Abstract
The microbiological diagnosis of periprosthetic joint infection (PJI) is crucial for successful antimicrobial treatment. Cultures have limited sensitivity, especially in patients receiving antibiotics. We evaluated the value of multiplex PCR for detection of microbial DNA in sonication fluid from removed orthopedic prostheses. Cases of PJI in which the prosthesis (or part of it) was removed were prospectively included. The removed implant was sonicated, and the resulting sonication fluid was cultured and subjected to multiplex PCR. Of 37 PJI cases (17 hip prostheses, 14 knee prostheses, 4 shoulder prostheses, 1 elbow prosthesis, and 1 ankle prosthesis), pathogens were identified in periprosthetic tissue in 24 (65%) cases, in sonication fluid in 23 (62%) cases, and by multiplex PCR in 29 (78%) cases. The pathogen was detected in 5 cases in sonication fluid only (Propionibacterium acnes in all cases; none of these patients had previously received antibiotics) and in 11 cases by multiplex PCR only (all of these patients had previously received antibiotics). After exclusion of 8 cases caused by P. acnes or Corynebacterium species, which cannot be detected due to the absence of specific primers in the PCR kit, sonication cultures were positive in 17 cases and multiplex PCR sonication cultures were positive in 29 cases (59% versus 100%, respectively; P < 0.01). Among 19 cases (51%) receiving antibiotics, multiplex PCR was positive in all 19 (100%), whereas sonication cultures grew the organism in 8 (42%) (P < 0.01). Multiplex PCR of sonication fluid is a promising test for diagnosis of PJI, particularly in patients who previously received antibiotics. With modified primer sets, multiplex PCR has the potential for further improvement of the diagnosis of PJI.
Modern medicine has developed a variety of artificial devices to assist in the performance of physiological functions as short-term devices (e.g., catheters and fracture fixation hardware) or permanent devices (e.g., artificial cardiac valves, pacemakers, and prosthetic joints). Orthopedic devices are used for the treatment of degenerative joint disease (osteoarthritis) and bone fracture (10). They are increasingly implanted in the growing population of the elderly. The risk of periprosthetic joint infections (PJI) is additionally increasing due to longer resident time of the implant in the body, which can become infected from a distant infectious focus by hematogenous route at any time after implantation (29). Thus, the number of PJI is expected to increase steadily in the next decades.
The identification of the infecting microorganism is crucial for successful treatment of PJI. Currently, cultures of synovial fluid and intraoperative periprosthetic tissue represent the standard method for diagnosing PJI. With newer techniques, such as sonication of removed implants, the sensitivity has been significantly increased (18, 22). However, in a significant proportion of patients with PJI, the infecting microorganism remains unknown, particularly when patients had previously received antimicrobial treatment (1, 22). Molecular methods may improve the diagnosis for PJI due to high sensitivity and culture independence.
Several researchers have evaluated the role of PCR in the diagnosis of osteoarticular infections, including septic arthritis (8, 14, 15) and PJI (1a, 4, 5, 12, 17, 25). The value of PCR was mainly investigated in synovial fluid or periprosthetic tissue specimens, whereas sonication fluid was evaluated only recently (18, 26). The limitations of these studies are the use of a specific PCR, which is typically able to detect only a single microorganism, or the use of a broad-range (16S ribosomal DNA [rDNA]) PCR, which can detect previously unknown organisms but has lower sensitivity and specificity than specific PCR, requires subsequent sequencing for bacterial identification, and fails to detect mixed infections.
Herein, we investigated a novel approach for diagnosis of PJI combining two complementary diagnostic methods, namely, sonication of removed implants and multiplex real-time PCR of the resulting sonication fluid. The removed prostheses were sonicated as previously described (18, 22), and the resulting sonication fluid was cultured and subjected to molecular detection with a real-time multiplex PCR test (SeptiFast; Roche Diagnostics, Basel, Switzerland). This PCR kit was designed for detection and identification of the most common bacterial and fungal pathogenic species in blood (Table 1) (2, 11, 13, 16, 28) and has not yet been investigated for the diagnosis of PJI. We hypothesized that the combined approach of sonication and multiplex PCR of sonication fluid will improve the sensitivity and specificity for diagnosing PJI.
TABLE 1.
Microorganisms that can be detected by multiplex PCR (SeptiFast)
| Group and organism |
|---|
| Gram-negative organisms |
| Escherichia coli |
| Klebsiella pneumoniae/oxytoca |
| Serratia marcescens |
| Enterobacter cloacae/aerogenes |
| Proteus mirabilis |
| Pseudomonas aeruginosa |
| Acinetobacter baumannii |
| Stenotrophomonas maltophilia |
| Gram-positive organisms |
| Staphylococcus aureus |
| Coagulase-negative staphylococcia |
| Streptococcus pneumoniae |
| Streptococcus spp.b |
| Enterococcus faecalis/faecium |
| Fungi |
| Candida albicans |
| Candida glabrata |
| Candida krusei |
| Candida tropicalis |
| Candida parapsilosis |
| Aspergillus fumigatus |
Includes Staphylococcus hominis subsp. novobiosepticus, S. pasteuri, S. warneri, S. cohnii subsp. urealyticum, S. hominis subsp. hominis, S. lugdunensis, S. cohnii subsp. cohnii, S. capitis subsp. ureolyticus, S. capitis subsp. capitis, S. caprae, S. saprophyticus, S. saprophyticus subsp. saprophyticus, S. xylosus, S. epidermidis, and S. haemolyticus.
