the supplemental material). These parent mixtures were further diluted serially (101-, 103-, and 105-fold) to obtain a total of 80 mixture samples containing MRSA. Similarly, non-MRSA mixtures were prepared by mixing an MSSA and an MRCoNS isolate at 105 or 106 CFU (n = 4; see Table S1 in the supplemental material) and inoculated at this single high load without further dilution. Each of the 84 mixture samples was plated on the five chromogenic media, as well as on mannitol salt agar with 4 μg/ml cefoxitin (MS-CFOX) and MH agar, the latter two media acting as growth controls.
(iii) Clinical samples.
Nasal and groin screening swabs (n = 213) were collected from 165 patients admitted to the University Hospital of Antwerp with a history of MRSA carriage. Samples were individually incubated overnight in a selective enriched broth (BHI with 7% NaCl and 4 μg/ml oxacillin), and 50 μl of the broth sample was inoculated onto each of the five chromogenic media and also onto MH agar.
Before the samples were plated on the chromogenic media, the MRSA status of all of the patient samples was established using culture-based and molecular methods. All of the pre-enriched broth samples were subcultured on mannitol salt agar, and putative S. aureus colonies were confirmed by tube coagulase and screened for methicillin resistance on MH agar supplemented with 6 μg/ml oxacillin and 4% saline. Furthermore, oxacillin and cefoxitin MICs were determined according to CLSI guidelines (5). Molecular screening of samples was performed utilizing Hyplex StaphyloResist PCR (BAG, Lich, Germany) and an “in-house” multiplex PCR (11). Based on these tests, MRSA strains were detected in 87 of the 213 samples. To obtain an overview of the prevalent SCCmec types, one MRSA isolate each from 46 of the 87 MRSA-positive broths was typed for SCCmec types I to V by real-time PCR (Applied Biosystems, Lennik, Belgium) as described previously (8). Of the 46 MRSA isolates typed, 40 (87%) harbored SCCmec type IV, 5 (11%) harbored SCCmec type I, and 1 (0.02%) harbored SCCmec type II.
Confirmatory testing of putative MRSA colonies recovered from chromogenic media.
One investigator also recovered putative MRSA colonies from the isolate mixtures and patient samples inoculated onto the five chromogenic media for confirmatory testing by latex agglutination (Pastorex Staph Plus; Bio-Rad, Belgium), tube coagulase test, and a subculture on MS-CFOX. Up to five colonies showing a characteristic morphology were collected from each chromogenic medium and purified on blood agar. Confirmatory testing was done in succession on these colonies and stopped once a colony was confirmed as MRSA. Colonies identified as non-MRSA were identified to the genus level by using standard biochemical tests (23, 25).
Investigator assessment of the chromogenic media.
Investigators scored the five media for ease of colony color differentiation between MRSA and non-MRSA and general user friendliness. Investigators also graded the five media in order of preference: the media were assigned choice-weighted scores, with the first through the last choices weighted 5 through 1.
Statistical analysis.
Investigator response and confirmation test results were collected together with the true status of each sample established on the basis of a combination of biochemical/phenotypic/genotypic tests. Using a logistic regression model, sensitivities and specificities averaged for five investigators were estimated for pure isolates, mixtures, and patient samples cultured on each chromogenic medium. A Bayesian paradigm was utilized assuming readouts of the five investigators as a random effect and the sample concentration, the incubation period, and the different chromogenic media as fixed effects, and the model was fitted using the R software (www.r-project.org). In such an approach, credible intervals (CIs) are calculated that are roughly equivalent to confidence intervals in classical (frequentist) statistics. Positive predictive value and negative predictive value predictions were made based on MRSA prevalence estimates ranging from 0 to 50%.
RESULTS
Detection of pure isolates.
