Abstract
The capsulation (cap) locus of Haemophilus influenzae type e (Hie) was characterized and sequenced. No IS1016 element was found to flank the locus. The 18.2-kb locus included 14 open reading frames (ORFs), which were grouped into three functional regions. Eight new ORFs (named ecs1 to ecs8) were identified in the Hie capsule-specific region II.
In the post-Haemophilus influenzae serotype b (Hib) vaccine era, concern about the potential emergence of non-vaccine-preventable strains has arisen (1, 17, 20, 23, 26). In encapsulated H. influenzae strains, the genes for the production of the polysaccharide capsules are organized in a capsulation (cap) locus, which consists of three different functional regions (11, 13). Regions I and III are common to all capsular types and contain genes necessary for transport and process of the capsular material, while region II contains serotype-specific biosynthesis genes (7, 10, 18, 19, 25).
Invasive disease caused by H. influenzae serotype e (Hie) strains has recently been observed in Italy, suggesting the importance of further molecular investigations on Hie cap locus (4, 5). It is recognized that the Hie capsule is a copolymer of the repeat unit of an N-acetylglucosamine and N-acetylmannosamine uronic acid (22, 24), but the genes involved in the polysaccharide biosynthesis have neither been identified nor characterized.
In the present study, we characterized the Hie cap locus for the first time. Eleven invasive Hie strains isolated in Italy during the period of January 2000 to December 2008 were analyzed. The strains were identified as type e by PCR capsular genotyping (6).
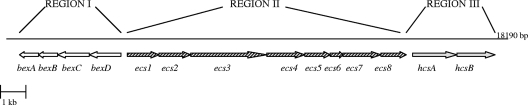
Location of the Hie cap locus within the chromosome.
PCR amplification of the 5′ and 3′ end junctions of the Hie cap locus was performed by using primer sets “capfSodC/bexBrev” and “hcsBfrw/HI1637,” respectively (Table 1). The resulting PCR products were sequenced and analyzed. All 11 Hie strains were found to have the cap locus in the identical chromosomal location as that of H. influenzae serotype f (Hif), associated with the same flanking genes (sodC at the 5′ end and HI1637 at the 3′ end), confirming previous investigations (19). Sequence analysis of the two end junctions also revealed that they contained no sequences reminiscent of the insertion element IS1016. It is well known that this element provides the molecular substrate for amplifications of the cap gene sequences (11). Most Hib strains, in which the cap locus lies between direct repeats of IS1016, possess a duplication of the capsule genes (11, 12). The finding that our Hie strains' lack of IS1016 sequences flanking the cap locus is remarkable, since reasonably the locus cannot be amplified.
TABLE 1.
PCR primers and products used for sequencing of the Hie capsulation locus
| Primer set | Nucleotide sequence (5′ to 3′) | Region amplified | Size (bp) | Source or reference |
|---|---|---|---|---|
| capfSodC | CATGCGCATTTTCCACGCCAGC | sodC-bexB | 1,581 | 19 |
| bexBrev | TAGCGATTCAAGGGAGGGT | sodC-bexB | 1,581 | This study |
| bexBfrw | ACGCCCATAACGAGAGACT | bexB-bexD | 2,004 | This study |
| bexDrev | TCGCAGGTAAGACACCAGAG | bexB-bexD | 2,004 | This study |
| bexDfrw | AAAGACACCTCGTGGGTCA | bexD-regionIII | 5,003 | This study |
| e2 | GCTTTACTGTATAAGTCTAG | bexD-regionIII | 5,003 | 6 |
| el | GGTAACGAATGTAGTGGTAG | regionIII-hcsA | 9,045 | 6 |
| hcsArev | ACTGACCGCACTTTACGACG | regionIII-hcsA | 9,045 | This study |
| hcsAfrw | GCACAAAGTGAGCGTCGTA | hcsA-hcsB | 1,703 | This study |
| hcsBrev | ATAGAAGTCTGCCTGGCGAG | hcsA-hcsB | 1,703 | This study |
| hcsBfrw | GATTGCTTATCGTGGCTCAGT | hcsB-HI1637 | 1,504 | This study |
| HI1637 | AAATTTCCATTATGGGAAACG | hcsB-HI1637 | 1,504 | 19 |
Sequencing of the Hie cap locus.