Includes Streptococcus agalactiae, S. anginosus, S. bovis/S. gallolyticus, S. constellatus, S. cristatus, S. gordonii, S. intermedius, S. milleri, S. mitis, S. mutans, S. oralis, S. parasanguinis, S. pneumoniae, S. pyogenes, S. salivarius, S. sanguinis, S. thermophilus, S. vestibularis, and S. viridans.
(Part of this research was presented at the European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], Helsinki, Finland, 16 to 19 May 2009 [poster Nr.1828.].)
MATERIALS AND METHODS
Study design.
A single-center prospective cohort study was conducted at the Schulthess Clinic, Zurich, Switzerland, a specialized orthopedic center with 150 beds and 7,500 surgical procedures performed per year, including primary implantations and revisions of joint prostheses. The study protocol was reviewed and approved by the institutional review board.
Study population.
We prospectively included all consecutive patients aged 18 years or more from August 2008 through March 2009 with a prosthetic joint infection in whom the prosthesis or part of it (metal fixed or polyethylene mobile component) was removed at diagnosis of infection. The removed prosthesis was sent for sonication to determine the presence or absence of adherent microorganisms. Subjects were excluded if obvious contamination of an explanted component occurred in the operating room or fewer than two periprosthetic tissue specimens were collected for culture. Medical records were evaluated for the following: demographic characteristics; clinical, radiographic, laboratory, histopathological, and microbiological data; type of surgical management; antimicrobial therapy; and information about the primary arthroplasty and subsequent revisions (if any). According to the study protocol, patients received no perioperative antimicrobial prophylaxis before surgery in order to optimize the diagnostic yield of intraoperative cultures. Patients who had received an intravenous antibiotic for at least 24 h in the 10 days before surgery were defined as having received previous antimicrobial therapy.
Study definitions.
PJI was considered if one of the following criteria was present (22, 23, 29): (i) visible purulence of a preoperative aspirate or intraoperative periprosthetic tissue (as determined by the surgeon), (ii) presence of a sinus tract communicating with the prosthesis, (iii) acute inflammation in intraoperative permanent periprosthetic tissue sections by histopathology (as determined by the pathologist), (iv) increased synovial fluid leukocyte count with >1,700 leukocytes/μl and/or >65% granulocytes (as described in reference 21), or (v) microbial growth in preoperative joint aspirate, intraoperative periprosthetic tissue, or sonication fluid of the removed implant. Low-virulence microorganisms, such as coagulase-negative staphylococci or Propionibacterium acnes, were considered pathogens, if at least one additional (culture-independent) criterion for PJI was fulfilled.
Synovial fluid and periprosthetic cultures.
Synovial fluid was aspirated preoperatively at the discretion of the operating surgeon. The aspirate was transferred into a sterile vial containing no additives. For all patients, at least two intraoperative periprosthetic tissue specimens were retrieved from the bone-cement or bone-prosthesis interface with the most obvious inflammatory changes. Tissue specimens were collected in sterile vials and were individually homogenized in 3 ml Trypticase soy broth for 1 min using a mortar and pestle. The synovial fluid and tissue homogenate samples were inoculated in 0.1-ml aliquots onto aerobic and anaerobic sheep blood agar plates and in 1-ml aliquots into thioglycolate broth. The cultures were incubated at 35°C ± 1°C for 10 days. A terminal subculture was performed from all thioglycolate broth specimens on blood agar plates and incubated at 35°C ± 1°C for 5 more days. Each unique colony of isolated microorganisms was identified, and their antimicrobial susceptibility was tested using standard microbiological techniques.
Sonication fluid cultures.
The removed prosthesis (or part of it) was aseptically removed in the operating room and transported to the microbiology laboratory in solid air-tight containers (Lock & Lock; Vetrag AG, Stäfa, Switzerland) within 48 h of removal. In the microbiological laboratory, sonication of the implant was performed as previously described (22). Briefly, 50 to 200 ml sterile Ringer solution (depending on the size of implant) was added to the container in a laminar airflow biosafety cabinet. The container with the implant was vortexed for 30 s, followed by sonication for 1 min (at a frequency of 40 ± 2 kHz and power density of 0.22 ± 0.04 W/cm2), as determined by a calibrated hydrophone (type 8103; Brüel and Kjær, Naerum, Denmark). For sonication, ultrasound bath BactoSonic (Bandelin GmbH, Berlin, Germany) (www.bactosonic.info) was used. No differences in frequency or power density were observed at various locations within the ultrasound bath during the study period. The container was subsequently vortexed for an additional 30 s to remove any residual microorganisms and to homogeneously distribute them in the sonication fluid, which was plated in 0.1-ml aliquots onto aerobic and anaerobic sheep blood agar plates; 1 ml was inoculated in thioglycolate broth. All cultures were incubated at 35°C for 7 days and inspected daily for bacterial growth. Microorganisms on plates were enumerated (i.e., number of CFU/ml sonication fluid) and classified by using routine microbiological techniques.
Multiplex PCR assay.
One milliliter of the sonication fluid was subjected to mechanical lysis and purification of DNA (SeptiFast lysis kit MGRADE; Roche Diagnostics) according to the manufacturer's instructions. The lysed specimens were incubated for 1 h with a protease and chaotropic lysis buffer. Primer mix was introduced into each sample before amplification. The real-time PCR amplification of target DNA was performed in parallel reactions for Gram-positive bacteria, Gram-negative bacteria, and fungi using Hot Start Taq polymerase and the LightCycler instrument (LightCycler 2.0; Roche Diagnostics). The internal transcribed spacer (ITS) region was selected as the target region for bacterial and fungal species differentiation. Internal controls for the amplification step were included with each assay run.