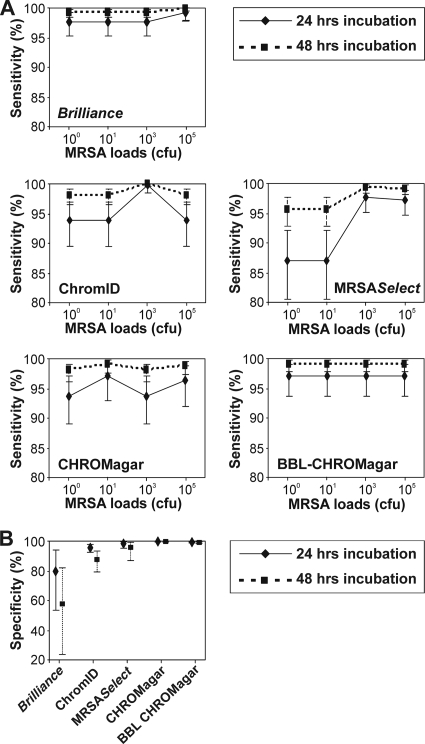
The sensitivities of Brilliance MRSA agar (97.6%; 95% CI, 95.13 to 99.10) and BBL-CHROMagar (97.0%; 95% CI, 93.70 to 98.97) were consistently high, while those of ChromID, MRSASelect, and CHROMagar varied across the four pure MRSA isolate loads tested (Fig. 2A, continuous lines). However, at 48 h of incubation, the sensitivities of all five of the media increased to 95 to 100% at all of the MRSA loads tested (Fig. 2A, dotted lines). All five of the chromogenic media could sustain growth of MRSA strains at loads as low as 10 CFU and even 1 (100) CFU, although at lower loads, a few strains were misinterpreted as non-MRSA by investigators. These included MRSA harboring SCCmec type III on MRSASelect, CHROMagar, and ChromID and SCCmec type IV on BBL-CHROMagar (strains 5, 7, and 12, Table 1). Brilliance MRSA agar showed the highest sensitivity among the five media at an MRSA load of 100 CFU, with only one SCCmec type I-harboring MRSA strain being misinterpreted by two investigators (strain 1, Table 1). However, the specificity of Brilliance MRSA agar was the lowest because of misinterpretation of four of the five MRCoNS strains tested and one strain each of MSSA and E. coli at 24 h of incubation by two investigators (Fig. 2B). At 48 h of incubation, all four of the enterococcal strains tested were also misinterpreted by four investigators, thus further decreasing the specificity of this medium to less than 60%. On the other hand, the specificities of CHROMagar and BBL-CHROMagar were more than 99% at both 24 and 48 h of incubation with extremely narrow CIs, indicating the high precision of these estimates (Fig. 2B). Interinvestigator variations in observations were negligible for pure isolate samples (σ̂R2= 0.03; 95% CI, 0.00 to 0.15).
FIG. 2.
Sensitivities and specificities of the five chromogenic media utilizing MRSA and non-MRSA as pure isolates. The x axes in panel A show the absolute MRSA loads inoculated onto the five media. All of the inoculated media were independently evaluated by five investigators.
Detection of defined strain mixtures.
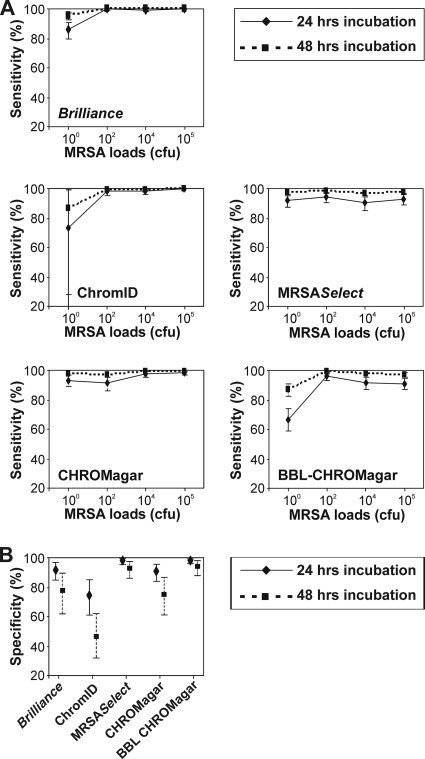
Of the 80 MRSA-positive mixture samples, Brilliance MRSA agar, ChromID, MRSASelect, CHROMagar, and BBL-CHROMagar failed to grow 1, 1, 4, 0, and 7 samples, respectively. While all five of the media showed high sensitivities for detection of MRSA in mixtures inoculated at 105, 104, and 102 CFU, the sensitivities of BBL-CHROMagar, ChromID, and Brilliance MRSA agar were decreased for the detection of 100 CFU of MRSA (Fig. 3A). Either these bacteria did not grow on these media or the colonies were misinterpreted as non-MRSA by investigators. At 24 h of incubation, the decrease in sensitivity was the most pronounced for BBL-CHROMagar (66.7%; 95% CI, 58.88 to 74.03), followed by ChromID (73.7%; 95% CI, 10.41 to 99.51), and Brilliance MRSA agar (85.8%; 95% CI, 79.44 to 91.05) (Fig. 3A, continuous lines). False-negative results were due to MRSA harboring SCCmec types III and IV on BBL-CHROMagar and ChromID and only SCCmec type IV on Brilliance MRSA agar. The specificities of both MRSASelect and BBL-CHROMagar were high and sustained at 48 h of incubation (Fig. 3B). However, with the exception of BBL-CHROMagar, at least one of the four non-MRSA mixtures tested was misinterpreted by one investigator on the other four media. Interinvestigator variations in observations were minor for the strain mixtures (σ̂R2= 0.02; 95% CI, 0.00 to 0.10).