The complete cap locus from the strain Hie 274 (isolated from the cerebrospinal fluid of a patient with meningitis) was sequenced. To cover the entire Hie cap locus, overlapping amplicons ranging from 1,500 bp to 9 kb were obtained by PCR analysis using several primer pairs based on the published sequences of the Hib and Hif cap loci (GenBank accession numbers AF549213 and AF549211, respectively) (Table 1). Each amplicon was then subcloned into pCR4-TOPO (TOPO TA cloning kit or TOPO XL PCR cloning kit; Invitrogen, Milan, Italy). Both strands of the insert from each plasmid were sequenced by the primer walking service at Eurofins MWG Operon (Ebersberg, Germany). The nucleotide sequences were assembled and analyzed using DNAMAN sequence analysis software (version 5.2; Lynnon Corp., Quebec, Canada). Nucleotide and deduced amino acid sequences were compared to other known sequences databases by using the National Center for Biotechnology Information BLAST programs. The entire Hie cap locus was shown to be 18.2 kb in length. It contained 14 open reading frames (ORFs) which, by analogy with other capsule loci, were grouped into three distinct regions (I, II, and III) (Fig. 1). A comparison of the deduced proteins from the Hie cap locus genes with the corresponding gene products from Hib and Hif as well as with proteins from other bacterial species is shown in Table 2.
FIG. 1.
Genetic organization of the Hie capsulation locus of strain 274. The arrows indicate genes. Region I contains four genes called bexDCBA, homologous to those found in Hib and Hif (white arrows). Region II includes eight serotype-specific genes designated ecs1 to ecs8 (right-hatched arrows). Region III comprises two genes named hcsA and hcsB, homologous to those found in Hib and Hif (gray arrows).
TABLE 2.
Comparison of the deduced proteins from the H. influenzae serotype e capsulation locus of strain 274
| Hie 274 deduced protein, no. of amino acids (aa) | Similar protein (source organism) | Accession no. | % Identity | % Similarity |
|---|---|---|---|---|
| Region I | ||||
| BexA, 217 aa | BexA (H. influenzae serotype f) | AF549211 | 94 | 97 |
| BexA (H. influenzae serotype b) | AF549213 | 94 | 97 | |
| CpxA (Actinobacillus pleuropneumoniae) | CP000687 | 82 | 91 | |
| CtrD (Neisseria meningitidis) | EU038216 | 80 | 89 | |
| CpxA (Mannheimia haemolytica) | AF170495 | 76 | 88 | |
| BexB, 265 aa | BexB (H. influenzae serotype f) | AF549211 | 97 | 99 |
| BexB (H. influenzae serotype b) | M33788 | 95 | 99 | |
| CpxB (Actinobacillus pleuropneumoniae) | CP000687 | 77 | 90 | |
| CpxB (Mannheimia haemolytica) | AF170495 | 74 | 86 | |
| CtrC (Neisseria meningitidis) | EU038216 | 69 | 83 | |
| BexC, 377 aa | BexC (H. influenzae serotype f) | AF549211 | 98 | 98 |
| BexC (H. influenzae serotype b) | AF549213 | 94 | 96 | |
| CpxC (Mannheimia haemolytica) | AF170495 | 76 | 88 | |
| CpxC (Actinobacillus pleuropneumoniae) | CP000687 | 74 | 87 | |
| CtrB (Neisseria meningitidis) | EU038216 | 59 | 79 | |
| BexD, 428 aa | BexD (H. influenzae serotype f) | AF549211 | 91 | 94 |
| BexD (H. influenzae serotype b) | AF549213 | 91 | 95 | |
| CpxD (Actinobacillus pleuropneumoniae) | CP001091 | 73 | 85 | |
| CpxD (Mannheimia haemolytica) | AF170495 | 71 | 84 | |
| CtrA (Neisseria meningitidis) | AF520902 | 55 | 74 | |
| Region II | ||||
| Orf1 (Ecs1), 374 aa | SacA (Neisseria meningitidis) | AL157959 | 71 | 85 |