Melting curve analysis was performed after completion of amplification with subsequent detection of PCR products by specific hybridization probes. The emitted fluorescence was simultaneously measured in four detection channels using a wavelength of 610 nm, 640 nm, 670 nm, and 705 nm. The data were automatically analyzed by the manufacturer's identification software program SIS (Roche Diagnostics) and reviewed manually for each detection channel. The crossing point of the amplification curve was recorded, and the emitted fluorescence signal was expressed as the H value, an integral of the fluorescence intensity and a semiquantitative measure of the initial quantity of target DNA. According to the manufacturer (Roche Diagnostics), the limit of detection in blood is 30 CFU/ml, except for coagulase-negative staphylococci, Streptococcus pyogenes, Streptococcus agalactiae, and Candida glabrata, for which the limit of detection is 100 CFU/ml (13, 14). A positive control and a negative control for multiplex PCR, supplied by the manufacturer, were included in each extraction series. All specimen handling preparations and the PCR setup were performed in a dedicated laminar airflow biosafety cabinet, equipped with an UV lamp, operated overnight. Dedicated equipment and unidirectional flow were used for the microbiological procedure.
Negative controls.
Ten consecutive explanted prostheses from patients, who were diagnosed with mechanical (aseptic) loosening of the prosthesis and had no history of previous infection, were included as controls. After the prosthesis was removed, it was subjected to sonication, followed by culture and multiplex PCR of sonication fluid, processed in the same way as prostheses from the study patients.
Statistical analysis.
Comparisons of individual diagnostic tests were performed using the McNemar test. For mixed infections, a diagnostic test was considered positive if all infecting organisms were detected. Differences were considered significant when P values were <0.05. All calculations were performed using statistical software package SAS (version 8.2; SAS Institute Inc., Cary, NC). For graphic analysis, OriginPro software (version 8; Origin Lab Corp., Northampton, MA) was used.
RESULTS
Demographic and clinical characteristics of PJI.
A total of 37 PJI cases were included in this study; the 37 cases included patients with hip (n = 17), knee (n = 14), shoulder (n = 4), ankle (n = 1), and elbow (n = 1) prostheses (Table 2). Three patients were included twice: two at the time of second debridement and one at the time of definite removal of the prosthesis after an initial salvage attempt. The median patient age at the time of infection was 71 years (range, 39 to 89 years), and 57% were males. The most common underlying joint disorder was osteoarthritis (70%), followed by rheumatoid arthritis (19%) and trauma (11%). Primary implantation was performed in 27% and revision surgery in 73% of the patients. Most infections (68%) occurred 3 to 24 months after surgery (delayed infections). The median time between the last surgical procedure and time of infection was 7.5 months (range, 0.3 to 349 months). Nineteen of the 37 (51%) patients had previously received antimicrobial therapy with a median duration of 9 days (range, 1 to 60 days).
TABLE 2.
Characteristics of 37 patients with PJI
| Characteristic | No. of patients or parameter value (%, unless range is specified) |
|---|---|
| Patient age, yr [median (range)] | 71 (39-89) |
| Male | 21 (57) |
| Location of PJI | |
| Hip | 17 (46) |
| Knee | 14 (38) |
| Shoulder | 4 (11) |
| Ankle | 1 |
| Elbow | 1 |
| Type of removed implant for sonication | |
| Total prosthesis | 26 (70) |
| Polyethylene mobile part | 10 (27) |
| Joint spacer | 1 |
| Underlying joint disorder | |
| Osteoarthritis | 26 (70) |
| Rheumatoid arthritis | 7 (19) |
| Trauma | 4 (11) |
| Type of arthroplasty | |
| Primary | 10 (27) |
| Revision | 27 (73) |
| Manifestation of infection according to surgery | |
| Early (<3 months) | 6 (16) |
| Delayed (3 to 48 months) | 25 (68) |
| Late (>48 months) | 6 (16.2) |
| Time between the last surgical procedure and occurrence of infection, mo [median (range)] | |
| 7.5 (0.3-349) | |
| Previous antimicrobial therapy received | 19 (51) |
Microbiology.
Table 3 summarizes the microbiological findings of 37 cases of PJI. A single causative organism was found in 31 (84%) and a polymicrobial infection in 6 (16%) cases. Most monobacterial infections were staphylococcal infections. Methicillin resistance was detected in 7 of 11 (63%) episodes of coagulase-negative staphylococci (not including those from polymicrobial infections); none of the nine Staphylococcus aureus isolates causing infection was methicillin resistant. In addition, coagulase-negative staphylococci from two patients showed rifampin resistance (one patient with Staphylococcus lugdunensis infection who previously had a fistula and one patient with S. aureus infection in which no debridement was done 1 year ago and only ciprofloxacin and rifampin were given). None of the 10 implants from control patients showed growth in sonication fluid culture, and all were negative by multiplex PCR.
TABLE 3.