FIG. 3.
Sensitivities and specificities of the five chromogenic media utilizing defined MRSA and non-MRSA mixtures at various concentrations. The x axes in panel A show the absolute MRSA loads inoculated as mixtures onto the five media. All of the inoculated media were independently evaluated by five investigators.
In addition, up to five putative MRSA colonies were collected by an investigator for further confirmatory testing by standard biochemical methods. Correct MRSA identifications in the first colony were the highest for CHROMagar (95%, n = 76), followed by BBL-CHROMagar (86%, n = 69), ChromID (85%, n = 68), Brilliance MRSA agar (79%, n = 63), and MRSASelect (73%, n = 58). However, with two colony analyses, Brilliance MRSA agar, ChromID, and MRSASelect also achieved correct MRSA identifications of >95%. All of the remaining samples were correctly identified by the third colony analysis for all of the media. Isolates giving the same colony color and morphology as MRSA on all of the media were identified as MRCoNS (S. epidermidis).
Validation of chromogenic media with screening specimens from patients.
Brilliance MRSA agar showed the highest sensitivities and lowest specificities with patients' samples incubated for 24 and 48 h (Table 2). The specificities of CHROMagar and BBL-CHROMagar were the highest at 24 h of incubation and sustained at 48 h of incubation. The sensitivities of all of the media increased by ∼8% at 48 h of incubation and, expectedly, paralleled a concomitant decrease in specificity, except for the two CHROMagars. Interinvestigator variations in observations were small for the patient samples (σ̂R2= 0.01; 95% CI, 0.00 to 0.06).
TABLE 2.
Sensitivities and specificities of the five chromogenic media tested with patient samplesa
| Chromogenic medium | 24 h |
48 h |
||||||
|---|---|---|---|---|---|---|---|---|
| % Sensitivity | 95% CI | % Specificity | 95% CI | % Sensitivity | 95% CI | % Specificity | 95% CI | |
| Brilliance MRSA agar | 89.9 | 87.3-92.2 | 86.9 | 84.9-88.7 | 96.4 | 95.3-97.3 | 69.0 | 65.9-72.1 |
| ChromID | 82.8 | 79.6-85.8 | 96.3 | 95.4-97.1 | 93.5 | 92.0-94.8 | 89.7 | 87.6-91.6 |
| MRSASelect | 80.7 | 77.6-83.8 | 97.2 | 96.4-97.9 | 92.6 | 91.0-94.0 | 92.1 | 90.2-93.8 |
| CHROMagar | 81.9 | 78.6-84.9 | 99.1 | 98.7-99.5 | 93.1 | 91.5-94.5 | 97.4 | 96.3-98.4 |
| BBL-CHROMagar | 82.9 | 79.8-85.8 | 99.2 | 98.8-99.5 | 93.5 | 92.0-94.8 | 97.8 | 96.7-98.6 |
All of the inoculated media were independently evaluated by five investigators.
The diagnostic accuracy of each medium assessed on the basis of investigator observations of patient samples is illustrated in the receiving operator characteristic plot (see Fig. S1 in the supplemental material). Areas under the curve were the greatest for BBL-CHROMagar and CHROMagar (0.93), followed by ChromID and MRSASelect (0.90) and Brilliance MRSA agar (0.86). The positive predictive values of BBL-CHROMagar and CHROMagar were the highest among the five media (see Fig. S2 in the supplemental material). At 24 h of incubation and for an arbitrary MRSA prevalence of 5%, the positive predictive values of BBL-CHROMagar and CHROMagar were 85% and 83%, respectively, which are substantially higher than those of the other three media. On the other hand, the negative predictive values for all five of the media at 24 h of incubation were uniformly high (≥92% with MRSA prevalence of up to 30%; see Fig. S2 in the supplemental material).
