Abstract
Achromobacter xylosoxidans is an emerging pathogen increasingly being isolated from respiratory samples of cystic fibrosis (CF) patients. Its role and clinical significance in lung pathogenesis have not yet been clarified. The aim of the present study was to genetically characterize A. xylosoxidans strains isolated from CF patients by use of randomly amplified polymorphic DNA (RAPD) profiles and to look for a possible correlation between RAPD profiles and the patients' clinical features, such as their spirometry values, the presence of concomitant chronic bacterial flora at the time of isolation, and the persistent or intermittent presence of A. xylosoxidans strains. A set of 106 strains of A. xylosoxidans were typed by RAPD analysis, and their profiles were analyzed by agglomerative hierarchical classification (AHC) and associated with the patient characteristics mentioned above by factorial discriminant analysis (FDA). The overall results obtained in this study showed that (i) there is a marked genetic relationship between strains isolated from the same patients at different times, (ii) characteristic RAPD profiles are associated with different predicted classes for forced expiratory volume in 1 s (FEV1%), (iii) some characteristic RAPD profiles are associated with different concomitant chronic flora (CCF) profiles, and (iv) there is a significant division of RAPD profiles into “persistent strains” and “intermittent strains” of A. xylosoxidans. These findings seem to imply that the lung habitats found in CF patients are capable of shaping and selecting the colonizing bacterial flora, as seems to be the case for the A. xylosoxidans strains studied.
Cystic fibrosis (CF) is the most common lethal genetic disease, causing a chronic infection of the respiratory tract, which in turn leads to progressive respiratory deficiency (6, 15). Pseudomonas aeruginosa is the most frequently found Gram-negative pathogen in the sputa of patients with CF, while Staphylococcus aureus is the most frequently found Gram-positive one. Recently, new pathogens have also emerged, such as Burkholderia cepacia complex, Stenotrophomonas maltophilia, and Achromobacter xylosoxidans (3, 9, 18, 19, 20, 24). Although the clinical significance of A. xylosoxidans is not yet clear, it is increasingly being isolated from the sputum cultures of CF patients. Tan et al. (22) found that 2.3% of CF patients had at least 3 positive cultures for A. xylosoxidans during a 6-month period. The U.S. Cystic Fibrosis Foundation's National Patient Registry reported an increase of 4.5%, from 1995 to 2002 (1, 2), in the frequency of isolation of this microorganism from CF patients. Recently, A. xylosoxidans has been considered a nosocomial pathogen, particularly in immunocompromised patients, causing a variety of infections, including bacteremia, meningitis, pneumonia, and peritonitis (8, 23, 25). Achromobacter spp. are aerobic, nonfermentative, Gram-negative bacilli (5, 7, 21) that are frequently misidentified by routine laboratory tests, thus seriously compromising control measures related to epidemiology studies. These microorganisms are often highly resistant to various antibiotics, including β-lactams, quinolones, aminoglycosides, and carbapenems, all commonly used for the management of lung infection in CF patients.
Considering the importance of bacterial lung infections in CF patients, our goals were (i) to assess the genetic relationships among isolated A. xylosoxidans strains by randomly amplified polymorphic DNA (RAPD) analysis and (ii) to use multivariate analysis techniques to look for possible correlations between A. xylosoxidans RAPD profiles and patients' clinical features (predicted classes for forced expiratory volume in 1 s [FEV1%], presence of concomitant chronic flora [CCF] during the isolation step, and persistent or intermittent presence of A. xylosoxidans). The study was conceived in order to give a picture of adaptive changes of A. xylosoxidans during lung infection in patients with CF and to improve our knowledge about this emerging pathogenic species, highlighting its potential role in CF disease.
MATERIALS AND METHODS
Ethics.
All patients were involved in the study after providing written consent. The study protocol was approved by the Committee on Ethical Practice of the Policlinico Umberto I, Rome, Italy.
Patients.
