Abstract
Six hundred ninety nonduplicate isolates of Acinetobacter species were identified using a combination of detection of blaOXA-51-like and rpoB sequence cluster analysis. Although most isolates were identified as A. baumannii (78%), significant numbers of other species, particularly A. lwoffii/genomic species 9 (8.8%), A. ursingii (4%), genomic species 3 (1.7%), and A. johnsonii (1.7%), were received, often associated with bacteremias.
The Acinetobacter genus consists of more than 30 species, of which A. baumannii, and to a lesser extent genomic species 3 and 13TU, are most associated with the clinical environment and nosocomial infections. Identification within the genus is difficult and requires molecular methods, and these organisms are rarely identified to the species level using appropriate methods (3, 4, 6, 24). While A. baumannii can relatively readily be identified by detection of blaOXA-51-like, the intrinsic carbapenemase gene in this species (22), the use of rpoB sequencing has facilitated identification across the genus (5, 8), and it is becoming clear that other species, such as A. ursingii (which has also been called A. septicus [13]) and A. haemolyticus, are also important nosocomial pathogens in some cases (3, 4, 5, 6, 9, 24). rpoB sequencing has advantages over such techniques as amplified ribosomal DNA restriction analysis (ARDRA), based on 16S rRNA gene sequences, and those based on the 16S-23S intergenic spacer region, since there is a relatively high degree of polymorphism in this gene among the Acinetobacter species, and sequences are available for all the currently described species (8), including those more recently described (11-14).
Our laboratory provides a typing and identification service for hospitals in the United Kingdom and Republic of Ireland for this organism, and here we describe the species found among 690 isolates of Acinetobacter, each from a different patient, submitted over a 20-month period during 2008/2009, from some 135 hospitals. While A. baumannii, which is frequently associated with outbreaks, is still by far the most common of the Acinetobacter species among clinical isolates, it is clear that lesser-known species, such as A. lwoffii, A. ursingii, and A. parvus, are regularly encountered, have been associated with serious infections, and may represent emerging pathogens.
The majority of isolates were received as Acinetobacter species for typing, identification, and/or susceptibility determinations and were subjected to a multiplex PCR for detection of blaOXA-58-like, blaOXA-23-like, blaOXA-51-like, blaOXA-40-like, and class 1 integrase genes, as described by Turton et al. (22), with the addition of primers for blaOXA-58-like (27). Detection of blaOXA-51-like was regarded as a positive identification of A. baumannii; the identity of a proportion of such isolates was also checked by rpoB sequence cluster analysis; in addition, many were shown by pulsed-field gel electrophoresis (PFGE) to be further representatives of strains previously identified as A. baumannii, and all gave amplicons in a PCR to amplify variable-number tandem repeat loci found in A. baumannii (23). The remaining isolates were identified by rpoB sequence cluster analysis using primers described by La Scola et al. (8). Briefly, a 903-bp portion of the rpoB gene covering two variable regions was amplified using the primers Ac696F and Ac1598R. Amplicons were treated with Exo-SAP-IT (USB Corporation, Cleveland, OH) according to the manufacturer's instructions, and four sequencing reactions were carried out, using the primers Ac696F, Ac1055F, Ac1093R, and Ac1598R, respectively. The resulting fragments were separated on a Beckman-Coulter CEQ8000 genetic analysis system or an Applied Biosystems 3730 DNA analyzer and aligned, and sequences of a 765-bp fragment corresponding to nucleotides (nt) 2964 to 3728 of the coding sequence were compared using the BioNumerics software program; a phylogenetic tree was constructed using the MEGA software program (http://www.megasoftware.net/mega41.html) (19) (Fig. 1). Sequences of reference isolates were also included, and isolates were identified both by BLAST searches and according to which species they clustered most closely with; as more isolates were added to the database, a measure of the extent of sequence diversity associated with each species was obtained, allowing determination of whether isolates clustered closely enough to be assigned to that species. Susceptibilities to at least 16 antibiotics were determined by agar dilution and interpreted using British Society of Antimicrobial Chemotherapy (BSAC) breakpoints (http://www.bsac.org.uk). Pulsed-field gel electrophoresis (PFGE) of ApaI-digested genomic DNA was carried out as described previously (21).