Comparison of periprosthetic tissue culture, sonication fluid culture, and multiplex PCR of sonication fluid in 37 cases of PJI
| Infection type and microorganism | No. of episodes | No. of episodes with positive result by diagnostic test: |
||
|---|---|---|---|---|
| Periprosthetic tissue culture | Sonication fluid culture | Multiplex PCR of sonication fluid | ||
| Single microorganism | 31 | 22 | 20 | 26 |
| Staphylococcus aureus | 9 | 5 | 5 | 9 |
| Coagulase-negative staphylococci | 11 | 9 | 8 | 11 |
| Streptococcus mitis | 1 | 1 | 1 | 1 |
| Streptococcus agalactiae | 1 | 1 | 1 | 1 |
| Streptococcus dysgalactiae | 1 | 1 | 0 | 1 |
| Streptococcus gallolyticus | 1 | 0 | 0 | 1 |
| Streptococcus pneumoniae | 1 | 0 | 0 | 1 |
| Propionibacterium acnes | 5 | 4 | 4 | 0 |
| Candida albicans | 1 | 1 | 1 | 1 |
| Polymicrobial infectiona | 6 | 2 | 3 | 3 |
| Total no. of episodes (%) | 37 (100) | 24 (65) | 23 (62) | 29 (78) |
Included coagulase-negative staphylococci, Klebsiella pneumoniae/oxytoca and Enterococcus faecalis (n = 1), P. acnes and coagulase-negative staphylococci (n = 1), S. aureus and coagulase-negative staphylococci (n = 2), Corynebacterium species and coagulase-negative staphylococci (n = 1), and P. acnes and S. aureus (n = 1).
Comparison of culture and multiplex PCR.
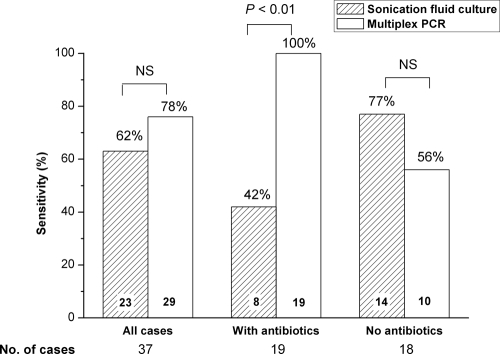
Among 37 cases of PJI, the causative organisms were identified in periprosthetic tissue culture in 24 (65%) cases, in sonication fluid culture in 23 (62%) cases, and by multiplex PCR in 29 (78%) cases (Table 3 and Fig. 1). In 21 cases (57%), the results of sonication fluid culture and multiplex PCR were concordant (positive results for 18 cases by both tests and negative results for 3 cases by both tests). Among 16 discordant cases, the causative organism was detected in 5 cases in sonication fluid culture only (P. acnes in all cases; none of these patients had previously received antibiotics) and in 11 cases by multiplex PCR only, including 4 S. aureus isolates, 3 coagulase-negative staphylococcal isolates, 2 Streptococcus species isolates, 1 Klebsiella pneumoniae/oxytoca isolate (part of polymicrobial infection), and 1 Streptococcus pneumoniae isolate (all of these patients had previously received antibiotics).
FIG. 1.
Sensitivity of culture and multiplex PCR of sonication fluid. The sensitivity values are shown overall (all cases) and stratified according to patients who had received antimicrobial therapy previously (n = 19) and patients who had not received antimicrobial therapy (n = 18). Eight pathogens (7 Propionibacterium acnes and 1 Corynebacterium species) were missed by multiplex PCR due to lack of specific primers for these species. The value at the bottom of each bar indicates the number of PJI cases in that group, and the percentage above each bar indicates the sensitivity. NS, not significant.
Among the 8 cases in which the organism was missed by multiplex PCR (concordant results in 3 cases and discordant results in 5 cases), 7 were caused by P. acnes and 1 was caused by a Corynebacterium species (as a single organism or as part of a polymicrobial infection), as detected by sonication culture (n = 2) or periprosthetic tissue culture only (n = 3) or both (n = 3). After exclusion of 8 PJI cases caused by P. acnes or Corynebacterium species that cannot be detected by our multiplex PCR, the sonication culture detected the causative pathogen in 17 of 29 cases (59%) and multiplex PCR detected the causative pathogen in all 29 cases (100%) (P < 0.01).
Influence of previous antimicrobial therapy.
Among 37 PJI cases, 19 of the patients (51%) had previously received antimicrobial therapy for a median duration of 9 days and 18 (49%) had received no antibiotics before diagnostic tests. Among 19 patients who had previously received antibiotics, sonication fluid cultures were positive in 8 cases (42%), compared to multiplex PCR, which gave positive results for all 19 (100%) (P < 0.01) (Fig. 1). In 18 patients who had not previously received antibiotic treatment, sonication fluid culture was positive in 14 cases (77%) and multiplex PCR was positive in 10 cases (56%). The 8 missed pathogens by multiplex PCR included 5 P. acnes isolates, 1 polymicrobial infection with Corynebacterium species, and 2 polymicrobial infections with P. acnes (Table 3).
Quantitative assessment of target DNA in sonication fluid.
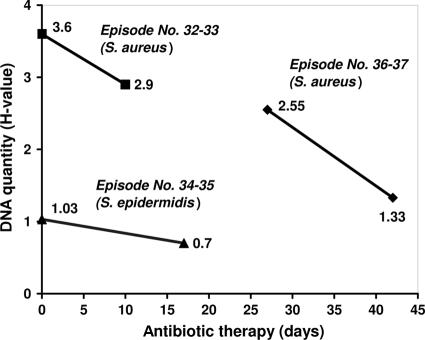
The DNA quantity (expressed as H value) showed no correlation with the CFU count in the sonication fluid (Table 4). However, in three patients for which serial testing of sonication fluid by multiplex PCR was available, a decrease of H value for the infecting microorganism was observed during antibiotic therapy (Fig. 2). The DNA of the initially infecting organism was identified up to 43 days after initiation of antimicrobial therapy.
TABLE 4.
