Abstract
PCR has been used as an aid in the diagnosis of invasive aspergillosis for almost 2 decades. A lack of standardization has limited both its acceptance as a diagnostic tool and multicenter clinical evaluations, preventing its inclusion in disease-defining criteria. In 2006, the European Aspergillus PCR Initiative was formed. The aim of the initiative was to provide optimal standardized protocols for the widespread clinical evaluation of the Aspergillus PCR to determine its diagnostic role and allow inclusion in disease diagnosis criteria. Quality control panels were developed and circulated to centers for evaluation of the existing methodology before recommendations based on the initial results were proposed for further panels. The centers were anonymously classified as “compliant” or “noncompliant,” according to whether they had followed the proposed recommendations before the performance parameters were determined and meta-regression analysis was performed. Most PCR amplification systems provided similar detection thresholds, although positivity was a function of the fungal burden. When PCR amplification was combined with DNA extraction, 50% of the centers failed to achieve the same level of detection. Meta-regression analysis showed positive correlations between sensitivity and extraction protocols incorporating the proposed recommendations and the use of bead beating, white cell lysis buffer, and an internal control PCR. The use of elution volumes above 100 μl showed a negative correlation with sensitivity. The efficiency of the Aspergillus PCR is limited by the extraction procedure and not by PCR amplification. For PCR testing of whole blood, it is essential that large blood volumes (≥3 ml) be efficiently lysed before bead beating to disrupt the fungal cell and performance of an internal control PCR to exclude false negativity. DNA should be eluted in volumes of <100 μl.
Despite the improved awareness of, and ability to diagnose, invasive aspergillosis (IA), definitive diagnosis of IA remains challenging. Clinical signs may be no more specific than fever of unknown origin refractory to treatment with broad-spectrum antibiotics, and classical mycology is seldom useful. With more people surviving protracted bouts of immunosuppression as a result of chemotherapy or preparation for stem cell transplantation, the incidence of disease will continue to rise. Mortality rates are high, approaching 100% for cerebral disease, but they can be reduced by the early initiation of antifungal therapy (14, 24). However, without an accurate diagnosis, clinicians frequently resort to empirical antifungal therapy, exposing patients to unnecessary treatment and its associated toxicity and side effects.
Health care providers also incur extra costs, with the annual cost of treating fungal infections in Europe running into millions of euros (26). Moreover, patients diagnosed with IA often require extended hospital care, leading to supplementary costs estimated to be 75,000 euros per patient, underlining the medical and socioeconomic importance of IA (26).
The introduction of nonculture techniques has enhanced the ability to diagnose IA in patients at high risk. In neutropenic patients, pulmonary abnormalities consistent with IA, such as nodules often surrounded by a “halo sign,” can be detected by high-resolution computed tomography (8). However, the halo sign is transient and is detectable only during early IA, after which radiological signs become nonspecific or appear too late to be therapeutically useful (3). Commercially available tests for the detection of galactomannan antigen and beta-d-glucan show variable performance characteristics and are influenced by the prevalence of the disease, the testing strategy used, age, the underlying condition, concomitant therapy with certain antimicrobials, and even food consumption (10, 19, 21, 27).
The successful PCR-aided diagnosis of IA has been reported in many publications (29). However, the widespread acceptance of the PCR diagnosis of IA has been hindered by a lack of standardization, compounded by limited knowledge of the disease process. A multicenter evaluation is required to overcome the statistical bias introduced by the relatively low frequency of IA, which is often 10% or less in high-risk populations. Paradoxically, it is unlikely that an appropriate evaluation will be achieved until a standard for PCR is agreed upon.
In 2006, a publication describing the design and clinical evaluation of an Aspergillus PCR technique was accompanied by an editorial highlighting the fact that even though a PCR for the detection of Aspergillus in human specimens has existed for almost 2 decades, the technique was not included in the European Organization for Research and Treatment of Cancer and the Mycoses Study Group (EORTC/MSG) consensus definitions for the diagnosis of invasive fungal diseases (IFDs) because of a lack of standardization (1, 4, 5, 31). Later that year, the United Kingdom Fungal PCR Consensus Group published an agreed-upon methodology for the PCR-aided diagnosis of IA within the United Kingdom and Ireland (30). Those reports galvanized a group of mainly European experts into forming the European Aspergillus PCR Initiative (EAPCRI) working group of the International Society for Human and Animal Mycoses (ISHAM), which involved more than 60 centers across Europe and which included centers in Australia and the Middle East. EAPCRI agreed to collaborate to develop a standard for an Aspergillus PCR methodology and to validate this in clinical trials so that PCR could be incorporated into future consensus definitions for the diagnosis of IFDs. EAPCRI consists of laboratory, clinical, and statistical working groups and a steering committee charged with focusing the overall direction of the group, providing a link between the working parties, and also raising the necessary funds (www.eapcri.eu).
Here we report on the process, findings, and methodological recommendations of the EAPCRI Laboratory Working Party (LWP) for a PCR used to test blood. The preferred function of the PCR was as a screening test rather than a confirmatory test. The aim was to ensure the highest sensitivity to attain the maximum negative predictive value, as this has been the most pressing issue in clinical practice, and to obtain an assay suited to the diagnosis of diseases with relatively low incidence rates.
MATERIALS AND METHODS
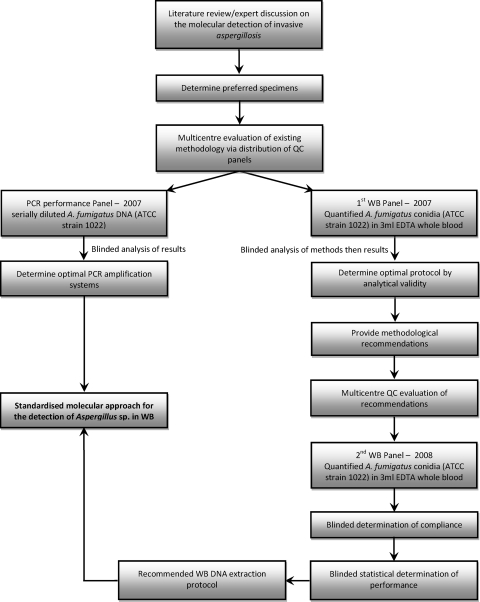
As the study described here was primarily an analytical validation of the Aspergillus PCR, the study followed the guidelines of the Standards for Reporting of Diagnostic Accuracy, where applicable (2). A summary of the EAPCRI laboratory standardization process is shown in Fig. 1.
FIG. 1.
Summary of the EAPCRI Aspergillus PCR standardization process.
Participants.
LWP comprised a core group of 8 laboratories and an extended group of an additional 16 laboratories. The centers had both clinical and scientific expertise in the field. All centers were designated by a numerical code, which allowing unbiased assessment of the results by the core group and the maintenance of impartiality throughout the analytical process.
Specimens.