Of the 87 MRSA-positive patient samples, 5, 8, 10, 11, and 10 samples failed to grow on Brilliance MRSA agar, ChromID, MRSASelect, CHROMagar, and BBL-CHROMagar, respectively. Confirmatory testing by one investigator on putative MRSA recovered from samples growing on the media showed that correct MRSA identification in samples on first colony screening was the highest for Brilliance MRSA agar (92%, n = 80), followed by BBL-CHROMagar and MRSASelect (87%, n = 76), ChromID (85%, n = 74), and CHROMagar (84%, n = 73). All of the remaining samples were correctly identified by the third colony analysis with all of the media.
Qualitative evaluation of media by investigators.
Four of the five investigators graded the media. Both CHROMagar and BBL-CHROMagar scored equal and maximum numbers of votes for clear colony coloration and ease of differentiation between non-MRSA and MRSA colonies (3/4 votes each). CHROMagar also obtained the maximum number of votes for user friendliness (3/4 votes), followed by BBL-CHROMagar and MRSASelect (2/4 votes each). CHROMagar was the most preferred medium (weighted score, 18), and BBL-CHROMagar the second most preferred medium (weighted score, 15). MRSASelect, Brilliance MRSA agar, and ChromID were third, fourth, and fifth, with weighted scores of 14, 10, and 5, respectively.
DISCUSSION
In the present study, BBL-CHROMagar (BD Diagnostics) and CHROMagar (CHROMagar Microbiology) gave the best overall results for the detection of MRSA, irrespective of the sample concentration, investigator, or incubation period. Table 3 shows a global comparison of the medium performance results obtained in the present study. All five of the media exhibited excellent negative predictive values and are well suited as screening tests for excluding MRSA carriage with a low risk of false-negative results. However, poor discriminative power or low specificities clearly demarcated Brilliance MRSA agar (Oxoid) and ChromID (bioMérieux) from the other three media. In a clinical setting, this might result in lost time and higher costs due to unnecessary patient isolation.
TABLE 3.
Summary of the comparison of the five chromogenic media
| Characteristic | Brilliance MRSA agar | ChromID | MRSASelect | CHROMagar | BBL CHROMagar |
|---|---|---|---|---|---|
| Cumulative avg sensitivity (%), 24 h of incubationa | 96.9 | 93.8 | 92.4 | 94.5 | 92.3 |
| Cumulative avg specificity (%), 24 h of incubation | 85.9 | 88.8 | 97.9 | 96.7 | 99.1 |
| Sensitivity (%) of MRSA detection at lowest pure isolate load (100 CFU) | 97.6 | 93.8 | 86.9 | 93.7 | 97.0 |
| Specificity (%) of media at highest non-MRSA loads in mixtures (105-106 CFU) | 92.1 | 74.5 | 97.8 | 90.9 | 98.2 |
| Positive predictive value (%) at 5% MRSA prevalence, 24 h of incubationb | 27 | 54 | 60 | 83 | 85 |
| Negative predictive value (%) at 30% MRSA prevalence, 24 h of incubationb | 95 | 93 | 92 | 93 | 93 |
| Correct identification (%) of MRSA-positive samples on first colony analysis | |||||
| Mixture samples | 79 | 85 | 73 | 95 | 86 |
| Patient samples pre-enriched in an enrichment/selective broth | 92 | 85 | 87 | 84 | 87 |
| Investigator evaluation of easy colony color differentiation (preference rank)c | 4 | 5 | 3 | 1 | 2 |
The cumulative average sensitivity or specificity is the average sensitivity or specificity of each medium for the three sample types (isolates, mixtures, and patient samples).
Positive and negative predictive values are shown for arbitrary “low” and “high” MRSA prevalences, respectively.
Media were graded according to choice-weighted scores.
To stress the importance of investigator perception and decision making based on colony characteristics, the performance of the chromogenic media was assessed on the basis of evaluations by five investigators. Work experience in a microbiology laboratory varied widely among these investigators, ranging from a few months to more than 25 years. Despite these differences, interinvestigator variations in observations were negligible, underscoring the ease of interpretation and the nonessentiality of technical expertise required for all five of the media.