Characteristics of diagnostic results in 37 cases with PJI
| Patient characteristics |
Diagnostic resulta |
Antibiotic treatment before sonication |
Comment | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Periprosthetic tissue culture |
Sonication fluid culture |
Multiplex PCR of sonication fluid |
|||||||||
| Case no.b | Patient age (yr) | Implant typec | Organism | No. of positive samples/total no. of samples | Organism | Quantity (CFU/ml) | Organism | H value | Antibiotic(s)d | No. of days of treatment | |
| 1 | 76 | Knee (T) | No growth | 0/6 | No growth | Streptococcus spp. | 0.85 | Amc | 6 | S. gallolyticus in tissue cultures 6 days before | |
| 2 | 67 | Knee (T) | No growth | 0/9 | CNS | >1,000 | CNS | 1.35 | No | 0 | |
| P. acnes | 7/9 | P. acnes | >1,000 | Negative | |||||||
| 3 | 71 | Hip (T) | S. epidermidis | 2/7 | CNS | >1,000 | CNS | 2.99 | Ipm | 14 | |
| 4 | 80 | Hip (P) | S. epidermidis | 5/7 | CNS | 20 | CNS | 1.27 | Van-Rif | 1 | |
| 5 | 49 | Ankle (T) | S. lugdunensis | 5/6 | CNS | 70 | CNS | 0.45 | No | 0 | |
| 6 | 86 | Hip (T) | S. aureus | 2/8 | S. aureus | >1,000 | S. aureus | 2.01 | Cli | 32 | |
| 7 | 54 | Hip (T) | P. acnes | 4/9 | P. acnes | In broth only | Negative | No | 0 | ||
| 8 | 84 | Knee (P) | S. agalactiae | 5/5 | S. agalactiae | >1,000 | Streptococcus spp. | 0.35 | Ipm-Rif | 0 | |
| 9 | 80 | Hip (T) | No growth | 0/7 | E. faecalis | >1,000 | E. faecalis | 0.90 | Cip | 2 | K. pneumoniae in blood, hip, synovial fluid, and urine cultures 1 month before |
| S. epidermidis | 6/7 | CNS | 260 | CNS | 0.38 | ||||||
| No growth | 0/7 | No growth | K. pneumoniae | 0.41 | |||||||
| 10 | 82 | Shoulder (T) | P. acnes | 5/6 | P. acnes | 200 | Negative | No | 0 | ||
| 11 | 52 | Hip (T) | S. epidermidis | 5/6 | CNS | >1,000 | CNS | 1.10 | No | 0 | |
| S. aureus | 4/6 | S. aureus | >1,000 | S. aureus | 1.4 | ||||||
| 12 | 85 | Hip (P) | S. aureus | 3/7 | No growth | S. aureus | 1.78 | Amc | 3 | ||
| 13 | 84 | Hip (T) | S. aureus | 1/5 | S. aureus | In broth only | S. aureus | 2.34 | Flx | 17 | |
| 14 | 63 | Shoulder (T) | S. epidermidis | 3/6 | CNS | >1,000 | CNS | 2.07 | No | 0 | |
| 15 | 77 | Hip (T) | S. epidermidis | 1/6 | CNS | 30 | CNS | 1.59 | No | 0 | |
| 16 | 79 | Hip (T) | S. mitis | 4/10 | S. mitis | 850 | Streptococcus spp. | 1.86 | Cro-Gen | 2 | Growth in blood cultures (all 6 samples) |
| 17 | 78 | Hip (T) | S. epidermidis | 6/6 | CNS | >1,000 | CNS | 1.96 | No | 0 | |
| Corynebacterium spp. | 3/6 | No growth | Negative | ||||||||
| 18 | 39 | Elbow (T) | S. aureus | 6/6 | S. aureus | >1,000 | S. aureus | 2.67 | No | 0 | |
| No growth | 0/6 | CNS | >1,000 | CNS | 0.70 | ||||||
| 19 | 77 | Knee (P) | No growth | 0/4 | No growth | S. pneumoniae | 4.18 | Cro | 9 | S. pneumoniae in knee synovial fluid and blood cultures 9 days before | |
| 20 | 68 | Hip (P) | S. dysgalactiae | 4/4 | No growth | Streptococcus spp. | 0.49 | Amc | 2 | ||
| 21 | 72 | Knee (T) | S. aureus | 1/3 | S. aureus | >1,000 | S. aureus | 3.5 | Ipm | 5 | |
| 22 | 89 | Shoulder (P) | P. acnes | 6/6 | P. acnes | >1,000 | Negative | No | 0 | ||
| 23 | 73 | Hip (T) | C. albicans | 1/9 | C. albicans | 20 | C. albicans | 1.94 | No | 0 | Candida albicans PJI diagnosed 6 months before |
| 24 | 61 | Hip (T) | No growth | 0/5 | S. aureus | 10 | S. aureus | 0.26 | No | 0 | |
| 25 | 59 | Hip (T) | S. epidermidis | 4/5 | CNS | >1,000 | CNS | 2.10 | No | 0 | |
| 26 | 64 | Hip (T) | P. acnes | 2/5 | No growth | Negative | No | 0 | |||
| 27 | 84 | Hip (T) | S. epidermidis | 6/6 | No growth | CNS | 1.82 | Flx | 28 | ||
| 28 | 66 | Knee (T) | S. epidermidis | 5/5 | CNS | >1,000 | CNS | 1.96 | Amc | 4 | |
| 29 | 57 | Knee (T) | No growth | 0/6 | P. acnes | 10 | Negative | No | 0 | Acute inflammation in histopathology | |
| 30 | 70 | Shoulder (P) | No growth | 0/6 | P. acnes | 160 | Negative | No | 0 | ||
| S. aureus | 5/6 | No growth | S. aureus | 0.26 | |||||||
| 31 | 71 | Knee (T) | S. epidermidis | 2/5 | No growth | CNS | 1.64 | Van | 4 | ||
| 32 | 58 | Knee (P) | S. aureus | 5/5 | S. aureus | >1,000 | S. aureus | 3.60 | 0 | ||
| 33 | Knee (P) | No growth | 5/5 | No growth | S. aureus | 2.90 | Flx-Rif | 10 | |||
| 34 | 82 | Knee (P) | S. epidermidis | 5/5 | CNS | In broth only | CNS | 1.03 | No | 0 | |
| 35 | Knee (T) | No growth | 0/5 | No growth | CNS | 0.71 | Van-Rif | 17 | |||
| 36 | 55 | Knee (T) | No growth | 0/3 | No growth | S. aureus | 2.55 | Ipm-Rif | 27 | S. aureus cultured 27 days before | |
| 37 | Knee (S) | No growth | 0/2 | No growth | S. aureus | 1.35 | Ipm-Rif | 42 | |||
CNS, coagulase-negative staphylococci. “In broth only” denotes growth only in thioglycolate medium.