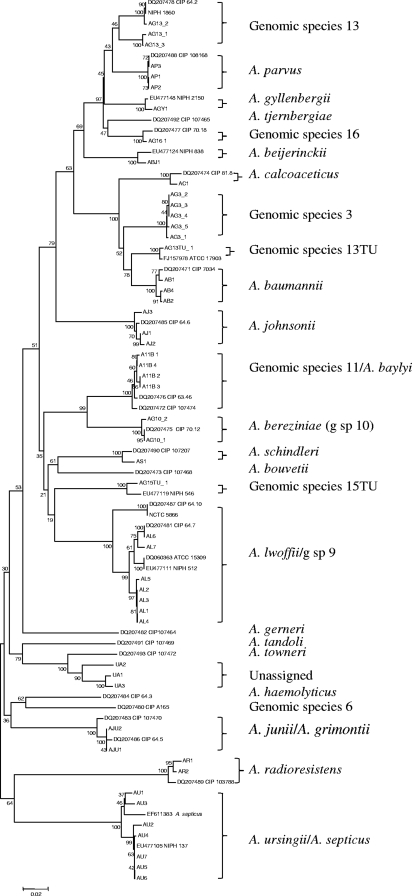
FIG. 1.
Phylogenetic tree of sequences corresponding to nt 2964 to 3728 of the rpoB coding sequence of isolates of Acinetobacter species. Clinical and reference isolates were included, with GenBank accession numbers being included with the latter. Phylogenetic analyses were conducted in MEGA4 (19) using the neighbor-joining method. One thousand replicates were used for bootstrap analysis.
Using this method, we were able to identify most isolates to the species level, although isolates of A. lwoffii and genomic species 9 clustered too closely to be distinguished from one another, as did those of A. baylyi and genomic species 11 (A. guillouiae). Similarly, as has been observed by others (26), A. grimontii and A. junii could not be distinguished and are likely to be a single species. Three isolates (UA1 to -3), two of which had highly similar rpoB sequences, did not cluster closely enough with any of the described species and may represent new species. For all three, the closest currently described species is A. towneri. Isolates that were identified as A. radioresistens were PCR positive for blaOXA-23-like, the naturally occurring carbapenemase gene in this species (17), consistent with the identification. Detection of OXA carbapenemase genes among species other than A. baumannii was rare, with the only other examples being two isolates, one of genomic species 3 and the other of genomic species 16, with blaOXA-58-like.
As expected, the majority (78.0%) of isolates were identified as A. baumannii; with A. lwoffii/genomic species 9 (8.8%), A. ursingii (4.0%), genomic species 3 (1.7%), A. johnsonii (1.7%), and A. parvus (1.3%) accounting for most of the rest (Table 1). In most cases, these non-A. baumannii isolates were from blood and were associated with bacteremia or septicemia. Of note is that some isolates (of A. johnsonii, genomic species 13, and A. beijerinckii) were implicated in endocarditis; this has been described for other Acinetobacter species (7, 18, 28) but not these and provides further evidence that these organisms can cause life-threatening infections. The relatively high incidence of A. ursingii, which exceeded those of both genomic species 3 and 13TU, was unexpected but agrees with observations from hospitals in the Netherlands and Northern Ireland (2, 24). It has previously been documented that this organism has the capacity to cause bloodstream infections in hospitalized patients (4, 9, 11), and it has been associated with a nosocomial outbreak of bloodstream infections in a neonatal intensive care unit, in which two babies died (6). The isolates in the present study were from 28 patients in 24 centers, suggesting that they were not epidemiologically related; nevertheless, three isolates, each from different centers, formed a cluster by PFGE (see Fig. S1 in the supplemental material). Similarly, A. lwoffii has previously been linked with catheter-related bloodstream infections (20), as has A. parvus (12).
TABLE 1.