The literature supports the use of EDTA-anticoagulated whole blood (WB), serum, and bronchoalveolar lavage (BAL) fluid (29). For the initial laboratory standardization (quality control [QC]), WB was selected as the target specimen, as it can be obtained by relatively noninvasive means, and large WB specimen volumes from all adult patients may be used for high-frequency screening. WB is a clinical sample whose use is more technically demanding and methodologically diverse than serum, and its use requires greater assay standardization. WB was obtained from healthy volunteers, and the samples were pooled.
Panels.
For the QC panels, 3-ml WB aliquots were provided and represented the midpoint volumes used in studies described in the literature. All WB material was tested for the presence of contaminating Aspergillus spp. before it was processed (30, 31). To avoid airborne contamination, the processing of all material took place in a category 2 laminar-flow hood. The QC panels were spiked with Aspergillus fumigatus conidia (strain ATCC 1022) quantified by use of a Fuchs-Rosenthal counting chamber. Negative controls consisting of fungus-free blood were included. Two WB panels were distributed in 2007 and 2008, respectively (Fig. 1).
To determine whether DNA extraction or PCR amplification was rate limiting, an additional DNA panel focusing solely on the performance of PCR amplification was also distributed alongside the WB panel distributed in 2007 (Fig. 1). DNA was extracted from 106 A. fumigatus conidia before it was serially diluted. The expediency of the panel was tested by an Aspergillus-specific real-time PCR (30, 31). The acceptable threshold for the PCR performance panel was determined, and the DNA concentration in the specimen was equivalent to approximately 50 A. fumigatus conidia eluted in 100 μl or 27 rRNA gene copies per μl eluate, when the mean number of copies of the rRNA genes is considered to be 54 per A. fumigatus genome (9). The mean cycle threshold (CT) value was 35.3 cycles (upper limit, 38.8 cycles) and corresponded to the typical CT values obtained by the core group when WB from patients with proven/probable IA was tested. It was essential that all PCR methods evaluated be able to attain this level of detection. PCR methods not performing to this standard were considered suboptimal. Additional dilution factors were included to identify methods with enhanced PCR performance (EPCR). Negative controls consisting of molecular-grade water were included.
The QC panels were designed to determine the detection limit and concentrated on clinically relevant low fungal burdens, with between 70 and 80% of the specimens in each WB panel being spiked with less than 100 conidia per specimen. To validate the panels, initial blind testing was performed by the core group (n = 8) before distribution of the panels and blind testing by all centers (n = 24). After the panels were developed, the distributing center froze all panels at −80°C for up to 1 week with no freeze-thaw intervals. The panels were circulated on dry ice for next-day delivery by courier. The receiving centers were asked to confirm receipt of the panel, comment on the state of the panel (frozen or thawed), and keep the specimens frozen at −80°C until they were tested. Any panels that were received thawed would be noted and the results would be excluded from further analysis. A control panel was kept by the distributing center and was tested for validity on confirmation of the receipt of the panels from all receiving centers.
DNA extraction.
For the 2007 WB panel, the distribution centers were encouraged to use the existing methodology. This typically involved the lysis of human blood cells prior to enzymatic or mechanical fungal cell lysis and then DNA purification and precipitation with a commercial kit. Following analysis of the first distribution panel, extraction recommendations (process the entire 3-ml specimen and use bead beating for fungal cell lysis) that all centers were encouraged to incorporate into their existing methods for testing of the 2008 WB panel were proposed.
PCR amplification.
Because there was no universally standardized PCR method, the participating centers were told to use their current amplification systems. For all panels, it was recommended that the DNA extracts be tested in duplicate and preferably in triplicate. The use of an internal control PCR was a prerequisite. It was decided that the use of nested PCR assays should be avoided, as these were considered too prone to producing false-positive results.
Defining compliance.
When the specimens in the 2008 WB panel were tested, the centers were graded as “compliant” or “noncompliant,” depending on whether they had followed the proposed recommendations: to use the entire specimen provided, use bead beating to lyse the fungal cells, use an internal control PCR to avoid false-negative results, and perform PCR analysis at least in duplicate.
Data analysis.
The centers were requested to return their results within a designated time frame and provide detailed protocols for their DNA extraction and PCR amplification systems. For the 2008 WB panel distribution, for which methodological recommendations had been proposed, the core group, blinded to the individual assay performance and center identity, scrutinized the procedures for compliance. The various protocols used for the 2008 panel are summarized in Tables 1 and 2. A data analysis subgroup was elected to confirm the classifications and to determine covariates for blinded statistical analysis by an independent party.
TABLE 1.
Summary of methods used to test the 2008 EAPCRI WB panela
| Center | Vol (ml) used | RCL | WCL | Fungal lysis | Method of fungal DNA purification | DNA strategy | Internal control PCR | No. of replicates | Compliant | PCR strategy |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | Yes | Yes | B. beating | QIAamp Blood kit (Qiagen) | DNA 1 | No | 3 | No | PCR 1 |
| 3 | 3 | Yes | No | B. beating | MagNA Pure LC Total NA kit (Roche) | DNA 2 | Yes | 3 | Yes | PCR 2 |
| 4 | 1-2 | Yes | No | Lyticase | Phenol-chloroform | DNA 3 | Yes | 1 | No | PCR 3 |
| 5 | 3 | Yes | Yes | Lyticase | DNAeasy Tissue (Qiagen) | DNA 4 | No | 3 | No | PCR 4 |
| 6 | 3 | Yes | No | B. beating | MagNA Pure LC DNA kit I (Roche) | DNA 5 | No | 3 | No | PCR 5 |
| 7 | 3 | Yes | No | B. beating | ZR Fungal/Bacterial DNA kit (Zymo research) | DNA 6 | Yes | 3 | Yes | PCR 6 |
| 8 | 1.5 | Yes | No | B. beating | SeptiFast kit (Roche) | DNA 7 | Yes | 3 | No | PCR 7 |
| 9 | 3 | Yes | Yes | B. beating | SeptiFast kit (Roche) | DNA 8 | Yes | 3 | Yes | PCR 7 |
| 10 | 3 | Yes | Yes | B. beating | MagNA Pure LC Total NA kit (Roche) | DNA 9 | Yes | 3 | Yes | PCR 1 |
| 11 | 3 | Yes | Yes | B. beating | Tissue kit, EZ1 (Qiagen) | DNA 10 | Yes | 3 | Yes | PCR 8 |
| 13 | 3 | Yes | Yes | B. beating | High Pure Template PCR kit (Roche) | DNA 11 | Yes | 3 | Yes | PCR 1 |
| 14 | 3 | Yes | Yes | B. beating | SeptiFast kit (Roche) | DNA 8 | Yes | 3 | Yes | PCR 9 |
| 15 | 3 | No | No | B. beating | QIAamp blood kit (Qiagen) | DNA 12 | Yes | 3 | Yes | PCR 1 |
| 16b | 3 | ND | ND | B. beating | QIAamp DNA minikit (Qiagen) | DNA 13 | Yes | 3 | Yes | PCR 10 |
| 17 | 3 | Yes | Yes | Lyticase | DNA Lego kit (Top Bio) | DNA 14 | No | 3 | No | PCR 11 |
| 18 | 3 | Yes | No | B. beating | MagNA Pure LC DNA kit I (Roche) | DNA 5 | No | 3 | No | PCR 5 |
| 19 | 3 | Yes | Yes | B. beating | GeneXpert (Cepheid) | DNA 15 | Yes | 3 | Yes | PCR 12 |
| 20 | 3 | Yes | Yes | B. beating | High Pure Template PCR kit (Roche) | DNA 11 | Yes | 1 | No | PCR 2 |
| 21 | 2 | Yes | No | B. beating | SeptiFast kit (Roche) | DNA 7 | Yes | 3 | No | PCR 1 |
| 22 | 3 | Yes | Yes | B. beating | High Pure Template PCR kit (Roche) | DNA 11 | Yes | 3 | Yes | PCR 13 |
| 23 | 3 | Yes | Yes | B. beating | bioMérieux MiniMagc | DNA 16 | Yes | 3 | Yes | PCR 1 |
The results for the centers that used methods that attained the detection threshold are indicated in boldface. These included 11 compliant centers and 3 noncompliant centers. Eleven different DNA extraction techniques were used together with nine different PCR protocols by the 14 centers attaining the threshold, 9 of which were compliant; 8 attained the PCR performance threshold, and 5 achieved enhanced PCR performance. No results were received from centers 1 and 12, whereas logistical limitations prohibited the participation of center 24. Abbreviations: RCL, red cell lysis; WCL, white cell lysis; B. beating, bead beating; ND, not disclosed.