One potential caveat in our study could be that patient samples were not derived from multiple hospitals or geographical areas, thereby limiting variability. Moreover, an overnight sample pre-enrichment/oxacillin selection would also decrease the likelihood of observing cross-reactivity due to cohabitant bacteria and generally enhance medium performance. We overcame these issues by also challenging the media with defined mixtures of bacteria that are likely to coexist with MRSA at various sampling sites on the human body in various proportions. Confirmatory testing of putative MRSA colonies revealed that cross-reactivity on the chromogenic media at 24 h of incubation was largely due to other staphylococcal species, especially S. epidermidis (MRCoNS). This would impact medium specificity while screening groin/perineal samples and, to a lesser extent, also nasal samples (1, 2, 24). Our confirmation results further suggested that a definitive identification of an MRSA-positive sample might necessitate the screening of up to two or three putative MRSA colonies from the chromogenic media evaluated here. On the other hand, all five of the media could easily detect MRSA strains classified as low level resistant based on oxacillin MICs of 1.5 to 2 μg/ml. This was because of the high cefoxitin MICs of these strains (32 to 64 μg/ml), as cefoxitin is a more efficient inducer of the mecA-encoded methicillin resistance protein PBP2a than is oxacillin (7), which allowed growth on the cefoxitin (or other cephamycin analogue)- containing chromogenic media utilized in the present study.
Issues with sensitivity were mainly evident at lower MRSA loads. Either some MRSA strains harboring SCCmec type III or IV did not yield characteristic colonies at 24 h of incubation at MRSA loads of 1 and 10 CFU or the colony color was not specific enough, causing some investigators to misinterpret these samples as MRSA negative. These findings can partly explain the high variability in medium performance generally observed between diagnostic studies, which might be influenced both by the predominant circulating MRSA types and by the differences in their colonization potential (19). For instance, the postenrichment sensitivities of ChromID and MRSASelect reported in the present study are similar to those found in another Belgian study (17), which are, on average, 13% lower than those reported in a Swiss study (4). A predominance of SCCmec type IV-harboring MRSA in the patient samples screened in the present study and also in the other Belgian study (Claire Nonhoff, Hôpital Erasme, Brussels, Belgium, personal communication) (17), might be responsible for the inferior performance of the media. Whether problems in detecting certain MRSA strains are related to the genetic background of the S. aureus strains that have acquired specific SCCmec types or are due to differences in fitness costs incurred by the carriage of these SCCmec types is not known. Interstrain differences in colonization potential have been previously reported. We have recently shown a 10,000-fold higher average MRSA yield from nasal swabs collected from patients at the University of Geneva Hospitals in comparison to that of a previous study in the United States (14, 26). While other variables cannot be discounted, the predominant MRSA clone recovered from the Geneva samples, ST228 (SCCmec type I), is an efficient colonizer, as shown by its aggressively persistent and long-term bronchial colonization of cystic fibrosis patients (15).
Active screening now forms an integral part of all effective strategies to control MRSA, and a search for the optimal “rapid” screening assay has motivated several studies comparing the utility of molecular and culture-based tests for MRSA detection. These studies show mixed results. One study modeled the impact of direct detection of MRSA carriage using a chromogenic medium or real-time PCR on capture of patient isolation days (PIDs) (21). Their results showed that a real-time PCR could capture up to 22.7% more PIDs, although it also generated more unnecessary PIDs due to a specificity slightly lower than that of a chromogenic medium (21). Another study found a real-time PCR and a chromogenic medium to perform similarly (3), while yet another study showed molecular testing to be superior in terms of rapidity of results and sensitivity (18). Major deterrents to the more widespread use of PCR-based assays have been their higher costs and, for some assays, also the need for technical expertise or sample batching. In contrast, costs of screening on a chromogenic medium are similar to those of conventional culture once technician time and downstream processing costs are considered, and the time until a result is obtained can vary from 1.4 days to 1.7 days, depending on the confirmatory tests required for each medium (16). While the choice between molecular and culture-based screening assays is governed by several factors, including hospital turnover, available isolation facilities, technical expertise, etc., the advantages afforded by chromogenic media should encourage laboratories currently screening for MRSA carriage by conventional methods to adopt these rapid culture-based tools.
