Abstract
The reliable differentiation of live Brucella vaccine strains from field isolates is an important element in brucellosis control programs. We describe the design, validation, and implementation of a novel single nucleotide polymorphism (SNP)-based typing platform that offers a rapid, reliable, and robust tool to achieve this with improved diagnostic accuracy compared to existing molecular tests. Furthermore, the assays described are designed such that they supplement, and can be run as an intrinsic part of, a previously described assay identifying Brucella isolates to the species level (K. K. Gopaul, C. J. Smith, M. S. Koylass, and A. M. Whatmore, BMC Microbiol. 8:86), giving a comprehensive molecular typing platform.
Brucellosis is a zoonotic disease caused by a number of species of the genus Brucella (25), which has global implications both economically and socially (8). The most important species in terms of both animal and human disease are Brucella melitensis (sheep and goats are the natural hosts) and Brucella abortus (cattle are the natural host). While treatment is available for cases of human brucellosis, in other animals, disease control is mediated through culling of infected individuals (“test and slaughter”) and/or vaccination of healthy livestock (8). Currently, the three vaccine strains recommended by the World Organization for Animal Health (Office International des Epizooties [OIE]) for brucellosis control are Brucella abortus strains S19 and RB51 and B. melitensis Rev1 (14, 18). All three vaccines, when administered correctly, can protect livestock from brucellosis but can still cause abortions when administered at the wrong time (4, 10, 21, 22). Furthermore, while the vaccines are considered sufficiently attenuated for animal use, they may still be pathogenic to humans. There are documented cases showing the pathogenic nature of strains Rev1 and S19 in humans (3). In the case of strain RB51, it has been noted that accidental inoculation via open wounds or needle-stick injuries have led to localized infections, but there is no evidence of accidental exposure to this strain leading to brucellosis (2, 6). Both Rev1 and RB51 are also resistant to drugs that may be used in frontline chemotherapy for the treatment of brucellosis in humans. Thus, for the effective monitoring of both brucellosis control programs and human disease it is important to have reliable tests to differentiate vaccine and field strains.
Existing molecular assays.
Previously molecular assays to distinguish each of the three live vaccine strains have been described. These target a 702-bp deletion within the eryCD locus for B. abortus S19 (19), an insertion sequence, IS711, in the wboA gene of B. abortus RB51 (23), and a single nucleotide polymorphism (SNP) within rpsL in B. melitensis Rev1 (7). However, currently the commonly used assays for these strains consist of time-consuming and labor-intensive individual conventional PCR or PCR-restriction fragment length polymorphism (RFLP) reactions. Furthermore, the marker used for S19 identification is reportedly absent in older variants of this strain, such as NCTC 8038 (19), and vaccines that are derived from these variants may still be in use in some parts of the world (1, 17).
We have previously described a multiple-outcome real-time PCR assay that can define Brucella isolates to the species level based on SNPs identified by multilocus sequencing studies (15, 24). In brief, these are competition assays using two differently labeled minor groove binding (MGB) probes targeting either the “species-specific” or “nonspecific” sequence. Depending on the presence or absence of the target SNP, one probe is cleaved in preference to the other, generating an increase in dye fluorescence from the cleaved oligonucleotide (15). To extend the scope of this assay as a genotyping tool, we have sought to incorporate targets that define the three Brucella vaccine strains, using data compiled from recently determined genome sequences for S19 (9), RB51 (B. Bricker et al., unpublished data), and Rev1 (http://www.broadinstitute.org/annotation/genome/brucella_group/MultiHome.html and unpublished data).
Target identification and validation.
As the derivation of vaccine strain B. abortus RB51 from its parent strain 2308 is well described (20), we chose to use only a single SNP, A667→C667 in BAB1_0649, for assay development. This SNP was identified by comparison of the RB51 draft genome with the sequences of B. abortus strain 2308, B. abortus strain 9-941, B. melitensis strain 16 M, B. ovis ATCC 25840, and B. suis strain 1330. In order to confirm the specificity of this target for RB51, we sequenced the loci in 68 strains of B. abortus corresponding to members of all 28 multilocus sequence analysis types identified in this species to date (unpublished data) and 26 other Brucella strains representing members of all other species and biovars (data not shown).