| Putative UDP-N-acetyl-d-glucosamine 2-epimerase | VIBHAR_00689(Vibrio harveyi) | CP000789 | 63 | 77 |
| VV0341 (Vibrio vulnificus) | BA000037 | 62 | 77 | |
| WecB (Klebsiella pneumoniae) | CP000647 | 61 | 75 | |
| WecB (Escherichia coli) | AE014075 | 61 | 76 | |
| Orf2 (Ecs2), 421 aa | WecB (Mannheimia haemolytica) | AF170495 | 67 | 79 |
| Putative UDP-N-acetyl-d-mannosaminuronic acid dehydrogenase | EcbB (Pasteurella multocida) | AF302466 | 64 | 77 |
| ORF6 (Pseudomonas aeruginosa) | AF498407 | 63 | 77 | |
| WecC (Escherichia coli) | CP000948 | 61 | 75 | |
| SeD_A4308 (Salmonella enterica) | CP001144 | 61 | 74 | |
| Orf3 (Ecs3), 991 aa | BcbC (Pasteurella multocida) | AF169324 | 50 | 69 |
| Putative glycosyltransferase | Msm_1297 (Methanobrevibacter smithii) | CP000678 | 34 | 55 |
| Msp_0219 (Methanosphaera stadtmanae) | CP000102 | 33 | 51 | |
| Eco1C (Escherichia coli) | CP000946 | 31 | 50 | |
| WaaV (Shigella sonnei) | CP000038 | 30 | 50 | |
| Orf4 (Ecs4), 486 aa | BcbD (Pasteurella multocida) | AF169324 | 46 | 65 |
| Unknown function | KfoD (Escherichia coli) | AB079602 | 36 | 55 |
| ORFA (Yersinia enterocolitica) | AY653208 | 21 | 43 | |
| EcbD (Pasteurella multocida) | AF302466 | 26 | 42 | |
| CMU_015760 (Cryptosporidium muris) | XM_002140142 | 26 | 41 | |
| Orf5 (Ecs5), 240 aa | ORF5 (Actinobacillus suis) | AY253301 | 68 | 85 |
| Unknown function | BcbE (Pasteurella multocida) | AF169324 | 65 | 82 |
| BcbE (Photobacterium damselae) | AB074293 | 49 | 67 | |
| SamA (Shewanella amazonensis) | CP000507 | 46 | 64 | |
| VC0395 (Vibrio cholera) | CP000627 | 45 | 63 | |
| Orf6 (Ecs6), 125 aa | ORF6 (Actinobacillus suis) | AY253301 | 77 | 91 |
| Unknown function | BcbF (Pasteurella multocida) | AF169324 | 73 | 89 |
| SamA (Shewanella amazonensis) | CP000507 | 60 | 79 | |
| VC0395 (Vibrio cholerae) | CP000627 | 60 | 78 | |
| BAG50482 (Vibrio parahaemolyticus) | AB353134 | 64 | 78 | |
| Orf7 (Ecs7), 519 aa | BcbG (Pasteurella multocida) | AF302466 | 65 | 77 |
| Unknown function | ORF7 (Actinobacillus suis) | AY253301 | 63 | 78 |
| BcbG (Photobacterium damselae) | AB074293 | 37 | 56 | |
| VC0395 (Vibrio cholerae) | CP000627 | 36 | 55 | |
| BcbG (Zymomonas mobilis) | AE008692 | 36 | 54 | |
| Orf8 (Ecs8), 325 aa | BcbI (Pasteurella multocida) | AF169324 | 64 | 78 |
| Unknown function | ORF2 (Mannheimia haemolytica) | AF170495 | 63 | 74 |
| Fphi_1179 (Francisella philomiragia) | CP000937 | 54 | 73 | |
| Neut_1976 (Nitrosomonas eutropha) | CP000450 | 40 | 63 | |
| NE1334 (Nitrosomonas europaea) | AL954747 | 43 | 63 | |
| Region III | ||||
| HcsA, 595 aa | HcsA (H. influenzae serotype b) | DQ368335 | 96 | 97 |
| HcsA (H. influenzae serotype f) | AF549211 | 95 | 97 | |
| LipA2 (Actinobacillus pleuropneumoniae) | CP000687 | 62 | 75 | |
| PhyA (Mannheimia haemolytica) | AF170495 | 59 | 72 | |
| LipA (Neisseria meningitidis) | AM421808 | 56 | 71 | |
| HcsB, 420 aa | HcsB (H. influenzae serotype b) | DQ368335 | 91 | 95 |
| HcsB (H. influenzae serotype f) | AF549211 | 90 | 94 | |
| PhyB (Actinobacillus pleuropneumoniae) | CP000687 | 65 | 78 | |
| PhyB (Pasteurella multocida) | AF067175 | 64 | 77 | |
| LipB (Neisseria meningitidis) | Z13995 | 55 | 68 |
Region I.