Cases 32 amd 33 are the same patient, cases 34 and 35 are the same patient, and cases 36 and 37 are the same patient.
The location of the implant is shown first. The specific part of the implant is shown in parentheses as follows: (P), polyethylene mobile part; (T), total prosthesis; (S), spacer consisting of polymethyl methacrylate impregnated with vancomycin.
Anc, amoxicillin-clavulanate; Ipm, imipenem; Van, vancomycin; Rif, rifampin; Cli, clindamycin; Cip, ciprofloxacin; Flx, floxacillin; Cro, ceftriaxone; Gen, gentamicin.
FIG. 2.
Serial determinations of DNA quantity by multiplex PCR according to the duration of antibiotic treatment in three patients. The quantity of DNA by multiplex PCR is expressed as H values on the y axis. The infecting microorganism was S. aureus in 2 cases and S. epidermidis in 1 case. Note that the H value is a measure of DNA quantity, defined as the integral of the fluorescence intensity and a semiquantitative measure of the initial quantity of target DNA.
DISCUSSION
Despite the fact that sonication of removed prostheses and subsequent culture of the sonication fluid has improved the sensitivity for detecting infection, a significant proportion of PJI still remains without identification of the organism causing the infection, especially in patients who previously received antimicrobial treatment (18, 22). Culture-independent (molecular) methods, such as PCR, have been proposed to improve the diagnosis of PJI.
Most commonly, specific or broad-range (16S rDNA) PCR was applied to synovial fluid or periprosthetic tissue (3-7, 9, 14, 17, 18, 26, 27). In a recent study, the sensitivities of specific PCR for detection of P. acnes and Staphylococcus spp. in sonication fluid were 89% and 97%, respectively (18). In contrast, broad-range PCR of tissue cultures of patients with PJI showed a sensitivity of only 50% (3). Unfortunately, contaminants detected with the broad-range PCR (false-positive results) belong to the same type of organisms as the microorganisms causing low-grade PJI, making the distinction of true-positive and false-positive PCR results difficult. For these reasons, PCR has not yet been integrated in the standard routine diagnostic procedure of PJI by most laboratories. With multiplex PCR, several disadvantages of the specific and broad-range PCR could be overcome. Up to now, multiplex PCR was used to investigate blood samples from patients with bloodstream infections and infective endocarditis (2, 11, 24). Another approach for molecular diagnosis of PJI used the messenger RNA-based reverse transcription-quantitative PCR using universal primers, which showed sensitivity equivalent to that of intraoperative cultures but exhibited detectable signals for 7 days after cultures turned negative (1a). To our knowledge, until this study, multiplex PCR had not yet been evaluated in periprosthetic tissue or sonication fluid samples.
In this study, the multiplex PCR of sonication fluid samples showed the potential for improved diagnosis of PJI. The sensitivity of multiplex PCR was better than the sensitivity of sonication fluid cultures (78% versus 62%), particularly in patients who had previously received antibiotic therapy (100% versus 42%; P < 0.01). Seven of 8 false-negative PCR results were caused by P. acnes, and one false-negative PCR result was caused by a Corynebacterium species, which cannot be detected by the multiplex PCR used, due to the absence of specific primers for this organism in the PCR kit. When P. acnes or Corynebacterium species were excluded from the analysis, all 19 infecting microorganisms were detected by multiplex PCR. P. acnes is a common pathogen in PJI, especially in shoulder prosthetic joint disease (18, 20). We found all P. acnes isolates in either periprosthetic tissue or synovial fluid cultures. A prolonged incubation time (10 to 14 days) of periprosthetic tissue samples and sonication fluid is mandatory to optimize the detection of this pathogen by culture (19).
Interestingly, we were able to show that the microbial DNA density (represented as H value) decreases with antimicrobial treatment but remains positive for up to 43 days of treatment. This provides the opportunity to detect the pathogen despite previous antibiotic treatment, a common clinical situation. With additional molecular tests, specific resistance genes, such as the genes conferring resistance to methicillin, quinolones, and rifampin, can be detected in addition. This information is crucial for efficient and targeted antimicrobial therapy in negative cultures. Interestingly, no correlation between the bacterial density in sonication fluid and the DNA quantity was observed. This observation could be the result if some microorganisms were killed by sonication (despite reduced acoustic energy used for this purpose) but the DNA was not affected. The sensitivity of the sonication fluid culture may be improved by an additional centrifugation step of the fluid and cultivation of the sediment with concentrated bacteria only. Importantly, a positive PCR result cannot be used for evaluation of treatment efficacy, unless serial determinations over time are performed and available. Nevertheless, this feature may be an advantage in PJI where etiological diagnosis is needed for treatment if conventional cultures were negative.