The origins of both B. melitensis Rev1 and B. abortus S19 are somewhat less clear, with no comparison to parental strains possible (11, 16). Thus, for additional confidence in the ability of the assay to distinguish vaccine and wild-type strains, two SNPs were selected for each of these assays. For B. abortus S19, SNPs G290→T290 in BruAB1_0060 and G450→C450 in BruAB1_1114 were chosen based on comparative genome analysis (9). These changes correspond to SNPs on line 9 and line 75, respectively, of Table S2 in the Crasta et al. paper describing the comparison of S19 with other B. abortus genomes (9). In order to confirm the specificity of these two targets for S19, we sequenced the region including these markers from the same panel of strains described above to confirm the specificity of the RB51 SNP target.
For Rev 1, we selected the SNP C271→T271 in BMEI0752 (rpsL), which forms the basis of a currently accepted PCR-RFLP assay (7). In addition, based on a whole-genome sequence comparison between B. melitensis 16 M and Rev1 (A. R. Wattam, personal communication) and subsequent confirmation from sequencing 95 Brucella isolates, we included an additional marker, G933→A933, in BMEI0208.
Assay development and validation.
The locations of primers, probes, and putative gene functions are shown in Table 1. We employed a “checkerboard” approach to optimize concentrations of primers and probes for each assay. Reaction volumes and cycling parameters were as previously published for our multiple-outcome species-defining assays (15) such that these additional assays can simply be included as additional wells in this test. Briefly, assays were set up in a reaction mixture volume of 12.5 μl made up of 6.25 μl TaqMan genotyping mix (Applied Biosystems, Warrington, United Kingdom) with the working concentrations of primers and probes listed in Table 1. For sensitivity determination, genomic DNA extracted by a standard phenol-chloroform method was quantified in triplicate by spectrophotometry (Smartspec Plus; Bio-Rad, Hemel Hempstead, United Kingdom) and diluted to the required concentration in DNA/RNA-free sterile water (Ambion). Reactions were run on an Agilent MX3005p platform (Agilent, La Jolla, CA) using cycling conditions of one hold at 95°C for 10 min followed by 40 cycles of 92°C for 15 s and 60°C for 1 min. Analysis was undertaken using the MxPro software provided with the MX3005p.
TABLE 1.
Targets, primers, and probes for the various vaccine MGB assays with associated working concentrationsa
| Vaccine | Target (position) | Gene description | Working concn (nM) of: |
|
|---|---|---|---|---|
| Probe | Primer | |||
| B. abortus S19 | BruAB1_0060 (290)b | Transcription regulator (lysR family) | VAC: 5′-CCGATAGGAGAACCG-3′ (150) | F: 5′-ACGCTCTGGATCGCATATTCATC-3′ (900) |
| NON: 5′-CCGATAGGCGAACCG-3′ (100) | R: 5′-GCACAGATCGGCGAACTG-3′ (100) | |||
| B. abortus S19 | BruAB1_1114 (450) | Chaperone (clpX) | VAC: 5′-CTGACGCAAGCCG-3′ (250) | F: 5′- CACATCTTCGCCGACATAGC-3′ (700) |
| NON: 5′-CTGACGGAAGCCG-3′ (50) | R: 5′-CATCGATGTGCCGTTCATCATG-3′ (100) | |||
| B. abortus RB51 | BAB1_0649 (667)b | Glutathione S- transferase | VAC: 5′-CTCCTCTTCCGTATTGG-3′ (250) | F: 5′-TGCTGGTTGGCAAGGAAGAG-3′ (100) |
| NON: 5′-CTCCTCTTCCTTATTGG-3′ (50) | R: 5′- CGATCAACGCGCCTTCTG-3′ (700) | |||
| B. melitensis Rev 1 | BMEI0752 (271)b | Ribosomal protein S12 (rpsL) | VAC: 5′-ACACCCAGCAAGTC-3′ (250) | F: 5′-TGACACCCTGGGTATCGAGAA-3′ (700) |
| NON: 5′-ACACCCGGCAAGTC-3′ (50) | R: 5′-GTGGCGGTCGTGTGAAG-3′ (100) | |||
| B. melitensis Rev 1 | BMEI0208 (933)b | Gamma-glutamyl phosphate reductase (proA) | VAC: 5′-CGCAGGTTTTGCC-3′ (50) | F: 5′-CGCGTCGAGATATTCGGTTGA-3′ (900) |
| NON: 5′-CGCAGGCTTTGCC-3′ (250) | R: 5′-GGTGCTGGCGCTCTATCC-3′ (100) | |||
The position of the SNP within each target is shown in parentheses. The target SNP is shown underlined and bold within the vaccine (VAC) and nonspecific (NON) probes.