Overall, region I exhibited 90% and 96% sequence identity to the previously described region I from Hib and Hif, respectively (18, 19). Region I included four ORFs, which were named bexA, bexB, bexC, and bexD. Although the putative proteins of genes bexABCD were nearly identical (from 91 to 98% identity) to the region I corresponding gene products from both Hib and Hif (Table 2), some polymorphism at nucleotide sequence level was observed. The bexA gene from the Hie cap locus exhibited 95% identity to bexA from Hif but only 84% identity to bexA from Hib, in agreement with a previous study demonstrating bexA nucleotide sequence diversity among different H. influenzae serotypes (27).
Region III.
Overall, region III showed 91% and 93% sequence identity to the previously described region III from Hib and Hif, respectively (18, 19). Region III contained two ORFs, which were named hcsA and hcsB. Their deduced amino acid sequences exhibited high identity (from 90 to 96% identity) with the corresponding products from both Hib and Hif region III (Table 2). Recently, both HcsA and HcsB proteins have been demonstrated to be crucial for transport of capsular polysaccharide from the periplasm to the bacterial surface across the outer membrane (21).
Region II.
Overall, region II showed no sequence identity to the previously described specific capsular regions from other H. influenzae serotypes (7, 18, 19). On the contrary, high overall sequence identity (67%) was found with the capsule biosynthetic-specific region II from Pasteurella multocida B:2 (accession number AF169324), indicating that the genetic organization of the whole region is similar (2, 3). The G+C content of the DNA in the Hie cap locus region II is 31.3%, significantly different from that of both regions I and III (38% and 39.4%, respectively) and from the overall background for the H. influenzae species (38%), suggesting that region II might be more recently acquired. However, since the G+C content of DNA of P. multocida cap locus region II is 35%, this microorganism was probably not the direct source of the region II for Hie. Although we cannot rule out a common evolutionary origin of the two polysaccharide biosynthetic regions followed by a partial diversification of their DNA content, no data are available to support this hypothesis. Region II contained 8 ORFs, which were named ecs1 to ecs8 (for serotype e capsule-specific genes) (Table 2). The deduced products of ecs1 and ecs2 had homology with putative UDP-N-acetyl-d-glucosamine 2-epimerase and UDP-N-acetyl-d-mannosaminuronic acid dehydrogenase enzymes, respectively, which catalyze the two-step conversion of UDP-N-acetyl-d-glucosamine to N-acetyl-d-mannosaminuronic acid, as previously demonstrated with Escherichia coli (14). The encoded protein by the ecs3 gene showed similarity to glycosyltransferases (Table 2), which are involved in polymerization of the sugar monomers in several bacterial species (8, 9). Considering that the structure of the Hie capsular polymer is composed of repeating units of N-acetylglucosamine and N-acetylmannosamine uronic acid (22, 24), it is likely that the products of the ecs1, ecs2, and ecs3 genes play an essential role in the biosynthesis of serotype e polysaccharide. No specific putative functions were assigned to the remaining 5 ORFs (ecs4 to ecs8), although similarity with other deduced products in the database was detected, including the predicted products of the genes bcbDEFGI from cap locus region II from P. multocida (2), (Table 2). Further studies of functional activities of the Hie cap locus region II genes are required.