In conclusion, multiplex PCR of sonication fluid can improve the diagnosis of PJI, particularly among patients who had previously received antibiotic therapy. All undetected organisms by PCR were P. acnes or Corynebacterium species, which cannot be detected due to the absence of specific primers in the PCR kit. We suggest that the specific primer set be modified to include the most common organisms causing PJI, including low-virulence pathogens, such as P. acnes, Corynebacterium species, Finegoldia magna, and Peptostreptococcus species. The potential of multiplex PCR in the diagnosis of PJI is especially high in patients who had previously been exposed to antibiotics and have a high probability of false-negative cultures. Additional laboratory and clinical studies with multiplex PCR are needed to define the appropriate primer design, processing procedure, and proper patient selection for further optimization of the diagnosis of PJI.
Acknowledgments
We thank Reno Frei, Heinz Hermann, and Nathalie Schmid for their excellent work and support in performing multiplex PCR.
This study was supported by the Swiss National Science Foundation (3200B0-112547), Gebert Rüf Stiftung, and by a grant of the Hans-Paul Wälchli Foundation for Research (Lugano, Switzerland). The multiplex PCR test kits (SeptiFast) and reagents were provided by Roche Diagnostics, Basel, Switzerland.
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Berbari, E. F., C. Marculescu, I. Sia, B. D. Lahr, A. D. Hanssen, J. M. Steckelberg, R. Gullerud, and D. R. Osmon. 2007. Culture-negative prosthetic joint infection. Clin. Infect. Dis. 45:1113-1119. [DOI] [PubMed] [Google Scholar]
- 1a.Bergin, P. F., J. D. Doppelt, W. G. Hamilton, G. E. Mirick, A. E. Jones, S. Sritulanondha, J. M. Helm, and R. S. Tuan. 2010. Detection of periprosthetic infections with use of ribosomal RNA-based polymerase chain reaction. J. Bone Joint Surg. Am. 92:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casalta, J. P., F. Gouriet, V. Roux, F. Thuny, G. Habib, and D. Raoult. 2009. Evaluation of the LightCycler SeptiFast test in the rapid etiologic diagnostic of infectious endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 28:569-573. [DOI] [PubMed] [Google Scholar]
- 3.De Man, F. H., P. Graber, M. Luem, W. Zimmerli, P. E. Ochsner, and P. Sendi. 2009. Broad-range PCR in selected episodes of prosthetic joint infection. Infection 37:292-294. [DOI] [PubMed] [Google Scholar]
- 4.Dempsey, K. E., M. P. Riggio, A. Lennon, V. E. Hannah, G. Ramage, D. Allan, and J. Bagg. 2007. Identification of bacteria on the surface of clinically infected and non-infected prosthetic hip joints removed during revision arthroplasties by 16S rRNA gene sequencing and by microbiological culture. Arthritis Res. Ther. 9:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dora, C., M. Altwegg, C. Gerber, E. C. Bottger, and R. Zbinden. 2008. Evaluation of conventional microbiological procedures and molecular genetic techniques for diagnosis of infections in patients with implanted orthopedic devices. J. Clin. Microbiol. 46:824-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fihman, V., D. Hannouche, V. Bousson, T. Bardin, F. Liote, L. Raskine, J. Riahi, M. J. Sanson-Le Pors, and B. Bercot. 2007. Improved diagnosis specificity in bone and joint infections using molecular techniques. J. Infect. 55:510-517. [DOI] [PubMed] [Google Scholar]
- 7.Gallo, J., M. Kolar, M. Dendis, Y. Loveckova, P. Sauer, J. Zapletalova, and D. Koukalova. 2008. Culture and PCR analysis of joint fluid in the diagnosis of prosthetic joint infection. New Microbiol. 31:97-104. [PubMed] [Google Scholar]
- 8.Hoeffel, D. P., S. H. Hinrichs, and K. L. Garvin. 1999. Molecular diagnostics for the detection of musculoskeletal infection. Clin. Orthop. Relat Res. 360:37-46. [DOI] [PubMed] [Google Scholar]
- 9.Koskela, A., A. Nilsdotter-Augustinsson, L. Persson, and B. Soderquist. 2009. Prevalence of the ica operon and insertion sequence IS256 among Staphylococcus epidermidis prosthetic joint infection isolates. Eur. J. Clin. Microbiol. Infect. Dis. 28:655-660. [DOI] [PubMed] [Google Scholar]
- 10.Kurtz, S. M., K. L. Ong, J. Schmier, F. Mowat, K. Saleh, E. Dybvik, J. Karrholm, G. Garellick, L. I. Havelin, O. Furnes, H. Malchau, and E. Lau. 2007. Future clinical and economic impact of revision total hip and knee arthroplasty. J. Bone Joint Surg. Am. 89(Suppl. 3):144-151. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann, L. E., K. P. Hunfeld, T. Emrich, G. Haberhausen, H. Wissing, A. Hoeft, and F. Stuber. 2008. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med. Microbiol. Immunol. 197:313-324. [DOI] [PubMed] [Google Scholar]