Probes based on the reverse complement of the target SNP region.
Sensitivity for each of the assays was established by testing titrations of target DNA. All vaccine MGB assays were shown to have a minimum discriminatory sensitivity of around 100 fg, a result equating to around 30 cells and in line with previous findings (15).
To confirm the specificity of the newly devised vaccine strain identification assays, two large panels of test strains were assembled. The panels consisted of a mix of purified genomic DNA and crude cell extracts (obtained by inactivating material either by immersion in 66% [vol/vol] methanol or by heat inactivation). For the Rev1 MGB assays the specificity of both markers was examined using a panel of 277 isolates consisting predominantly of a worldwide collection of B. melitensis field isolates (n = 180) and including an additional 34 Rev1 isolates. For the S19 and RB51 MGB assays, we examined a panel of 498 Brucella isolates representing all Brucella species and biovars but consisting predominantly of a worldwide collection of B. abortus field isolates (n = 245). Furthermore, given previous evidence of the inability of existing conventional assays to identify all S19 isolates (17), we tested all 498 isolates with both the S19 and RB51 conventional PCR tests accepted by the OIE (18).
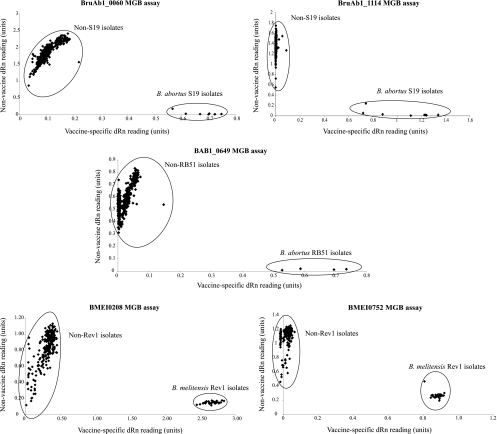
The results of this validation exercise are shown in Fig. 1. In brief, the endpoint fluorescence readings for vaccine-specific and nonspecific probes for each sample were plotted with the former on the x axis and the latter on the y axis. In all assays the discrimination of vaccine and nonvaccine isolates was clear-cut, with well-defined vaccine and nonvaccine populations. SNP-based typing and the conventional PCR test (18, 23) identified the same four isolates as RB51. However, there was discord between the conventional S19 PCR and MGB assays. The conventional ΔeryCD PCR identified only five strains as being S19, while the SNP discrimination assay identified an additional three isolates as S19. These isolates were NCTC 8038, a United Kingdom field isolate from 1964 (64/42), and a Chinese strain (55005) supplied to the VLA in the 1980s. NCTC 8038 was deposited in 1949, before the United States Department of Agriculture (USDA) decided to replace the diverse vaccine stocks circulating at the time with an erythritol-sensitive isolate in 1956 (12). The provenance of the remaining strains is less clear, but the phenotype is consistent with S19, as they have previously been biotyped as CO2-independent B. abortus biovar 1 (L. Perrett, personal communication). To prove the strains were indeed derivatives of S19, we amplified and sequenced gene fragments corresponding to BAbS19_I10690, BAbS19_I10750, BAbS19_I12470, BAbS19_I14270, and BAbS19_Ι14530, all of which we knew from pilot studies harbored additional SNPs unique to S19 (data not shown). In all five gene fragments from NCTC 8308, 64/42, and 55005, these additional SNPs, consistent with S19, were present, providing strong evidence that they represent S19 derivatives.
FIG. 1.