The specificity of the assay was poor (11.1%).
For the extraction reagents, any possible contaminating DNA sources were removed by magnetic silica treatment.
TABLE 2.
Details of the PCR systems and performance by testing of the PCR performance panela
| Center | PCR format | Platform | Gene target | Detection threshold (no. of copies/μl)b | PCR strategy |
|---|---|---|---|---|---|
| 1 | RT hydrolysis (TaqMan) probe | Rotorgene | 28S gene | 27 | PCR 1 |
| 2 | RT hydrolysis (TaqMan) probe | LightCycler | 28S gene | 2.7 | PCR 1 |
| 3 | RT hydrolysis (TaqMan) probe | TaqMan | 18S gene | 0.27 | PCR 2 |
| 4 | Nested PCR | Conventional | 18S gene | 27 | PCR 3 |
| 5 | RT hybridization (FRET) probes | LightCycler | 18S gene | No positive results | PCR 4c |
| 6 | RT hybridization (FRET) probes | LightCycler | 18S gene | 27 | PCR 5c |
| 7 | RT hydrolysis (TaqMan) probe | Rotorgene | ITS2 gene | 2.7 | PCR 6 |
| 8 | RT hybridization (FRET) probes | LightCycler | ITS gene | 27 | PCR 7 |
| 9 | RT hybridization (FRET) probes | LightCycler | ITS gene | 2.7 | PCR 7 |
| 11 | RT hydrolysis (TaqMan) probe | TaqMan | 18S gene | 27 | PCR 8 |
| 12 | RT hydrolysis (TaqMan) probe | LightCycler | 28S gene | 27 | PCR 1 |
| 14 | RT hybridization (FRET) probes | LightCycler | ITS gene | 27 | PCR 9 |
| 15 | RT hydrolysis (TaqMan) probe | Mx3000P | 28S gene | 2.7 | PCR 1 |
| 16 | RT FAM molecular beacons | LightCycler | ITS1 gene | 270d | PCR 10 |
| 17 | Nested PCR | Conventional | 18S gene | 2.7 | PCR 11 |
| 18 | RT hybridization (FRET) probes | LightCycler | 18S gene | 2.7 | PCR 5c |
| 19 | RT hydrolysis (TaqMan) probe | Smartcycler | 18S gene | 27 | PCR 12 |
| 20 | RT hydrolysis (TaqMan) probe | TaqMan | 18S gene | 27 | PCR 2 |
| 22 | RT hybridization (FRET) probe | LightCycler | ITS gene | 2.7 | PCR 13 |
| 23 | RT hydrolysis (TaqMan) probe | Rotorgene | 28S gene | 2.7 | PCR 1 |
| 24 | Nested PCR | Conventional | 18S gene | 270e | PCR 3 |
A total of 13 different PCR strategies were used by the 21 centers that returned results. Assays with enhanced PCR performance for the detection of conidia below the threshold of detection are indicated in boldface. PCR 1 is the assay of White et al. (31). Three of five centers that used that assay achieved enhanced PCR performance. Five of five centers that used that assay attained the threshold. The assay was also one of two assays deemed to be optimal by the United Kingdom Fungal PCR Consensus Group (30). PCR 2 is the assay of Kami et al. (12). One of two centers that used that assay achieved enhanced PCR performance. Two of two centers that used that assay attained the threshold. The assay was also one of two assays deemed to be optimal by the United Kingdom Fungal PCR Consensus Group (30). Note that the assay should not be performed on the Roche LightCycler system (30). PCR 5 is the assay described by Klingspor and Jalal (13) and Loeffler et al. (17). One of two centers that used that assay achieved enhanced PCR performance. Two of two centers that used that assay attained the threshold. PCR 6 is the assay of Lengerova et al. (15). The one center that used that assay achieved enhanced PCR performance. PCR 7 is the Roche Septifast test. One of two centers that used that assay achieved enhanced PCR performance. Two of two centers that used that assay attained the threshold. PCR 13 is the assay of Loeffler et al. (16). The one center that used that assay achieved enhanced PCR performance. As PCR 11 is a nested PCR system, its use is not recommended by the EAPCRI. Centers 10, 13, and 21 were unable to return results. Abbreviations: RT, real-time; FRET, fluorescence resonance energy transfer; FAM, 6-carboxyfluorescein; Rotorgene, Corbett Rotorgene apparatus; LightCycler, Roche LightCycler apparatus, various models; TaqMan, Applied Biosystems TaqMan apparatus, various models; Mx 3000P, Stratagene Mx3000P apparatus; Smartcycler, Cepheid Smartcycler apparatus.
As determined by the PCR performance panel.
The PCR 4 and PCR 5 assays use the same probes but use slightly different primer sequences.
PCR 10 was able to reproducibly detect <0.27 copies/μl but failed to detect 2.7 and 27 copies/μl.
The panel was delayed in transit and results excluded from analysis.
Statistical analysis.