From January 2005 to January 2007, our laboratory cultured respiratory samples from 450 patients attending the Cystic Fibrosis Centre of the Pediatric Department of Policlinico Umberto I of Rome. A. xylosoxidans was isolated from the sputum cultures of 40 of these 450 patients. For the present study, we selected the 16 patients among these 40 for whom A. xylosoxidans strains were isolated more than once. We divided the 16 selected patients into the following three FEV1% classes (following the European Respiratory Society's criteria): class 3, mild obstruction or normal (≥70%); class 2, moderate obstruction (>40% and <70%); and class 1, severe obstruction (≤40%). A further subdivision was based on the persistent or intermittent presence of A. xylosoxidans. Table 1 summarizes the patients' demographic and clinical features.
TABLE 1.
CF patient characteristics
| Patienta | Strains | Age (yr) | FEV1% class | Intraindividual similarity (%) (mean ± SD) |
|---|---|---|---|---|
| 1M | 123, 124, 145 | 33 | 1 | 34.34 ± 3.37 |
| 2M | 2, 126, 143 | 19 | 3 | 53.74 ± 13.35 |
| 3F | 28, 55, 127 | 5 | 3b | 65.69 ± 12.60 |
| 4F | 99, 122 | 40 | 2 | 83.33 ± 1.40 |
| 5M | 7, 37, 114 | 24 | 1 | 80.45 ± 6.11 |
| 6F | 6, 35, 52, 63, 73, 75, 79, 86, 92, 108, 120, 135 | 33 | 1 | 31.99 ± 16.33 |
| 7M | 12, 34, 56, 78, 98, 139 | 34 | 2 | 41.50 ± 46.19 |
| 8F | 5, 26, 47, 77 | 32 | 1/2 | 53.58 ± 8.31 |
| 9M | 25, 46, 71, 83, 90 | 25 | 2 | 34.99 ± 21.16 |
| 10M | 13, 18, 36, 53, 65, 87 | 30 | 3 | 36.43 ± 24.44 |
| 11F | 30, 49, 50, 57, 60, 91, 134 | 14 | 3 | 54.29 ± 22.84 |
| 12F | 11, 22, 27, 45, 64, 89, 133 | 22 | 3 | 56.57 ± 13.88 |
| 13F | 23, 40g, 40p, 59, 80, 100, 112, 118 | 27 | 1 | 58.97 ± 14.86 |
| 14M | 4, 14, 19, 29, 38, 48, 96, 104, 115, 138 | 38 | 1 | 42.22 ± 16.48 |
| 15M | 3, 32, 43, 51, 74, 76, 97, 102, 110, 111, 136, 137 | 21 | 1 | 41.59 ± 20.02 |
| 16F | 1, 8, 9, 10, 15, 21, 31, 41, 81, 82, 93, 94, 95, 119, 121 | 27 | 1 | 32.69 ± 22.65 |
M, male patient; F, female patient.
The FEV1% class was arbitrarily assigned.
Microbiological methods.
All samples were cultured by using appropriate media, including Burkholderia cepacia selective agar (BCSA) (bioMérieux, Marcy l'Etoile, France) for B. cepacia complex isolates. All Gram-negative isolates were identified with an API 20NE system (bioMérieux, Marcy l'Etoile, France) and with an automated Vitek2 system (bioMérieux, Marcy l'Etoile, France). The biochemical results of the API 20NE system (bioMérieux, Marcy l'Etoile, France) were read after 48 and 72 h of incubation at 30°C. Oxidase activity was checked with dimethyl-paraphenylenediamine disks (bioMérieux, Marcy l'Etoile, France). The results of the API 20NE tests and oxidase reaction were further interpreted with the Apilab Plus software package (bioMérieux, Marcy l'Etoile, France). All Achromobacter sp. strains were cryopreserved at −80°C before use.
DNA extraction.