Submissions of Acinetobacter sp. other than A. baumannii received during the study period and associated clinical informationa
| Species | No. of patients (% of total) | No. of hospitals | Source |
Clinical information (combined) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Blood | Sputum | CSFb | Eye | Wound swab | Urine | Other/not given | ||||
| A. lwoffii/genomic sp. 9 | 61 (8.8) | 48 | 41 | 1 | 4 | 2 | 4 | 1 | 8 | Sepsis, bacteremia, pyrexia, meningitis, post hemorrhagic hydrocephalus, rigors, pneumonia, cellulitis, rash, ophthalmia neonatum, UTI,c abscess |
| A. ursingii | 28 (4.0) | 24 | 17 | 1 | 1 | 2 | 2 | 5 | Septicemia, line infection, pyrexia, pneumonia, chest infection, sticky eye, pancreatitis | |
| Genomic species 3 | 12 (1.7) | 9 | 3 | 5 | 1 | 4 | Septic shock, bacteremia, pyrexia | |||
| A. johnsonii | 11 (1.6) | 10 | 6 | 3 | 2 | Septicemia, endocarditis, abscess, UTIc | ||||
| A. parvus | 9 (1.3) | 9 | 9 | Bacteremia, line infection, pyrexia | ||||||
| Genomic species 13 | 6 (0.9) | 5 | 4 | 1 | 1 | Septic shock, endocarditis, pyrexia | ||||
| A. radioresistens | 4 (0.6) | 4 | 2 | 2 | No information provided | |||||
| A. baylyi/genomic sp. 11 | 3 (0.4) | 3 | 2 | 1 | Bacteremia, corneal ulcer | |||||
| A. calcoaceticus | 3 (0.4) | 3 | 1 | 2 | Infected burn, ear discharge | |||||
| Unassigned | 3 (0.4) | 3 | 2 | 1 | Bacteremia, pyrexia | |||||
| Genomic species 13TU | 2 (0.3) | 2 | 1 | 1 | Pyrexia, pneumonia | |||||
| A. haemolyticus | 2 (0.3) | 2 | 1 | 1 | Meningitis following neurosurgery | |||||
| A. junii | 2 (0.3) | 2 | 2 | Sepsis | ||||||
| A. beijerinckii | 1 (0.1) | 1 | 1 | Endocarditisd | ||||||
| A. bereziniae | 1 (0.1) | 1 | 1 | Chronic obstructive pulmonary disease | ||||||
| A. gyllenbergii | 1 (0.1) | 1 | 1 | No information provided | ||||||
| A. schindleri | 1 (0.1) | 1 | 1 | No information provided | ||||||
| Genomic species 15TU | 1 (0.1) | 1 | 1 | Pneumonia | ||||||
| Genomic species 16 | 1 (0.1) | 1 | 1 | No information provided | ||||||
Percentages given are of all 690 submissions of Acinetobacter sp., of which 538 (78%) were A. baumannii. Only one isolate per patient was included, with the exception of a single case in which isolates of two different species were received from one patient.
Cerebrospinal fluid.
UTI, urinary tract infection.
Patient also had A. johnsonii.
Antibiotic susceptibility investigation of isolates of A. ursingii (n = 14), A. lwoffii/genomic species 9 (n = 5), A. johnsonii (n = 2), genomic species 3 (n = 3), 13 (n = 5), and 13TU (n = 1), A. calcoaceticus (n = 2), A. schindleri (n = 1), A. haemolyticus (n = 1), A. bereziniae (n = 1), A. gyllenbergii (n = 1), and A. beijerinckii (n = 1), largely requested by the sending laboratories, revealed that it was those of A. lwoffii/genomic species 9 and A. schindleri that were susceptible to the greatest number of antibiotics, with isolates of genomic species 3, 13, and 13TU, A. bereziniae, and A. gyllenbergii exhibiting resistance (full or intermediate) to 8 or more of the 17 antibiotics tested (see Table S1 in the supplemental material). An isolate of genomic species 3 (genomic species 3_3) displayed resistance to 13 antibiotics, including the carbapenems; it was PCR positive for blaOXA-58-like, explaining the latter resistance. However, most of the non-A. baumannii isolates tested were susceptible to amikacin, gentamicin, the carbapenems, sulbactam, ciprofloxacin, and minocycline, in stark contrast to the situation with A. baumannii. Isolates of A. ursingii were resistant to up to eight of the antibiotics tested. There were some notable differences between the species; all isolates tested of genomic species 13 and that of A. beijerinckii were resistant to colistin, one of the last antibiotics useful for treating A. baumannii infections; isolates of the other species tested were susceptible. A reference strain of genomic species 13 (ATCC 17905) was also found to be resistant to colistin, suggesting this resistance is intrinsic in this species, in agreement with recent observations by others (15).