The data produced by the responding centers were qualitative and quantitative. Qualitative data were coded as true positive (positive PCR result obtained with blood spiked with Aspergillus conidia), false positive (positive PCR result obtained with blood that did not contain Aspergillus conidia), true negative (negative PCR result obtained with blood that did not contain Aspergillus conidia), and false negative (negative PCR result obtained with blood spiked with Aspergillus conidia). The diagnostic odds ratio (DOR), sensitivity, and specificity were calculated. For the evaluation of the diagnostic accuracy indices, the specimen with 10 conidia per 3 ml was excluded due to the possible influence of burden variability between replicates. The sensitivity, specificity, and DOR were also transformed to their logits in order to obtain a normal distribution of values. In order to manage the heterogeneity arising from the different centers (random effects), the effects exerted by several categorical (binary) covariates on sensitivity, specificity, and DOR were evaluated by meta-regression methods. The covariates are shown in Table 3. The random-effects inverse variance weighting method was used for calculations related to meta-analytic pooling and meta-regression (25).
TABLE 3.
Categorical binary covariates investigated by regression methods
| Covariate | Explanation |
|---|---|
| Extraction method | Compliant with extraction recommendations: yes or no |
| Blood vol useda | Entire blood vol used: yes or no |
| Beadsa | Beads for mechanical cell wall disruption used: yes or no |
| Internal controla | Internal control PCR used: yes or no |
| RCLB | Red cell lysis buffer used: yes or no |
| WCLB | White cell lysis buffer used: yes or no |
| NaOH | NaOH used as lytic agent: yes or no |
| Purification | A protocol for DNA purification was used after bead beating |
| More than nine | |
| manual steps | Number of manual steps exceeds nine: yes or no |
| ITS | Internal transcribed spacer (ITS) target used for Aspergillus PCR: yes or no |
| 18S | 18S rRNA gene target used for Aspergillus PCR: yes or no |
| 28S | 28S rRNA gene target used for Aspergillus PCR: yes or no |
| Elution vol | Elution vol below 100 μl: yes or no |
| Low specificity | Less than 100% specificity: yes or no |
Criteria used to determine compliance.
Linear regression analysis was performed for the centers providing CT values. The relationship between this quantitative outcome and the number of Aspergillus conidia/3 ml was investigated by use of a mixed-effects restricted-maximum-likelihood regression procedure to take into account the heterogeneity at the center level.
The efficiency of the PCR was calculated by using the equation −1 + 10(−1/slope).
RESULTS
The panels were distributed to a total of 24 centers on two separate occasions (in 2007 and 2008). Of these centers, 83.3%, 91.7%, and 87.5% returned results within the allocated time frame for the PCR performance panel, the 2007 WB panel, and the 2008 WB panel, respectively.
Determination of Aspergillus PCR performance.
Of the 24 centers receiving the panels, 23 actively tested the PCR performance panel, although 2 centers were unable to return results due to technical difficulties and an additional center experienced distribution delays. Eighteen of the 20 centers achieved the required detection limit of approximately 27 rRNA copies/μl eluate and used 11 different PCR protocols (Table 2). Nine of the 18 centers using seven different PCR protocols showed EPCR and consistently detected lower fungal burdens equivalent to 5 A. fumigatus conidia at a concentration of 2.7 rRNA copies/μl eluate (Table 2). There was no statistically significant difference between the mean PCR efficiency for the different platforms used. The specificity of the PCR methods tested was 95.2%.
The impact of DNA extraction on PCR performance.
Centers meeting the required PCR performance threshold should achieve a similar performance when they test spiked WB, provided an efficient DNA extraction method is used. The WB threshold was 50 conidia loaded into a 3-ml specimen. The use of any PCR system coupled with an inefficient extraction system would fail to achieve these standards.
The 2007 WB panel.
Nine of the 22 centers returning results for the 2007 WB panel achieved the required WB detection threshold. All nine centers achieved the prerequisite PCR performance threshold using seven different PCR protocols (PCRs 1, 3, 5, 6, 7, 9, and 13), and six centers had EPCR for the detection of lower fungal burdens by the use of five PCR protocols (PCRs 1, 5, 6, 7, and 13). Eight of the nine centers, including all six centers with EPCR, used the entire WB specimen for extraction, and the other center used 75% of the blood supplied. Eight of the nine centers, including all six centers with EPCR, used bead beating to disrupt the fungal cell. One center used lyticase to remove the fungal wall. Two additional centers that did not reach the WB extraction threshold also used the entire specimen and lyticase. Six centers that failed to reach the required WB threshold disrupted the fungal cell with bead-beating-based methods but did not use the entire specimen. Three centers that failed to reach the WB threshold did not use a recognized fungal cell lysis step. One center had major contamination issues (specificity, 0%), raising doubt as to the significance of the results, and another center experienced distribution delays. Eight of the 11 centers that failed to achieve the WB threshold achieved the prerequisite PCR performance threshold and three centers had EPCR, indicating the importance of the use of an efficient DNA extraction technique.
These findings formed the basis for the DNA extraction recommendations for testing of the 2008 WB panel. By conventional meta-analytical pooling, the overall sensitivity for the first WB panel was 64.5% (95% confidence interval [CI], 49.0 to 77.5%) and the overall specificity was 89.6% (95% CI 79.1 to 95.2%). The DOR was 15.1 (95% CI, 4.4 to 52.2).
The 2008 WB panel.
Twelve compliant centers followed the recommendations when they tested the 2008 panel, whereas nine centers did not and, consequently, were deemed noncompliant (Table 1). The number of centers achieving the required WB detection threshold was 14, and those centers used nine different PCR protocols (Table 1). Twelve of the 14 centers reaching the WB threshold used the entire specimen, and all used bead beating to lyse the fungal cell. Eleven of these 14 centers were classified as compliant. Twelve of the 14 centers (9/11 compliant centers) attaining the WB threshold were also capable of detecting the lower fungal burdens by using seven different PCR protocols (PCRs 1, 2, 6, 7, 10, 12, and 13). However, one compliant center had major contamination problems (specificity, 11.1%). Of the seven centers that failed to reach the required threshold, six centers were noncompliant and one center was compliant.