Allele discrimination plots generated by the individual MGB assays examining either 498 (in the case of strains S19 and RB51) or 277 (in the case of strain Rev1) isolates. In all cases, two distinct populations can be seen: vaccine (closest to the x axis) and nonvaccine (closest to the y axis).
Conclusions.
The SNP-based assays we have described here can be seamlessly combined with our Brucella species identification assays, giving a comprehensive genotyping tool that can rapidly, simply, and unambiguously characterize any Brucella isolate to the species level and concurrently determine whether it represents a vaccine isolate. The tool works well from crude bacterial extracts, avoiding the need for extensive bacterial processing. In terms of assay performance, we have shown that the BAB1_0649 assay for RB51 provides the same level of discrimination as the existing IS711 wboA PCR. The BruAB1_0060 and BruAB1_1114 assays for S19 proved more discriminatory than the existing ΔeryCD PCR, which is also the target used in some multiplex conventional PCR tests (5, 13), detecting S19 strains that lack this deletion. This will be particularly useful, as some countries may have generated their own S19 vaccines from stocks apparently originating before the occurrence of the eryCD deletion (1, 17). We have also demonstrated that BMEI0208 is a valid target in the differentiation of Rev1, giving the same discrimination as the OIE-accepted BMEI0752 marker. Thus, the assays described here give improved discriminatory capacity relative to conventional PCR tests and provide a unified platform, avoiding the need for multiple different conventional PCR and PCR-RFLP formats. As with all such assays, although we have validated them extensively, further studies with even more extensive collections of field isolates worldwide will undoubtedly be valuable in confirming the specificity of the markers selected for vaccine strains.
Acknowledgments
We thank Mark Koylass and James Edwards-Smallbone for their assistance in the sequencing work done at the Veterinary Laboratories Agency, Weybridge, United Kingdom. We thank Alice R. Wattam from the Virginia Bioinformatics Institute, Blacksburg, VA, for her comparison data of B. melitensis 16 M and B. melitensis Rev1 genomes. We also thank Félix Sangari, Cinzia Marianelli, Sevil Erdenlig, Ignacio López-Goñi, and Clara Marín for sending genomic DNA from various Brucella strains to validate our work.
This work was supported by the United Kingdom Department of Environment, Food and Rural Affairs (Defra) under projects SE0311 and SE0313.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Acha, P. N., and B. Szyfres. 2001. Brucellosis, p. 40-66. In Zoonoses and communicable diseases common to man and animals, vol. 3. Pan American Health Organization, Washington, DC. [Google Scholar]
- 2.Ashford, D. A., J. di Pietra, J. Lingappa, C. Woods, H. Noll, B. Neville, R. Weyant, S. L. Bragg, R. A. Spiegel, J. Tappero, and B. A. Perkins. 2004. Adverse events in humans associated with accidental exposure to the livestock brucellosis vaccine RB51. Vaccine 22:3435-3439. [DOI] [PubMed] [Google Scholar]
- 3.Berkelman, R. L. 2003. Human illness associated with the use of veterinary vaccines. Clin. Infect. Dis. 37:407-414. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, J. M. 1997. A review of the use of B. melitensis Rev 1 vaccine in adult sheep and goats. Prev. Vet. Med. 31:275-283. [DOI] [PubMed] [Google Scholar]
- 5.Bricker, B. J., and S. M. Halling. 1995. Enhancement of the Brucella AMOS PCR assay for differentiation of Brucella abortus vaccine strains S19 and RB51. J. Clin. Microbiol. 33:1640-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1998. Human exposure to Brucella abortus strain RB51-Kansas, 1997. MMWR Morb. Mortal. Wkly. Rep. 47:172-175. [PubMed] [Google Scholar]
- 7.Cloeckaert, A., M. Grayon, and O. Grépinet. 2002. Identification of Brucella melitensis vaccine strain Rev 1 by PCR-RFLP based on a mutation in the rpsL gene. Vaccine 20:2546-2550. [DOI] [PubMed] [Google Scholar]