- 12.Levine, M. J., B. A. Mariani, R. S. Tuan, and R. E. Booth, Jr. 1995. Molecular genetic diagnosis of infected total joint arthroplasty. J. Arthroplasty 10:93-94. [DOI] [PubMed] [Google Scholar]
- 13.Mancini, N., D. Clerici, R. Diotti, M. Perotti, N. Ghidoli, D. De Marco, B. Pizzorno, T. Emrich, R. Burioni, F. Ciceri, and M. Clementi. 2008. Molecular diagnosis of sepsis in neutropenic patients with haematological malignancies. J. Med. Microbiol. 57:601-604. [DOI] [PubMed] [Google Scholar]
- 14.Mariani, B. D., M. J. Levine, R. E. Booth, Jr., and R. S. Tuan. 1995. Development of a novel, rapid processing protocol for polymerase chain reaction-based detection of bacterial infections in synovial fluids. Mol. Biotechnol. 4:227-237. [DOI] [PubMed] [Google Scholar]
- 15.Mariani, B. D., D. S. Martin, M. J. Levine, R. E. Booth, Jr., and R. S. Tuan. 1996. Polymerase chain reaction detection of bacterial infection in total knee arthroplasty. Clin. Orthop. Relat Res. 331:11-22. [DOI] [PubMed] [Google Scholar]
- 16.Mussap, M., M. P. Molinari, E. Senno, P. Gritti, B. Soro, S. Mannelli, and C. Fabris. 2007. New diagnostic tools for neonatal sepsis: the role of a real-time polymerase chain reaction for the early detection and identification of bacterial and fungal species in blood samples. J. Chemother. 19(Suppl.):31-34. [DOI] [PubMed] [Google Scholar]
- 17.Panousis, K., P. Grigoris, I. Butcher, B. Rana, J. H. Reilly, and D. L. Hamblen. 2005. Poor predictive value of broad-range PCR for the detection of arthroplasty infection in 92 cases. Acta Orthop. 76:341-346. [PubMed] [Google Scholar]
- 18.Piper, K. E., M. J. Jacobson, R. H. Cofield, J. W. Sperling, J. Sanchez-Sotelo, D. R. Osmon, J. M. Steckelberg, J. N. Mandrekar, S. M. Fernandez, and R. Patel. 2009. Microbiologic diagnosis of prosthetic shoulder infection using implant sonication. J. Clin. Microbiol. 47:1878-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schafer, P., B. Fink, D. Sandow, A. Margull, I. Berger, and L. Frommelt. 2008. Prolonged bacterial culture to identify late periprosthetic joint infection: a promising strategy. Clin. Infect. Dis. 47:1403-1409. [DOI] [PubMed] [Google Scholar]
- 20.Topolski, M. S., P. Y. Chin, J. W. Sperling, and R. H. Cofield. 2006. Revision shoulder arthroplasty with positive intraoperative cultures: the value of preoperative studies and intraoperative histology. J. Shoulder Elbow Surg. 15:402-406. [DOI] [PubMed] [Google Scholar]
- 21.Trampuz, A., A. D. Hanssen, D. R. Osmon, J. Mandrekar, J. M. Steckelberg, and R. Patel. 2004. Synovial fluid leukocyte count and differential for the diagnosis of prosthetic knee infection. Am. J. Med. 117:556-562. [DOI] [PubMed] [Google Scholar]
- 22.Trampuz, A., K. E. Piper, M. J. Jacobson, A. D. Hanssen, K. K. Unni, D. R. Osmon, J. N. Mandrekar, F. R. Cockerill, J. M. Steckelberg, J. F. Greenleaf, and R. Patel. 2007. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 357:654-663. [DOI] [PubMed] [Google Scholar]
- 23.Trampuz, A., and W. Zimmerli. 2008. Diagnosis and treatment of implant-associated septic arthritis and osteomyelitis. Curr. Infect. Dis. Rep. 10:394-403. [DOI] [PubMed] [Google Scholar]
- 24.Tsalik, E. L., D. Jones, B. Nicholson, L. Waring, O. Liesenfeld, L. P. Park, S. W. Glickman, L. B. Caram, R. J. Langley, J. C. van Velkinburgh, C. B. Cairns, E. P. Rivers, R. M. Otero, S. F. Kingsmore, T. Lalani, V. G. Fowler, and C. W. Woods. 2010. Multiplex PCR to diagnose bloodstream infections in patients admitted from the emergency department with sepsis. J. Clin. Microbiol. 48:26-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tunney, M. M., S. Patrick, M. D. Curran, G. Ramage, D. Hanna, J. R. Nixon, S. P. Gorman, R. I. Davis, and N. Anderson. 1999. Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J. Clin. Microbiol. 37:3281-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tunney, M. M., S. Patrick, S. P. Gorman, J. R. Nixon, N. Anderson, R. I. Davis, D. Hanna, and G. Ramage. 1998. Improved detection of infection in hip replacements. A currently underestimated problem. J. Bone Joint Surg. Br. 80:568-572. [DOI] [PubMed] [Google Scholar]
- 27.Vandercam, B., S. Jeumont, O. Cornu, J. C. Yombi, F. Lecouvet, P. Lefevre, L. M. Irenge, and J. L. Gala. 2008. Amplification-based DNA analysis in the diagnosis of prosthetic joint infection. J. Mol. Diagn. 10:537-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vince, A., S. Z. Lepej, B. Barsic, D. Dusek, Z. Mitrovic, R. Serventi-Seiwerth, and B. Labar. 2008. LightCycler SeptiFast assay as a tool for the rapid diagnosis of sepsis in patients during antimicrobial therapy. J. Med. Microbiol. 57:1306-1307. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerli, W., A. Trampuz, and P. E. Ochsner. 2004. Prosthetic-joint infections. N. Engl. J. Med. 351:1645-1654. [DOI] [PubMed] [Google Scholar]