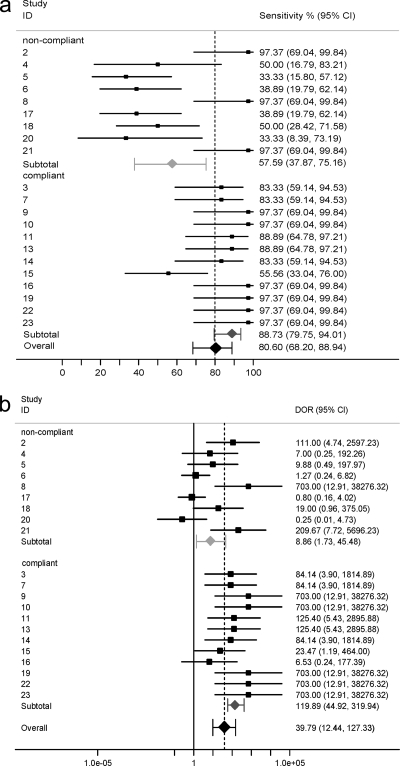
The overall sensitivity was statistically significantly greater for the second WB panel and the specificity was not compromised, resulting in an improved DOR. By conventional meta-analytical pooling, an overall sensitivity of 80.60% (95% CI, 68.20 to 88.94%), a specificity of 86.29% (95% CI, 76.06 to 92.58%), and a DOR of 39.79 (95% CI, 12.44 to 127.33) were calculated (Table 4). The mean positivity rate was a function of the conidial concentration, which was the greatest for the higher fungal burdens, and the rates for the 2008 panel were better than those for the 2007 panel (Table 5 and Fig. 2). The sensitivity and DOR showed considerable interlaboratory variability, resulting in two distinct populations, although most centers achieved satisfactory specificity (Table 4 and Fig. 3). Upon further investigation, it was shown that the sensitivity for compliant centers was significantly greater than that for noncompliant centers (Table 4 and Fig. 3). Six centers failed to achieve 100% specificity, and five of these centers had been classified as noncompliant. To exclude the possibility that positive results arose from contamination and the calculation of false associations, a further meta-analysis was performed without the results from those six centers.
TABLE 4.
Mean performance statistics for protocols used in the 2008 EAPCRI WB panela
| Extraction protocol | Sensitivity |
Specificity |
DOR |
|||
|---|---|---|---|---|---|---|
| % | 95% CI | % | 95% CI | Value | 95% CI | |
| All | 80.6 | 68.2-88.9 | 86.3 | 76.1-92.6 | 39.8 | 12.4-127.3 |
| Compliant | 88.7 | 79.8-94.0 | 91.6 | 79.1-96.9 | 119.9 | 44.9-319.9 |
| Noncompliant | 57.6 | 37.9-75.2 | 77.2 | 61.2-87.9 | 8.9 | 1.7-45.5 |
Twelve centers that used 10 different DNA extraction protocols and 9 different PCR amplification procedures were compliant. Nine centers that used seven different DNA extraction protocols and seven different PCR amplification procedures were noncompliant. The differences between the compliant and the noncompliant centers were statistically significant for sensitivity (P = 0.008) and DOR (P = 0.006).
TABLE 5.
Positivity rates for the two EAPCRI WB panels and PCR performance panela
| Fungal load (total no. of conidia) | Potential total no. of rRNA copiesb | No. of copies/μlc | Positivity rate (% [no. of PCR tests]) |
||
|---|---|---|---|---|---|
| PCR panel (n = 52) | 2007 panel (n = 61) | 2008 panel (n = 59) | |||
| 2.7 × 105 | 94.2 (49) | NT | NT | ||
| 2.7 × 104 | 94.2 (49) | NT | NT | ||
| >10,000 | 5.4 × 105 | 5.4 × 103 | NT | 77.0 (47) | NT |
| 2.7 × 103 | 92.3 (48) | NT | NT | ||
| 1,000 | 5.4 × 104 | 5.4 × 102 | NT | 75.4 (46) | 98.3 (58) |
| 500 | 2.7 × 104 | 2.7 × 102 | 94.2 (49) | 57.4 (35) | 98.3 (58) |
| 100 | 5.4 × 103 | 5.4 × 101 | NT | 34.4 (21) | 78.0 (46) |
| 75 | 4.1 × 103 | 4.1 × 101 | NT | 36.1 (22) | 67.8 (40) |
| 50 | 2.7 × 103 | 2.7 × 101 | 86.5 (45) | 37.7 (23) | 74.6 (44) |
| 20-25 | 1.1-1.4 × 103 | 1.1-1.4 × 101 | NT | 29.5 (18) | 62.7 (37) |
| 10 | 5.4 × 102 | 5.4 | NT | 27.9(17) | 49.2 (29) |
| 2.7 | 40.4 (21) | NT | NT | ||
| 0d | 0 | 0 | 2.9 (3)e | 4.9 (6)f | 11.3 (20)g |
A total of 22 centers contributed results for the PCR performance panel and the 2007 WB panel, although 1 center returned results only for the PCR panel, while another returned results only for the WB panel. For the 2008 WB panel, 21 centers returned the results. The theoretical threshold is indicated in boldface. NT, not tested in this panel.
Based on 54 copies of rRNA copies per genome (9).
The calculations were based on 100% extraction efficiency and an elution volume of 100 μl.
The 1st panel contained two negative control specimens. The 2nd panel contained three negative control specimens.
As the panel contained two negative specimens, the total number of tests was 104 and not 52.
For one center, both negative specimens were PCR positive. As the panel contained two negative specimens, the total number of tests was 122 and not 61.
As the panel contained three negative specimens, the total number of tests was 177 and not 59.
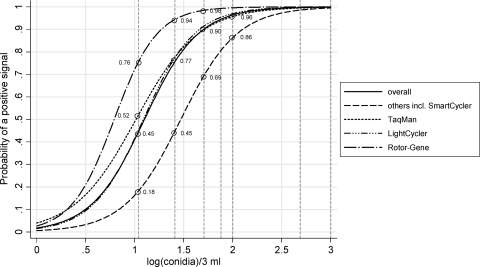
FIG. 2.
The probability of a positive Aspergillus PCR signal when the 2008 WB panel is tested. The vertical dashed reference lines indicate (log-transformed) conidial loads of 10, 25, 50, 75, 100, 500, and 1,000. They intersect the various platform-specific logistic curves at the indicated probability levels. Twenty-one centers performed 59 tests on each specimen. Ten centers used the Roche LightCycler apparatus to perform a total of 30 replicates for each specimen, five centers used the Applied Biosystems TaqMan apparatus to perform a total of 13 replicates, two centers used the Corbett Rotorgene apparatus to perform a total of 6 replicates, and four centers used other platforms to perform a total of 10 replicates.
FIG. 3.
Forest plot of sensitivity (a) and DOR (b) for each center participating in testing of the 2008 EAPCRI WB panel. Individual centers are indicated by numbers. The centers are grouped according to their compliance with the extraction protocol. The effect size sensitivity (%) and DOR. The closed rhomboids indicate the average values of sensitivity/DOR at the group level.
There was a positive correlation by bivariate meta-regression analysis between sensitivity and the compliant protocols, the use of bead beating, and the use of an internal control PCR (Table 6). Multivariate regression analysis determined positive correlations between sensitivity and the use of white cell lysis buffer (t = 2.95, P = 0.018), bead beating (t = 2.57, P = 0.033), and an internal control PCR (t = 2.25, P = 0.054) but a negative correlation between sensitivity and the use of elution volumes greater than 100 μl (t = −2.53, P = 0.035).
TABLE 6.