Although Hie strains belong to the phylogenetic division I of the encapsulated H. influenzae strains (15), the Hie cap locus shares two remarkable features of the division II cap loci: chromosomal location and lack of association with the IS1016 insertion element, confirming the previously described genetic distance of Hie from all other division I H. influenzae strains (16). The availability of the Hie cap locus sequences may be regarded as a powerful tool to be used in further investigations on molecular detection and characterization of the Hie isolates.
Nucleotide sequence accession number.
The nucleotide sequence for the Hie cap locus from this study has been deposited in the EMBL nucleotide sequence database under the accession number FM882247.
Acknowledgments
This work was partially supported by Ministry of Health-CCM project 116 “Surveillance of Invasive Bacterial Diseases.”
We are very grateful to Tonino Sofia for editorial assistance.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Adderson, E. E., C. L. Byington, L. Spencer, A. Kimball, M. Hindiyeh, K. Carroll, S. Mottice, S E. K. Korgenski, J. C. Christenson, A. T. Pavia, and L. Spencer. 2001. Invasive serotype a Haemophilus influenzae infections with a virulence genotype resembling Haemophilus influenzae type b: emerging pathogen in the vaccine era? Pediatrics 108:E18. [DOI] [PubMed] [Google Scholar]
- 2.Boyce, J. D., J. Y. Chung, and B. Adler. 2000. Genetic organisation of the capsule biosynthetic locus of Pasteurella multocida M1404 (B:2). Vet. Microbiol. 72:121-134. [DOI] [PubMed] [Google Scholar]
- 3.Boyce, J. D., J. Y. Chung, and B. Adler. 2000. Pasteurella multocida capsule: composition, function and genetics. J. Biotechnol. 83:153-160. [DOI] [PubMed] [Google Scholar]
- 4.Cerquetti, M., M. L. Ciofi degli Atti, R. Cardines, S. Salmaso, G. Renna, P. Mastrantonio, and the Hi Study Group. 2003. Invasive type e Haemophilus influenzae disease in Italy. Emerg. Infect. Dis. 9:258-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerquetti, M., M. L. Ciofi degli Atti, R. Cardines, M. Giufré, A. Romano, and P. Mastrantonio. 2004. Haemophilus influenzae serotype e meningitis in an infant. Clin. Infect. Dis. 38:1041. [DOI] [PubMed] [Google Scholar]
- 6.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Follens, A., M. Veiga-Da-Cunha, R. Merckx, E. Van Schaftingen, and J. Van Eldere. 1999. acs1 of Haemophilus influenzae type a capsulation locus region II encodes a bifunctional ribulose 5-phosphate reductase-CDP-ribitol pyrophosphorylase. J. Bacteriol. 181:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 9.Kolkman, M. A., W. Wakarchuk, P. J. Nuijten, and B. A. van der Zeijst. 1997. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol. Microbiol. 26:197-208. [DOI] [PubMed] [Google Scholar]
- 10.Kroll, J. S., B. Loynds, L. N. Brophy, and E. R. Moxon. 1990. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol. Microbiol. 4:1853-1862. [DOI] [PubMed] [Google Scholar]
- 11.Kroll, J. S., B. M. Loynds, and E. R. Moxon. 1991. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol. Microbiol. 5:1549-1560. [DOI] [PubMed] [Google Scholar]