Each strain was grown overnight at 37°C in brain heart infusion broth (Becton Dickinson, NJ), and DNA extraction was performed by use of a Wizard genomic DNA purification kit (Promega Corporation, Madison, WI). DNA was resuspended in RNA-free deionized water and quantified by spectrophotometry.
Species-specific PCR assay.
Species-specific PCR was performed as described elsewhere (11). Negative-control PCRs were employed for every experiment. PCR products were separated by electrophoresis in a 2% agarose gel (Invitrogen Corporation, CA), stained with ethidium bromide (EtBr; Invitrogen Corporation), and captured with a DigiDoc-It (UVP, Cambridge, United Kingdom) photographic system.
RAPD typing.
The RAPD amplification mixture and cycling conditions were as described elsewhere (13). The primer used was primer 270 (5′-TGCGCGCGGG-3′). RAPD products were separated by electrophoresis in a 1.5% agarose gel (Invitrogen Corporation, CA). Molecular size markers (Invitrogen Corporation) and a negative control were included in all gels. The gels were stained with EtBr (Invitrogen Corporation) and captured with a DigiDoc-It (UVP) photographic system.
Data analysis. (i) AHC.
Agglomerative hierarchical classification (AHC), an unsupervised method, was performed on RAPD profiles by means of a binary matrix generated by the presence/absence of RAPD bands, using Doc-It LS software (UVP), and the subsequent dendrogram was generated with XLStat 7.5 (Addinsoft), using a Euclidean distance dissimilarity matrix and the agglomeration method of Ward.
(ii) PCA.
Using XLStat 7.5 software, linearly dependent data (absence/presence of RAPD bands) were transformed into independent variables (factorial axes; F1, F2…Fn) through principal component analysis (PCA), an unsupervised method. The coordinates of the observations on the factorial axes were taken into account as the new variables for the subsequent factorial discriminant analysis (FDA).
(iii) FDA.
FDA, a supervised method closely linked to multivariate analysis of variance, was employed by means of XLStat 7.5 software. Explanatory variables were automatically verified to be linearly independent by calculating the multiple correlation of each variable with all the others. Wilks's lambda test was used to compare the patients' clinical features with A. xylosoxidans RAPD profiles, and a P value of ≤0.05 was considered statistically significant.
RESULTS
A. xylosoxidans isolates.
During the period of study (January 2005 to January 2007), 40 patients among the 450 patients attending the Regional Centre of Cystic Fibrosis of the Policlinico Umberto I of Rome showed the presence of A. xylosoxidans in sputum samples. In this study, we followed the outcomes of 16 patients showing repeated isolations of A. xylosoxidans, from July 2005 to January 2007. The isolation range was 2 to 15 bacterial strains per patient, with a total of 106 strains of A. xylosoxidans. Among the isolated strains, identified by API 20NE, 56 (52.8%) showed a very good identification, 38 (35.8%) a good identification, and 12 (11.4%) a low level of discrimination. The A. xylosoxidans strains were often associated with CCF. The most frequently isolated concomitant strains were P. aeruginosa mucoid and rough strains and S. aureus. The CCF profiles found during the culturing step are reported in Table 2. As Table 2 shows, we obtained six CCF profiles: ppp, ppa, paa, apa, aap, and aaa.
TABLE 2.
Concomitant chronic bacterial flora profiles
| Profilea | Presence of bacterium |
||
|---|---|---|---|
| Pseudomonas aeruginosa mucoid strain | Pseudomonas aeruginosa rough strain | Staphylococcus aureus | |
| ppp | + | + | + |
| ppa | + | + | − |
| paa | + | − | − |
| apa | − | + | − |
| aap | − | − | + |
| aaa | − | − | − |
p, presence; a, absence.
Species-specific PCR assay.
A species-specific PCR assay was performed to validate the chemical identification of A. xylosoxidans isolates. For this purpose, oligonucleotides AX-F1 and AX-B1 were used. A 163-bp PCR product was detected for all 106 A. xylosoxidans isolates (data not shown). The results obtained by species-specific PCR assay were in agreement with biochemical identification performed with bioMérieux systems.