Bivariate meta-regression analysis between logit sensitivity and the additional covariates for centers with 100% specificitya
| Covariate | t | P |
|---|---|---|
| Compliant extraction method | 2.69 | 0.018 |
| Blood vol used | 0.22 | 0.829 |
| RCLB | 1.12 | 0.285 |
| WCLB | 2.08 | 0.058 |
| NaOH | 1.59 | 0.137 |
| Beads | 2.59 | 0.023 |
| Purification | 1.14 | 0.274 |
| More than nine manual steps | 0.46 | 0.650 |
| ITS | 1.54 | 0.148 |
| 18S | −1.89 | 0.082 |
| 28S | 0.48 | 0.637 |
| Internal control PCR | 2.85 | 0.015 |
| Elution vol | −0.75 | 0.469 |
n = 15 centers. t, slope/standard error of the slope. A positive t value indicates a favorable correlation and vice versa. Significant correlations are indicated by a P value of <0.05 and are indicated in boldface.
DISCUSSION
The results from this study showed that most PCR amplifications systems assessed (18/20, 90%) efficiently detected the equivalent of 50 A. fumigatus conidia and that 45% (9/20) were able to detect the equivalent of 5 A. fumigatus conidia (Table 2). However, half the centers suffered dramatic reductions in detection thresholds when they tested the 2007 WB panel, highlighting the impact of the WB DNA extraction procedures used. Indeed, the extraction procedure appears to be the rate-limiting step and focused the investigations accordingly. Detailed assessment of the extraction methods used with the 2007 WB panel showed that protocols that used the entire specimen and bead beating were associated with better performance and were recommended for use for analysis of the 2008 WB panel, resulting in improved positivity rates (Table 5). It is accepted that the spiking of blood with conidia does not accurately reflect disease, as angioinvasion occurs via hyphal penetration. However, it is not possible to accurately quantify hyphae that may also be multinucleate, resulting in interspecimen variations. The recommended beat-beating protocol is equally effective at disrupting hyphae (28).
Three noncompliant centers (centers 2, 8, and 21) achieved excellent analytical sensitivity, and evaluation of their protocols revealed one center (center 2) that had been deemed to be noncompliant for not using an internal control PCR but otherwise had followed the recommendations (Table 1). The other two centers used only half of the specimen provided but otherwise followed the recommendations. However, two of the three noncompliant centers (centers 2 and 21) with excellent sensitivities also generated false-positive results, casting doubt over the relevance of the putatively true-positive results from that center. From a diagnostic perspective, any positive results in which the negative control also tests positive should be disregarded.
The sensitivities of the compliant systems were >30% higher than those of the noncompliant systems (Table 4). Only one compliant center (center 15) generated poor sensitivities. Detailed examination of the protocol used by that center showed that even though the center used the entire specimen, bead beating, and an internal control PCR, it made no attempt to remove the human blood cells (Table 1). This would have reduced the sensitivity, as white cell lysis is positively associated with sensitivity. Only one compliant center (center 16) had poor specificity, and this was reflected in a reduced diagnostic odds ratio (Fig. 3).
A high degree of analytical sensitivity is essential, as the fungal burden circulating in the bloodstream is low (17, 20). The use of larger sample volumes allows the detection of lower concentrations of fungal burden because both the overall amount of target and the opportunity for retrieval are increased (23). Paradoxically, too much extrinsic DNA may inhibit the PCR process, so it is essential that an internal control be used to prevent the reporting of false-negative results.
Statistical analysis between sensitivity and the other covariates showed that the recommendations (use of the entire specimen, bead beating, and an internal control PCR) and the individual stage of white cell lysis were all positively associated with sensitivity. Conversely, negative correlations were associated with larger elution volumes (>100 μl), which reduced the final DNA concentration.
There were no significant correlations between sensitivity and the PCR target. However, multicopy targets should be employed to enhance performance with some strains of A. fumigatus possessing almost 102 copies of the 18S rRNA gene (9). All rRNA gene targets (the 18S, 5.8S, 28S, and ITS regions) have the same copy number within individual organisms but may share significant similarity between fungal species and the homologous human genes, and both false positivity and false negativity may be a result of inappropriate oligonucleotide design (30, 32).
Recommendations of EAPCRI.
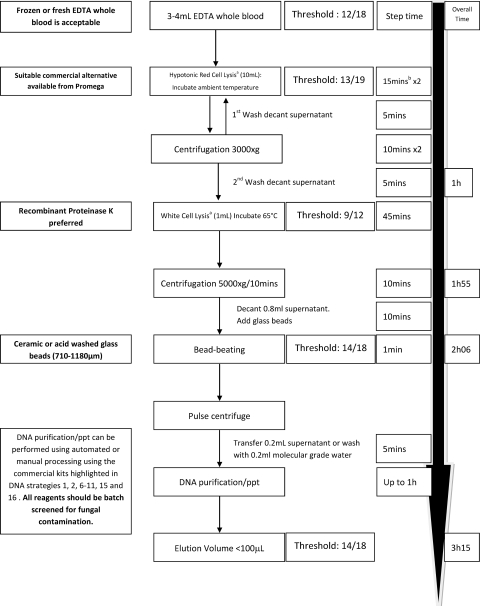
The recommended protocol for extracting fungal DNA from EDTA-anticoagulated whole-blood specimens was determined by comparing the protocols described in Table 1, their associated analytical performance characteristics, and the statistical association between the individual steps of the extraction process and sensitivity. The protocol is illustrated in Fig. 4.
FIG. 4.
Recommended protocol for extracting Aspergillus DNA from whole blood. Boldface text is expert opinion. The threshold is the proportion of centers reaching the threshold when the step is used. ppt, precipitation; a, the lysis buffer described previously (6); b, involves 5 min of processing time and 10 min of incubation time; when frozen specimens are processed, incubation is not necessary.
We recommend that EDTA be the only anticoagulant used for blood specimens, as heparin and sodium citrate have been associated with inhibition and contamination, respectively (7, 33). Relatively large blood volumes (≥3 ml) are required, and it is important to efficiently lyse both red and white blood cells prior to bead beating to disrupt the fungal cell. Fungal lysis through recombinant lyticase digestion should be replaced by bead beating, which removes considerable expense and additional processing time (30 to 60 min). After the fungal cell is lysed, commercial protocols can be used to purify, capture, and elute the DNA. Then different DNA extraction protocols shown in Table 1 were associated with good analytical sensitivity and specificity. Reagent contamination can be problematic when fungal PCR is performed (18, 22). It is good practice to screen both commercial and in-house reagents for contamination by using negative extraction controls. Likewise, positive extraction controls with consistent loads and representative of typical IA should routinely be used to monitor the performance of the extraction procedure. The use of DNA elution volumes greater than 100 μl should be avoided.