Three patients carried a single strain of genomic species 3, and a further strain was shared by two patients (see Fig. S1 in the supplemental material), suggesting that transmission between patients may have occurred.
As identification of species of Acinetobacter other than A. baumannii becomes more widely undertaken, their prevalence and the nature of infections associated with them will become clearer. Evidence from isolates submitted to our laboratory suggests that some of these species are associated with clinical infections in significant numbers of cases. Some isolates exhibited multiresistance, which may impact on therapy; carbapenem resistance in isolates of genomic species 3 (1) and colistin resistance in isolates of genomic species 13TU (16) have previously been described. In addition, some species, particularly genomic species 3, 13TU, and A. ursingii, have been associated with outbreaks (6, 10, 24, 25), suggesting that they may become increasingly important.
Nucleotide sequence accession number.
The partial rpoB sequence of isolate UA1, which could not be assigned to a currently described species, is deposited in GenBank under the accession number GU245962.
Supplementary Material
Acknowledgments
We are grateful to colleagues in hospital laboratories for sending these isolates and to the Genomic Services Unit at Centre for Infections, Colindale, for sequencing services.
Footnotes
Published ahead of print on 24 February 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Boo, T. W., F. Walsh, and B. Crowley. 2009. Molecular characterization of carbapenem-resistant Acinetobacter species in an Irish university hospital: predominance of Acinetobacter genomic species 3. J. Med. Microbiol. 58:209-216. [DOI] [PubMed] [Google Scholar]
- 2.Boo, T. W., and B. Crowley. 2009. Detection of blaOXA-58 and blaOXA-23-like genes in carbapenem-susceptible Acinetobacter clinical isolates: should we be concerned? J. Med. Microbiol. 58:839-841. [DOI] [PubMed] [Google Scholar]
- 3.Chen, T. L., C. L. Chuang, L. K. Siu, C. P. Fung, and W. L. Cho. 2007. Genomic species identification is important to delineate the pathological characteristics of Acinetobacter in tunnelled, cuffed haemodialysis catheter-related bacteraemia. Nephrol. Dial. Transplant. 22:936-938. [DOI] [PubMed] [Google Scholar]
- 4.Dortet, L., P. Legrand, C. J. Soussy, and V. Cattoir. 2006. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J. Clin. Microbiol. 44:4471-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gundi, V. A., L. Dijkshoorn, S. Burignat, D. Raoult, and B. La Scola. 2009. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 155:2333-2341. [DOI] [PubMed] [Google Scholar]
- 6.Kilic, A., H. Li, A. Mellmann, A. C. Basustaoglu, M. Kul, Z. Senses, H. Aydogan, C. W. Stratton, D. Harmsen, and Y. W. Tang. 2008. Acinetobacter septicus sp. nov. association with a nosocomial outbreak of bacteremia in a neonatal intensive care unit. J. Clin. Microbiol. 46:902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar, S. S., L. Vengadassalapathy, and T. Menon. 2008. Prosthetic valve endocarditis caused by Acinetobacter baumannii complex. Indian J. Pathol. Microbiol. 51:573. [DOI] [PubMed] [Google Scholar]
- 8.La Scola, B., V. A. Gundi, A. Khamis, and D. Raoult. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loubinoux, J., L. Mihaila-Amrouche, A. Le Fleche, E. Pigne, G. Huchon, P. A. Grimont, and A. Bouvet. 2003. Bacteremia caused by Acinetobacter ursingii. J. Clin. Microbiol. 41:1337-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald, A., S. G. Amyes, and R. Paton. 1999. The persistence and clonal spread of a single strain of Acinetobacter 13TU in a large Scottish teaching hospital. J. Chemother. 11:338-344. [DOI] [PubMed] [Google Scholar]
- 11.Nemec, A., T. De Baere, I. Tjernberg, M. Vaneechoutte, T. J. van der Reijden, and L. Dijkshoorn. 2001. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 51:1891-1899. [DOI] [PubMed] [Google Scholar]