- 8.Corbel, M. J. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 3:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crasta, O. R., O. Folkerts, Z. Fei, S. P. Mane, C. Evans, S. Martino-Catt, B. Bricker, G. Yu, L. Du, and B. W. Sobral. 2008. Genome sequence of Brucella abortus vaccine strain S19 compared to virulent strains yields candidate virulence genes. PLoS One 3:e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, D. S., J. W. Templeton, T. A. Ficht, J. D. Huber, R. D. Angus, and L. G. Adams. 1991. Brucella abortus in bison. II. Evaluation of strain 19 vaccination of pregnant cows. J. Wildl. Dis. 2:258-264. [DOI] [PubMed] [Google Scholar]
- 11.Elberg, S. S., and K. Faunce, Jr. 1957. Immunization against Brucella infection. VI. Immunity conferred on goats by a nondependent mutant from a streptomycin-dependent mutant strain of Brucella melitensis. J. Bacteriol. 73:211-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Lobo, J. M., and F. J. Sangari. 2004. Erythritol metabolism and virulence in Brucella. In I. Lopez-Goni and I. Moriyon (ed.), Brucella: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 13.García-Yoldi, D., C. M. Marín, M. J. de Miguel, P. M. Muñoz, J. L. Vizmanos, and I. López-Goñi. 2006. Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin. Chem. 52:779-781. [DOI] [PubMed] [Google Scholar]
- 14.Garin-Bastuji, B., and J. M. Blasco. 2008. Caprine and ovine brucellosis (excluding Brucella ovis), p. 974-982. In Manual of diagnostic tests and vaccines for terrestrial animals, 6th ed. OIE, Paris, France.
- 15.Gopaul, K. K., C. J. Smith, M. S. Koylass, and A. M. Whatmore. 2008. Rapid identification of Brucella isolates to the species level by real time PCR based single nucleotide polymorphism (SNP) analysis. BMC Microbiol. 8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graves, R. R. 1943. The story of John M. Buck's and Matilda's contribution to the cattle industry. J. Am. Vet. Med. Assoc. 102:193-195. [Google Scholar]
- 17.Mukherjee, F., J. Jain, M. J. Grilló, J. M. Blasco, and M. Nair. 2005. Evaluation of Brucella abortus S19 vaccine strains by bacteriological tests, molecular analysis of ery loci and virulence in BALB/c mice. Biologicals 33:153-160. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen, K., and D. R. Ewalt. 2008. Bovine brucellosis, p. 624-660. In Manual of diagnostic tests and vaccines for terrestrial animals, 6th ed. OIE, Paris, France.
- 19.Sangari, F. J., J. M. García-Lobo, and J. Agüero. 1994. The Brucella abortus vaccine strain B19 carries a deletion in the erythritol catabolic genes. FEMS Microbiol. Lett. 121:337-342. [DOI] [PubMed] [Google Scholar]
- 20.Schurig, G. G., R. M. Roop, I. I. T. Bagchi, S. Boyle, D. Buhrman, and N. Sriranganathan. 1991. Biological properties of RB51; a stable rough strain of Brucella abortus. Vet. Microbiol. 28:171-188. [DOI] [PubMed] [Google Scholar]
- 21.Thomas, E. L., C. D. Bracewell, and M. J. Corbel. 1981. Characterisation of Brucella abortus strain 19 cultures isolated from vaccinated cattle. Vet. Record. 108:90-93. [DOI] [PubMed] [Google Scholar]
- 22.Van Metre, D. C., G. A. Kennedy, S. C. Olsen, G. R. Hansen, and D. R. Ewalt. 1999. Brucellosis induced by RB51 vaccine in a pregnant heifer. J. Am. Vet. Med. Assoc. 215:1491-1493. [PubMed] [Google Scholar]
- 23.Vemulapalli, R., J. R. McQuiston, G. G. Schurig, N. Sriranganathan, S. M. Halling, and S. M. Boyle. 1999. Identification of an IS711 element interrupting the wboA gene of Brucella abortus vaccine strain RB51 and a PCR assay to distinguish strain RB51 from other Brucella species and strains. Clin. Diagn. Lab. Immunol. 6:760-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whatmore, A. M., L. L. Perrett, and A. P. MacMillan. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing BMC Microbiol. 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whatmore, A. M. 2009. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 9:1168-1184. [DOI] [PubMed] [Google Scholar]