Most PCR amplification systems work efficiently, the variability in current performance appears to be a direct consequence of the extraction technique used, and it is not necessary to define an individual PCR assay. However, the six real-time PCR protocols (PCRs 1, 2, 5, 6, 7, and 13) described in Table 2 had EPCR and may provide improved performance when their use is coupled with the DNA extraction technique described in Fig. 4. In general, all PCR testing should be performed at least in duplicate, with further testing being performed as required to interpret contradictory results that may arise as result of the low fungal burden or the limits of real-time PCR detection (20). An internal control system should also be used to avoid false-negative results. The internal control target should be present at levels similar to those that result in positive results by a typical Aspergillus PCR (CT values, 35 to 40 cycles).
These results confirm the analytical validity of the Aspergillus PCR for the screening and detection of Aspergillus. Further external validity and the testing accuracy of the proposed method will be strategically determined by other laboratories that have expressed an interest in this EAPCRI (11). Studies into dynamic linearity may also be necessary to determine the performance of the PCR across various fungal burdens, which is particularly important in monitoring therapeutic responses. Studies with animal models are being conducted to compare the performance of the PCR by using the different specimen options. Clinical validity also needs to be established in large-scale prospective trials designed to determine the performance and utility of the Aspergillus PCR with specimens from high-risk populations.
Acknowledgments
The EAPCRI of ISHAM comprises the steering committee and three working groups. The members of the EAPCRI steering committee are J. Peter Donnelly (chair of the foundation), Radboud University Nijmegen Medical Centre, Nijmegen, Netherlands; Juergen Loeffler (secretary), Wuerzburg University, Wuerzburg, Germany; and Rosemary A. Barnes (treasurer), Cardiff University, Cardiff, United Kingdom. The members of the EAPCRI Laboratory Working Group are Juergen Loeffler (leader of the Laboratory Working Group, Data Analysis Group); P. Lewis White, NPHS Microbiology, Cardiff, United Kingdom (panel development/distribution, Data Analysis Group); Stephane Bretagne, Henri Mondor Hospital, Creteil, France (Data Analysis Group); Willem Melchers, Radboud University Nijmegen Medical Centre, Nijmegen, Netherlands; Lena Klingspor, Karolinska University Hospital, Stockholm, Sweden; Niklas Finnstrom, Cepheid AB, Toulouse, France; Elaine McCulloch and Craig Williams, Royal Hospital for Sick Children, Glasgow, United Kingdom; and Bettina Schulz, Charite University Hospital, Berlin, Germany. The members of the EAPCRI Clinical Working Group are Rosemary A. Barnes (leader of the Clinical Working Group); Catherine Cordonnier, Henri Mondor Hospital, Creteil, France; Johan Maertens, University Hospital Gasthuisberg, Leuven, Belgium; Lena Klingspor; Werner Heinz, Wuerzburg University, Wuerzburg, Germany; and Brian Jones, Glasgow Royal Infirmary, Glasgow, United Kingdom. The members of the EAPCRI Statistical Working Group are Carlo Mengoli, University of Padua, Padua, Italy; Mario Cruciani, University of Verona, Verona, Italy; Juergen Loeffler; Rosemary A. Barnes; and J. Peter Donnelly. The participants and the participating laboratories include Richard Barton and Richard Hobson, Leeds General Infirmary, Leeds, United Kingdom; Dietet Buchheidt, Mannheim University Hospital, Mannheim, Germany; Daniel Codl, Charles University, Prague, Czech Republic; Manuel Cuenca-Estrella, National Microbiology Institute, Madrid, Spain; Malcolm Guiver, HPA Northwest, Manchester, United Kingdom; Catriona Halliday and Sue Sleiman, Westmead Hospital, Westmead, NSW, Australia; Petr Haml, Palacky University, Olomouc, Czech Republic; Cornelia Lass-Floerl, University of Medicine, Innsbruck, Austria; Chris Linton and Elizabeth Johnson, United Kingdom Mycology Reference Laboratory, HPA Southwest, Bristol, United Kingdom; Clare Ling and Chris Kibbler, Royal Free Hospital, London, United Kingdom; Martina Lengerova, Central Molecular Biology, Gene Therapy and Haematology Clinic, Brno, Czech Republic; Jose Palomares, University Hospital of Valme, Seville, Spain; Tom Rogers and Stephane Duval, Trinity College, Dublin, Ireland; Boualem Sendid, Lille University, Lille, France; and Birgit Willinger, University of Medicine, Vienna, Austria.
EAPCRI thanks Andreas Opitz (Institute of Transfusional Medicine, Wuerzburg University, Wuerzburg, Germany) for drawing and screening the blood kindly donated by our volunteers.
Footnotes
Published ahead of print on 10 February 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, T. J. Walsh, and the Invasive Fungal Infections Cooperative Group of the European Organization for Research and Treatment of Cancer, Mycoses Study Group of the National Institute of Allergy and Infectious Diseases. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Bossuyt, P. M., J. B. Reitsma, D. E. Bruns, C. A. Gatsonis, P. P. Glasziou, L. M. Irwig, J. G. Lijmer, D. Moher, D. Rennie, H. C. de Vet, and the STARD Group. 2003. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Fam. Pract. 21:4-10. [DOI] [PubMed] [Google Scholar]
- 3.Caillot, D., J. F. Couaillier, A. Bernard, O. Casasnovas, D. W. Denning, L. Mannone, J. Lopez, G. Couillault, F. Piard, O. Vagner, and H. Guy. 2001. Increasing volume and changing characteristics of invasive pulmonary aspergillosis on sequential thoracic computed tomography scans in patients with neutropenia. J. Clin. Oncol. 19:253-259. [DOI] [PubMed] [Google Scholar]
- 4.De Pauw, B., T. J. Walsh, J. P. Donnelly, D. A. Stevens, J. E. Edwards, T. Calandra, P. G. Pappas, J. Maertens, O. Lortholary, C. A. Kauffman, D. W. Denning, T. F. Patterson, G. Maschmeyer, J. Bille, W. E. Dismukes, R. Herbrecht, W. W. Hope, C. C. Kibbler, B. J. Kullberg, K. A. Marr, P. Muñoz, F. C. Odds, J. R. Perfect, A. Restrepo, M. Ruhnke, B. H. Segal, J. D. Sobel, T. C. Sorrell, C. Viscoli, J. R. Wingard, T. Zaoutis, J. E. Bennett, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, and National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnelly, J. P. 2006. Polymerase chain reaction for diagnosing invasive aspergillosis: getting closer but still a ways to go. Clin. Infect. Dis. 42:487-489. [DOI] [PubMed] [Google Scholar]
- 6.Einsele, H., H. Hebart, G. Roller, J. Löffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia, M. E., J. L. Blanco, J. Caballero, and D. Gargallo-Viola. 2002. Anticoagulants interfere with PCR used to diagnose invasive aspergillosis. J. Clin. Microbiol. 40:1567-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene, R. E., H. T. Schlamm, J. W. Oestmann, P. Stark, C. Durand, O. Lortholary, J. R. Wingard, R. Herbrecht, P. Ribaud, T. F. Patterson, P. F. Troke, D. W. Denning, J. E. Bennett, B. E. de Pauw, and R. H. Rubin. 2007. Imaging findings in acute invasive pulmonary aspergillosis: clinical significance of the halo sign. Clin. Infect. Dis. 44:373-379. [DOI] [PubMed] [Google Scholar]
- 9.Herrera, M. L., A. C. Vallor, J. A. Gelfond, T. F. Patterson, and B. L. Wickes. 2009. Strain-dependent variation in 18S ribosomal DNA copy numbers in Aspergillus fumigatus. J. Clin. Microbiol. 47:1325-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hope, W. W., T. J. Walsh, and D. W. Denning. 2005. Laboratory diagnosis of invasive aspergillosis. Lancet Infect. Dis. 5:609-622. [DOI] [PubMed] [Google Scholar]
- 11.Irwig, L., P. Bossuyt, P. Glasziou, C. Gatsonis, and J. Lijmer. 2002. Evidence base of clinical diagnosis: designing studies to ensure that estimates of test accuracy are transferable. BMJ 321:669-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kami, M., T. Fukui, S. Ogawa, Y. Kazuyama, U. Machida, Y. Tanaka, Y. Kanda, T. Kashima, Y. Yamazaki, T. Hamaki, S. Mori, H. Akiyama, Y. Mutou, H. Sakamaki, K. Osumi, S. Kimura, and H. Hirai. 2001. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clin. Infect. Dis. 33:1504-1512. [DOI] [PubMed] [Google Scholar]
- 13.Klingspor, L., and S. Jalal. 2006. Molecular detection and identification of Candida and Aspergillus spp. from clinical samples using real-time PCR. Clin. Microbiol. Infect. 12:745-753. [DOI] [PubMed] [Google Scholar]
- 14.Kontoyiannis, D. P., and K. A. Marr. 2009. Therapy of invasive aspergillosis: current consensus and controversies, p. 491-502. In J.-P. Latge and W. J. Steinbach (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC.