Data analysis.
In this study, 106 strains of A. xylosoxidans were typed by RAPD analysis. The profiles obtained were subsequently analyzed by AHC and FDA. These approaches were employed to look for genetic relationships between isolated strains and for putative associations between RAPD profiles and different CF patient clinical features.
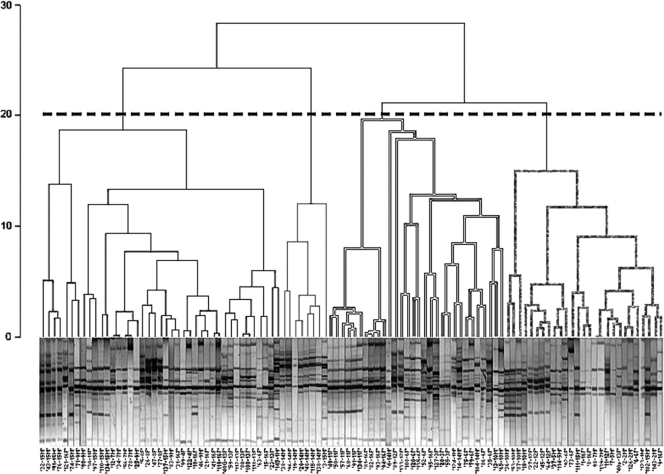
The RAPD profiles were first analyzed by XLStat software, using hierarchical cluster analysis. Four major well-defined clusters (Fig. 1) were obtained, as shown in the resulting dendrogram. The first cluster (A) grouped 41/106 bacterial strains, while the second one (B) grouped 8/106 bacterial strains, all from the same patients, the third (C) grouped 30/106 bacterial strains, and the fourth (D) grouped 27/106 bacterial strains. Intraindividual similarity was calculated by means of the Dice index, with values ranging from 31.99% to 83.33% (Table 1), with a mean value of 40.86% ± 22.81% (95% confidence interval [CI] = 2.21).
FIG. 1.
AHC. Genetic relationships between A. xylosoxidans strains are shown, as estimated by clustering analysis of genomic RAPD profiles. The Euclidean distance dissimilarity method and the agglomeration method of Ward were employed. The threshold defining a cluster was set at 80% similarity. A comprehensive text string with strain number and patient ID was added at the bottom of each RAPD profile.
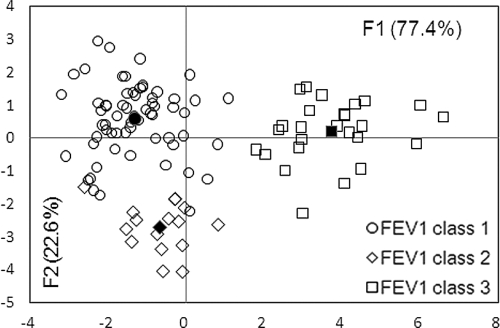
FDA of RAPD profiles showed that characteristic genomic profiles were associated with predicted FEV1% classes (Fig. 2). Predicted FEV1% classes 1 and 3, as well as classes 2 and 3, were significantly separated (P < 0.0001), indicating that there were significantly different RAPD profiles associated with these spirometry classes. Of particular interest are strains 47 and 137, isolated from two patients, of FEV1% class 2 and FEV1% class 1, respectively. FDA reclassified these strains as FEV1% class 1 (a posteriori probability, 73%) and FEV1% class 2 (a posteriori probability, 54%), respectively.
FIG. 2.