Supplementary Material
Acknowledgments
We thank Oxoid, bioMérieux, Bio-Rad Laboratories, CHROMagar Microbiology, and BD Diagnostics for providing chromogenic media. We thank MOSAR WP2 Study Team partners Waleria Hryniewicz, Jordi Vila, Marek Gniadkowski, and Claire Poyart for providing strains.
This work and W.S. are supported by funding from the European Community (MOSAR network contract LSHP-CT-2007-037941). S.M.-K. is funded by the Research Foundation Flanders (FWO-V), Belgium. G.M., M.A., and J.C.A. gratefully acknowledge support from FWO-V, research grant G.0151.05, and Belgian IUAP/PAI network P6/03, Statistical Techniques and Modelling for Complex Substantive Questions with Complex Data, of the Belgian Government (Belgian Science Policy).
S.M.-K. has received a speaker's honorarium from BD Diagnostics. We declare no other conflict of interest.
The sponsors of this study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. S.M.-K. had full access to all of the data in this study and had final responsibility for the decision to submit this report for publication.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Aires De Sousa, M., I. Santos Sanchez, M. L. Ferro, and H. de Lencastre. 2000. Epidemiological study of staphylococcal colonization and cross-infection in two West African hospitals. Microb. Drug Resist. 6:133-141. [DOI] [PubMed] [Google Scholar]
- 2.Becker, K., I. Pagnier, B. Schuhen, F. Wenzelburger, A. W. Friedrich, F. Kipp, G. Peters, and C. von Eiff. 2006. Does nasal cocolonization by methicillin-resistant coagulase-negative staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J. Clin. Microbiol. 44:229-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischof, L. J., L. Lapsley, K. Fontecchio, D. Jacosalem, C. Young, R. Hankerd, and D. W. Newton. 2009. Comparison of chromogenic media to BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR for detection of MRSA in nasal swabs. J. Clin. Microbiol. 47:2281-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherkaoui, A., G. Renzi, P. Francois, and J. Schrenzel. 2007. Comparison of four chromogenic media for culture-based screening of meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 56:500-503. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing. Fifteenth informational supplement, M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Coello, R., J. R. Glynn, C. Gaspar, J. J. Picazo, and J. Fereres. 1997. Risk factors for developing clinical infection with methicillin-resistant Staphylococcus aureus (MRSA) amongst hospital patients initially only colonized with MRSA. J. Hosp. Infect. 37:39-46. [DOI] [PubMed] [Google Scholar]
- 7.Felten, A., B. Grandry, P. H. Lagrange, and I. Casin. 2002. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): a disk diffusion method with cefoxitin and moxalactam, the Vitek 2 system, and the MRSA-screen latex agglutination test. J. Clin. Microbiol. 40:2766-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francois, P., G. Renzi, D. Pittet, M. Bento, D. Lew, S. Harbarth, P. Vaudaux, and J. Schrenzel. 2004. A novel multiplex real-time PCR assay for rapid typing of major staphylococcal cassette chromosome mec elements. J. Clin. Microbiol. 42:3309-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbarth, S. 2006. Control of endemic methicillin-resistant Staphylococcus aureus—recent advances and future challenges. Clin. Microbiol. Infect. 12:1154-1162. [DOI] [PubMed] [Google Scholar]
- 10.Harbarth, S., C. Masuet-Aumatell, J. Schrenzel, P. Francois, C. Akakpo, G. Renzi, J. Pugin, B. Ricou, and D. Pittet. 2006. Evaluation of rapid screening and pre-emptive contact isolation for detecting and controlling methicillin-resistant Staphylococcus aureus in critical care: an interventional cohort study. Crit. Care 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huletsky, A., R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Bastien, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leclercq, R. 2009. Epidemiological and resistance issues in multidrug-resistant staphylococci and enterococci. Clin. Microbiol. Infect. 15:224-231. [DOI] [PubMed] [Google Scholar]
- 13.Malhotra-Kumar, S., K. Haccuria, M. Michiels, M. Ieven, C. Poyart, W. Hryniewicz, and H. Goossens. 2008. Current trends in rapid diagnostics for methicillin-resistant Staphylococcus aureus and glycopeptide-resistant enterococcus species. J. Clin. Microbiol. 46:1577-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mermel, L., J. Lonks, S. Gordon, T. Perl, S. Fadem, A. Muralidhar, D. Dacus, G. E. Maxey, J. M. Cartony, D. Morse, and P. Mach. 2008. Quantitative analysis methicillin-resistant Staphylococcus aureus (MRSA) in clinical nasal swab samples collected from US patient populations, abstr. 313. Society for Healthcare Epidemiology of America 18th Annual Scientific Meeting, Orlando, FL.