- 12.Nemec, A., L. Dijkshoorn, I. Cleenwerck, T. De Baere, D. Janssens, T. J. van der Reijden, P. Jezek, and M. Vaneechoutte. 2003. Acinetobacter parvus sp. nov., a small-colony-forming species isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 53:1563-1567. [DOI] [PubMed] [Google Scholar]
- 13.Nemec, A., M. Musílek, M. Vaneechoute, E. Falsen, and L. Dijkshoorn. 2008. Lack of evidence for “Acinetobacter septicus” as a species different from Acinetobacter ursingii? J. Clin. Microbiol. 46:2826-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemec, A., M. Musílek, M. Maixnerová, T. De Baere, T. J. van der Reijden, M. Vaneechoutte, and L. Dijkshoorn. 2009. Acinetobacter beijerinckii sp. nov. and Acinetobacter gyllenbergii sp. nov., haemolytic organisms isolated from humans. Int. J. Syst. Evol. Microbiol. 59:118-124. [DOI] [PubMed] [Google Scholar]
- 15.Nemec, A., and L. Dijkshoorn. 2010. Variations in colistin susceptibility among different species of the genus Acinetobacter. J. Antimicrob. Chemother. 65:367-369. [DOI] [PubMed] [Google Scholar]
- 16.Park, Y. K., S. I. Jung, K. H. Park, H. S. Cheong, K. R. Peck, J. H. Song, and K. S. Ko. 2009. Independent emergence of colistin-resistant Acinetobacter spp. isolates from Korea. Diagn. Microbiol. Infect. Dis. 64:43-51. [DOI] [PubMed] [Google Scholar]
- 17.Poirel, L., S. Figueiredo, V. Cattoir, A. Carattoli, and P. Nordmann. 2008. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52:1252-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starakis, I., A. Blikas, D. Siagris, M. Marangos, C. Karatza, and H. Bassaris. 2006. Prosthetic valve endocarditis caused by Acinetobacter lwoffii: a case report and review. Cardiol. Rev. 14:45-49. [DOI] [PubMed] [Google Scholar]
- 19.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 20.Tega, L., K. Raieta, D. Ottaviani, G. L. Russo, G. Blanco, and A. Carraturo. 2007. Catheter-related bacteremia and multidrug-resistant Acinetobacter lwoffii. Emerg. Infect. Dis. 13:355-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turton, J. F., M. E. Kaufmann, M. Warner, J. M. Coelho, L. Dijkshoorn, T. van der Reijden, and T. L. Pitt. 2004. A prevalent, multiresistant, clone of Acinetobacter baumannii in South East England. J. Hosp. Infect. 58:170-179. [DOI] [PubMed] [Google Scholar]
- 22.Turton, J. F., N. Woodford, J. Glover, S. Yarde, M. E. Kaufmann, and T. L. Pitt. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turton, J. F., J. Matos, M. E. Kaufmann, and T. L. Pitt. 2009. Variable number tandem repeat loci providing discrimination within widespread genotypes of Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 28:499-507. [DOI] [PubMed] [Google Scholar]
- 24.van den Broek, P. J., T. J. van der Reijden, E. van Strijen, A. V. Helmig-Schurter, A. T. Bernards, and L. Dijkshoorn. 2009. Endemic and epidemic Acinetobacter species in a university hospital, an eight years' survey. J. Clin. Microbiol. 47:3593-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Dessel, H., T. E. Kamp-Hopmans, A. C. Fluit, S. Brisse, A. M. de Smet, L. Dijkshoorn, A. Troelstra, J. Verhoef, and E. M. Mascini. 2002. Outbreak of a susceptible strain of Acinetobacter species 13 (sensu Tjernberg and Ursing) in an adult neurosurgical intensive care unit. J. Hosp. Infect. 51:89-95. [DOI] [PubMed] [Google Scholar]
- 26.Vaneechoutte, M., T. De Baere, A. Nemec, M. Musílek, T. J. van der Reijden, and L. Dijkshoorn. 2008. Reclassification of Acinetobacter grimontii Carr et al. 2003 as a later synonym of Acinetobacter junii Bouvet and Grimont 1986. Int. J. Syst. Evol. Microbiol. 58:937-940. [DOI] [PubMed] [Google Scholar]
- 27.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. B. Amyes, and D. M. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]
- 28.Yu-Hsien, L., C. Te-Li, C. Chien-Pei, and T. Chen-Chi. 2008. Nosocomial Acinetobacter genomic species 13 TU endocarditis following an endoscopic procedure. Intern. Med. 47:799-802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.