A. xylosoxidans RAPD profiles grouped by spirometric class. The percentages of variation described by the factorial axes (F1 and F2) are given in parentheses. The center of gravity for each group is reported with a filled symbol. The Mahalanobis distances (D2) between the three centers of gravity were as follows: for FEV1% class 1 versus FEV1% class 2, 11.2; for FEV1% class 1 versus FEV1% class 3, 25.6; and for FEV1% class 3 versus FEV1% class 2, = 27.8. Comparisons of the aforementioned distances were statistically significant (Fisher tests; P < 0.0001) for FEV1% class 1 versus FEV1% class 3 and FEV1% class 2 versus FEV1% class 3. The predictability of the model is 96.2%, and Wilks's lambda value = 0.075.
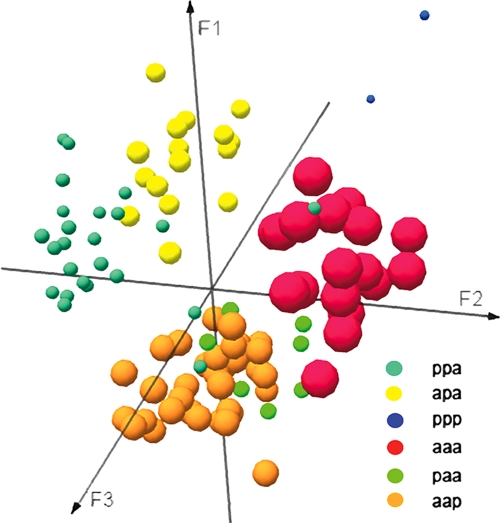
FDA of RAPD profiles also showed a correlation between the RAPD profiles and some of the CCF profiles (Table 2). RAPD profiles significantly differed between almost all the CCF profiles, as shown in Fig. 3.
FIG. 3.
A. xylosoxidans RAPD profiles grouped by the presence of chronic concomitant bacterial flora. Profiles were colored as follows: ppp = blue, aaa = red, apa = yellow, paa = green, aap = orange, and ppa = light cyan. The Mahalanobis distances (D2) between the six centers of gravity were as follows: for ppp versus ppa, 124.9; for ppp versus paa, 152.5; for ppp versus apa, 130.2; for ppp versus aap, 160.4; for ppp versus aaa, 116.1; for ppa versus paa, 32.2; for ppa versus apa, 19.5; for ppa versus aap, 25.5; for ppa versus aaa, 26.6; for paa versus apa, 34.4; for paa versus aap, 13.7; for paa versus aaa, 24.6; for apa versus aap, 28.5; for apa versus aaa, 30.1; and for aap versus aaa, 25.9. Comparisons of the aforementioned distances were statistically significant (Fisher tests; P < 0.0001) for all the groups, except for profile aap versus ppa, aap versus paa, apa versus ppa, and aaa versus paa (P = 0.78). The predictability of the model is 93.4%, and Wilks's lambda value = 0.003.
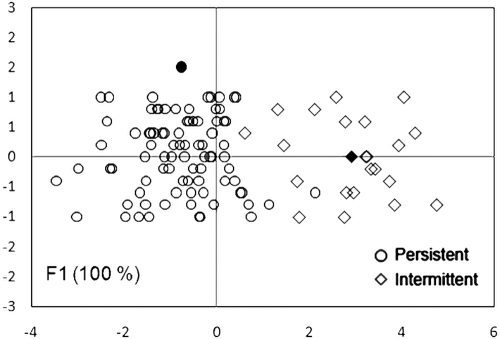
A. xylosoxidans RAPD profiles were also analyzed by FDA in order to study a possible relationship between the genotypes of the strains and their persistence or intermittence in CF patients. The classification of A. xylosoxidans RADP profiles showed a significant division (P < 0.0004) into two groups, with the first one grouping RAPD profiles of persistent strains and the second one grouping those of intermittent strains (Fig. 4). Strain 137, isolated from a persistently colonized patient, was reclassified as intermittent.
FIG. 4.