- 15.Molina, A., R. Del Campo, L. Maiz, M. I. Morosini, A. Lamas, F. Baquero, and R. Canton. 2008. High prevalence in cystic fibrosis patients of multiresistant hospital-acquired methicillin-resistant Staphylococcus aureus ST228-SCCmecI capable of biofilm formation. J. Antimicrob. Chemother. 62:961-967. [DOI] [PubMed] [Google Scholar]
- 16.Nahimana, I., P. Francioli, and D. S. Blanc. 2006. Evaluation of three chromogenic media (MRSA-ID, MRSA-Select and CHROMagar MRSA) and ORSAB for surveillance cultures of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 12:1168-1174. [DOI] [PubMed] [Google Scholar]
- 17.Nonhoff, C., O. Denis, A. Brenner, P. Buidin, N. Legros, C. Thiroux, M. Dramaix, and M. J. Struelens. 2009. Comparison of three chromogenic media and enrichment broth media for the detection of methicillin-resistant Staphylococcus aureus from mucocutaneous screening specimens: comparison of MRSA chromogenic media. Eur. J. Clin. Microbiol. Infect. Dis. 28:363-369. [DOI] [PubMed] [Google Scholar]
- 18.Paule, S. M., M. Mehta, D. M. Hacek, T.-M. Gonzalzles, A. Robicsek, and L. R. Peterson. 2009. Chromogenic media vs real-time PCR for nasal surveillance of methicillin-resistant Staphylococcus aureus: impact on detection of MRSA-positive persons. Am. J. Clin. Pathol. 131:532-539. [DOI] [PubMed] [Google Scholar]
- 19.Peacock, S. J., I. de Silva, and F. D. Lowy. 2001. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 9:605-610. [DOI] [PubMed] [Google Scholar]
- 20.Perry, J. D., and A. M. Freydiere. 2007. The application of chromogenic media in clinical microbiology. J. Appl. Microbiol. 103:2046-2055. [DOI] [PubMed] [Google Scholar]
- 21.Robicsek, A., S. M. Paule, D. M. Hacek, K. L. Kaul, and L. R. Peterson. 2008. Impact of test characteristics on an MRSA surveillance program, abstr. K-1704. Infectious Disease Society of America 46th Annual Meeting.
- 22.Rohr, U., A. Kaminski, M. Wilhelm, L. Jurzik, S. Gatermann, and G. Muhr. 2009. Colonization of patients and contamination of the patients' environment by MRSA under conditions of single-room isolation. Int. J. Hyg. Environ. Health 212:209-215. [DOI] [PubMed] [Google Scholar]
- 23.Ruoff, K. L. 2007. Algorithms for identification of aerobic gram-positive cocci, p. 262-263. In P. R. Murray, E. J. Baron, J. Jorgensen, M. Landry, and M. Pfaller (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, DC.
- 24.Ruppé, E., F. Barbier, Y. Mesli, A. Maiga, R. Cojocaru, M. Benkhalfat, S. Benchouk, H. Hassaine, I. Maiga, A. Diallo, A. K. Koumare, K. Ouattara, S. Soumare, J. B. Dufourcq, C. Nareth, J. L. Sarthou, A. Andremont, and R. Ruimy. 2009. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 53:442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreckenberger, P. C., J. M. Janda, J. D. Wong, and E. J. Baron. 2007. Algorithms for identification of aerobic gram-negative bacteria, p. 438-441. In P. R. Murray, E. J. Baron, J. Jorgensen, M. Landry, and M. Pfaller (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, DC.
- 26.Van Heirstraeten, L., J. C. Abrahantes, C. Lammens, A. Lee, S. Harbarth, G. Molenberghs, M. Aerts, H. Goossens, and S. Malhotra-Kumar. 2009. Impact of a short pre-enrichment on detection and bacterial loads of methicillin-resistant Staphylococcus aureus from screening specimens. J. Clin. Microbiol. 47:3326-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.