- 12.Kroll, J. S., and E. R. Moxon. 1988. Capsulation and gene copy number at the cap locus of Haemophilus influenzae type b. J. Bacteriol. 170:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroll, J. S., S. Zamze, B. Loynds, and E. R. Moxon. 1989. Common organization of chromosomal loci for production of different capsular polysaccharides in Haemophilus influenzae. J. Bacteriol. 171:3343-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier-Dieter, U., R. Starman, K. Barr, H. Mayer, and P. D. Rick. 1990. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J. Biol. Chem. 265:13490-13497. [PubMed] [Google Scholar]
- 15.Musser, J. M., J. S. Kroll, E. R. Moxon, and R. K. Selander. 1988. Clonal population structure of encapsulated Haemophilus influenzae. Infect. Immun. 56:1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, and G. Hammond. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro, G. S., J. N. Reis, S. M. Cordeiro, J. B. T. Lima, E. L. Gouveia, M. Peterson, K. Salgado, H. R. Silva, R. Cobo Zanella, S. C. Grassi Almeida, M. C. Brandileone, M. G. Reis, and A. I. Ko. 2003. Prevention of Haemophilus influenzae type b (Hib) meningitis and emergence of serotype replacement with type a strains after introduction of Hib immunization in Brazil. J. Infect. Dis. 187:109-116. [DOI] [PubMed] [Google Scholar]
- 18.Satola, S. W., P. L. Schirmer, and M. M. Farley. 2003. Complete sequence of the cap locus of Haemophilus influenzae serotype b and nonencapsulated b capsule-negative variants. Infect. Immun. 71:3639-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satola, S. W., P. L. Schirmer, and M. M. Farley. 2003. Genetic analysis of the capsule locus of Haemophilus influenzae serotype f. Infect. Immun. 71:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slack, M. P., H. J. Azzopardi, R. M. Hargreaves, and M. E. Ramsay. 1998. Enhanced surveillance of invasive Haemophilus influenzae disease in England, 1990 to 1996: impact of conjugate vaccines. Pediatr. Infect. Dis. J. 17:S204-S207. [DOI] [PubMed] [Google Scholar]
- 21.Sukupolvi-Petty, S., S. Grass, and J. W. St Geme III. 2006. The Haemophilus influenzae type b hcsA and hcsB gene products facilitate transport of capsular polysaccharide across the outer membrane and are essential for virulence. J. Bacteriol. 188:3870-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutton, A., R. Schneerson, S. Kendall-Morris, and J. B. Robbins. 1982. Differential complement resistance mediates virulence of Haemophilus influenzae type b. Infect. Immun. 35:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang, R. 2008. Changing epidemiology of invasive Haemophilus influenzae disease. Lancet Infect. Dis. 8:737. [DOI] [PubMed] [Google Scholar]
- 24.Tsui, F. P., R. Schneerson, and W. Egan. 1981. Structural studies of the Haemophilus influenzae type e capsular polysaccharide. Carbohydr. Res. 88:85-92. [DOI] [PubMed] [Google Scholar]
- 25.Van Eldere, J., L. Brophy, B. Loynds, P. Celis, I. Hancock, S. Carman, J. S. Kroll, and E. R. Moxon. 1995. Region II of the Haemophilus influenzae type b capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol. Microbiol. 15:107-118. [DOI] [PubMed] [Google Scholar]
- 26.Waggoner-Fountain, L. A., J. O. Hendley, E. J. Cody, V. A. Perriello, and L. G. Donowitz. 1995. The emergence of Haemophilus influenzae types e and f as significant pathogens. Clin. Infect. Dis. 21:1322-1324. [DOI] [PubMed] [Google Scholar]
- 27.Zhou, J., D. K. Law, M. L. Sill, and R. S. Tsang. 2007. Nucleotide sequence diversity of the bexA gene in serotypeable Haemophilus influenzae strains recovered from invasive disease patients in Canada. J. Clin. Microbiol. 45:1996-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]