A. xylosoxidans RAPD profiles grouped by persistent or intermittent presence of the strains in CF lung patients. The percentages of variation described by the factorial axes (F1 and F2) are given in parentheses. The center of gravity for each group is reported as a filled symbol. The Mahalanobis distance (D2) between the two centers of gravity was 13.5. Comparison of the distances was statistically significant (Fisher tests; P = 0.004) between the two groups of patients. The predictability of the model is 97.2%, and Wilks's lambda value = 0.306. Data were manually scattered along the y axis to avoid overlapping points.
DISCUSSION
This is the first study focused on the genetic characterization of A. xylosoxidans strains isolated from CF patients and its possible correlation with their clinical features. The overall results obtained in this study, although the patient number was low, showed a genetic relationship among A. xylosoxidans strains isolated from the same patient and a strong association connecting the RAPD profiles of A. xylosoxidans and CF patients' clinical characteristics. Our findings could be related to the particular lung habitat present in CF patients, which is capable of shaping and selecting the colonizing bacterial flora (14), generating significantly different RAPD profiles of indigenous bacterial flora. The reclassification of some strains, such as strains 137 and 47, supports our results.
Strain 137 was reclassified as belonging to FEV1% predicted class 2 and as an intermittent strain, in contrast to the other strains isolated from the same patient. This could be explained by the fact that strain 137 was acquired by the patient at a later time than the other ones, and thus its pathoadaptive evolution could be different from that of the other (older) colonizing strains.
Strain 47, isolated from patient 8F, was reclassified as a FEV1% class 1 strain. This patient had a borderline predicted FEV1% value of 35% to 45% during the study period, so it was not surprising that this RAPD profile was reclassified.
Finally, strains 28, 55, and 127, isolated from patient 3F, who was too young to be subjected to spirometry assay, were correctly classified as FEV1% class 3 strains (mild obstruction or normal status), in agreement with the patient's clinical conditions, as she was not hospitalized or subjected to intravenous therapy. This also seems to support the discriminatory power of RAPD profiles and FDA.
The clinical characteristics selected for the study are factors influencing and clinically characterizing the pulmonary habitat. The capability of different habitats to produce different forces and selective pressures, promoting colonization of some bacterial species or strains in place of others, has been reported in many recent studies (10, 12, 17). Strains with related RAPD profiles share common genetic traits, and it is possible that these common traits are phenotypic characters important for their survival in CF patients' lung tissue. In the future, it would be of great interest to understand, through DNA sequencing, the phenotypic traits that allow these bacteria to survive and to be at an advantage in these habitats. The survival of species depends on a balance between fidelity of DNA replication and repair, and the generation of variation allows adaptation to novel environmental challenges. The obtained partition between RAPD profiles of isolated persistent strains and RAPD profiles of isolated intermittent strains could be explained by different pathoadaptive evolutionary rates (4).
Individuals with CF are particularly susceptible to lung infection by a limited spectrum of microbial pathogens, among which A. xylosoxidans is an important emergent one (16). Chronic airway bacterial infection in patients with CF leads to progressive damage of lung tissue and, ultimately, to the death of patients. During long-standing persistence of particular bacterial strains, such as A. xylosoxidans, the environmental pressures are likely to select bacterial mutants more suited to the inflamed lung tissue. Our results could be explained by adaptive changes in the physiology of A. xylosoxidans during chronic lung infection. These changes could result in a genome adaptive shift to support the growth under nutritional and microaerobic conditions created by the suppurative secretions in the lungs of patients with CF.
We can suppose that the biotypes found associated with different degrees of lung function could be the result of alternative genome shaping due to various lung habitats. Moreover, the different biotypes could also play an important role in lung inflammation/damage. It was recently reported (9) that A. xylosoxidans can cause a level of inflammation similar to that induced by P. aeruginosa in chronically infected CF patients, supporting the observation that this bacterial species is an emerging pathogen in CF.
These results provide important clues about the persistence strategies used by A. xylosoxidans during progressive CF lung disease. Further biomolecular studies will be performed to gain more comprehensive knowledge about the involvement of A. xylosoxidans in CF disease.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Cystic Fibrosis Foundation National Patient Registry. 1995. Annual report. Cystic Fibrosis Foundation, Bethesda, MD.