- 15.Lengerova, M., K. Hrncirova, I. Kocmanova, Z. Racil, B. Weinbergerova, J. Vorlicek, and J. Mayer. 2008. Detection of Aspergillus fumigatus in bronchoalveolar lavage samples from immunocompromised patients using real-time PCR with fungal DNA preamplification, p.630. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 16.Loeffler, J., N. Schultze, H. Hebart, and H. Einsele. 2005. Detection of genomic DNA from Aspergillus species using the LightCycler technology, poster M-721. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 17.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the LightCycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeffler, J., H. Hebart, R. Bialek, L. Hagmeyer, D. Schmidt, F. P. Serey, M. Hartmann, J. Eucker, and H. Einsele. 1999. Contaminations occurring in fungal PCR assays. J. Clin. Microbiol. 37:1200-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mennink-Kersten, M. A., J. P. Donnelly, and P. E. Verweij. 2004. Detection of circulating galactomannan for the diagnosis and management of invasive aspergillosis. Lancet Infect. Dis. 4:349-357. [DOI] [PubMed] [Google Scholar]
- 20.Millon, L., R. Piarroux, E. Deconinck, C. E. Bulabois, F. Grenouillet, P. Rohrlich, J. M. Costa, and S. Bretagne. 2005. Use of real-time PCR to process the first galactomannan-positive serum sample in diagnosing invasive aspergillosis. J. Clin. Microbiol. 43:5097-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeiffer, C. D., J. P. Fine, and N. Safdar. 2006. Diagnosis of invasive aspergillosis using a galactomannan assay: a meta-analysis. Clin. Infect. Dis. 42:1417-1427. [DOI] [PubMed] [Google Scholar]
- 22.Rimek, D., A. P. Garg, W. H. Haas, and R. Kappe. 1999. Identification of contaminating fungal DNA sequences in zymolyase. J. Clin. Microbiol. 37:830-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz, B., K. Weber, C. Radecke, C. Scheer, and M. Ruhnke. 2009. Effect of different sample volumes on the DNA extraction of Aspergillus fumigatus from whole blood. Clin. Microbiol. Infect. 15:686-688. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz, S., and M. Ruhnke. 2009. Aspergillus sinusitis and cerebral aspergillosis, p. 301-318. In J.-P. Latge and W. J. Steinbach (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC.
- 25.Sterne, J. A. C., M. J. Bradburn, and M. Egger. 2001. Meta-analysis in Stata™, p. 347-369. In M. Egger, G. Davey Smith, and D. G. Altman (ed.), Systematic reviews in health care: meta-analysis in context, 2nd ed. BMJ Publishing Group, London, United Kingdom.
- 26.Tong, K. B., C. J. Lau, K. Murtagh, A. J. Layton, and R. Seifeldin. 2009. The economic impact of aspergillosis: analysis of hospital expenditures across patient subgroups. Int. J. Infect. 13:24-36. [DOI] [PubMed] [Google Scholar]
- 27.Upton, A., K. A. Kirby, P. Carpenter, M. Boeckh, and K. A. Marr. 2007. Invasive aspergillosis following hematopoietic cell transplantation: outcomes and prognostic factors associated with mortality. Clin. Infect. Dis. 44:531-540. [DOI] [PubMed] [Google Scholar]
- 28.Van Burik, J. A., H. R. W. Schreckhise, T. C. White, R. A. Bowden, and D. Myerson. 1998. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med. Mycol. 36:299-303. [PubMed] [Google Scholar]
- 29.White, P. L., and R. A. Barnes. 2009. Aspergillus PCR, p. 373-392. In J.-P. Latge and W. J. Steinbach (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC.
- 30.White, P. L., R. Barton, M. Guiver, C. J. Linton, S. Wilson, M. Smith, B. L. Gomez, M. J. Carr, P. T. Kimmitt, S. Seaton, K. Rajakumar, T. Holyoake, C. C. Kibbler, E. Johnson, R. P. Hobson, B. Jones, and R. A. Barnes. 2006. A consensus on fungal polymerase chain reaction diagnosis?: a United Kingdom-Ireland evaluation of polymerase chain reaction methods for detection of systemic fungal infections. J. Mol. Diagn. 8:376-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White, P. L., C. J. Linton, M. D. Perry, E. M. Johnson, and R. A. Barnes. 2006. The evolution and evaluation of a whole blood polymerase chain reaction assay for the detection of invasive aspergillosis in hematology patients in a routine clinical setting. Clin. Infect. Dis. 42:479-486. [DOI] [PubMed] [Google Scholar]
- 32.White, P. L., and R. A. Barnes. 2006. Aspergillus PCR—platforms, strengths and weaknesses. Med. Mycol. 44(Suppl.):191-198. [DOI] [PubMed] [Google Scholar]
- 33.Williamson, E. C. M. 2001. Molecular approaches to fungal infections in immunocompromised patients. MD thesis. University of Bristol, Bristol, United Kingdom.