- 2.Cystic Fibrosis Foundation National Patient Registry. 2002. Annual report. Cystic Fibrosis Foundation, Bethesda, MD.
- 3.Dakin, C. J., A. H. Numa, H. Wang, J. R. Morton, C. C. Vertzyas, and R. L. Henry. 2002. Inflammation, infection, and pulmonary function in infant and young children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 165:904-910. [DOI] [PubMed] [Google Scholar]
- 4.D'Argenio, D. A., M. Wu, L. R. Hoffman, H. D. Kulasekara, E. Déziel, E. E. Smith, H. Nguyen, R. K. Ernst, T. J. Larson Freeman, D. H. Spencer, M. Brittnacher, H. S. Hayden, S. Selgrade, M. Klausen, D. R. Goodlett, J. L. Burns, B. W. Ramsey, and S. I. Miller. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64:512-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, J. C., and B. K. Rubin. 2007. Emerging and unusual gram-negative infections in cystic fibrosis. Semin. Respir. Crit. Care Med. 28:312-321. [DOI] [PubMed] [Google Scholar]
- 6.Davis, P. B., M. Drumm, and M. W. Konstar. 1996. Cystic fibrosis. Am. J. Respir. Crit. Care Med. 154:1229-1256. [DOI] [PubMed] [Google Scholar]
- 7.De Baets, F., P. Schelstraete, S. Van Daele, F. Haerynck, and M. Vaneechoutte. 2007. Achromobacter xylosoxidans in cystic fibrosis: prevalence and clinical relevance. J. Cyst. Fibros. 6:75-78. [DOI] [PubMed] [Google Scholar]
- 8.Gómez-Cerezo, J., I. Suárez, J. J. Ríos, P. Peña, M. J. García de Miguel, M. de José, O. Monteagudo, P. Linares, A. Barbado-Cano, and J. J. Vázquez. 2003. Achromobacter xylosoxidans bacteremia: a 10-year analysis of 54 cases. Eur. J. Clin. Microbiol. Infect. Dis. 22:360-363. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, C. R., T. Pressler, K. G. Nielsen, P. Ø. Jensen, T. Bjarnsholt, and N. Høiby. 2009. Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J. Cyst. Fibros. doi: 10.1016/j.jcf. 2009.10.005. [DOI] [PubMed]
- 10.Hoboth, C., R. Hoffmann, A. Eichner, C. Henke, S. Schmoldt, A. Imhof, J. Heesemann, and M. Hogardt. 2009. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 200:118-130. [DOI] [PubMed] [Google Scholar]
- 11.Hogardt, M., C. Hoboth, S. Schmoldt, C. Henke, L. Bader, and J. Heesemann. 2007. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 195:70-80. [DOI] [PubMed] [Google Scholar]
- 12.Krzewinski, J. W., C. D. Nguyen, J. M. Foster, and J. L. Burns. 2001. Use of random amplified polymorphic DNA PCR to examine epidemiology of Stenotrophomonas maltophilia and Achromobacter (Alcaligenes) xylosoxidans from patients with cystic fibrosis. J. Clin. Microbiol. 39:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambiase, A., V. Raia, M. Del Pezzo, A. Sepe, V. Carnovale, and F. Rossano. 2006. Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect. Dis. doi: 10.1186/1471-2334-6-4. [DOI] [PMC free article] [PubMed]
- 14.Liu, L., T. Coenyet, J. L. Burns, P. W. Whitby, T. L. Stull, and J. J. LiPuma. 2002. Ribosomal DNA-directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J. Clin. Microbiol. 40:1210-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mena, A., E. E. Smith, J. L. Burns, D. P. Speert, S. M. Moskowitz, J. L. Perez, and A. Oliver. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 190:7910-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen, D., and P. K. Singh. 2006. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc. Natl. Acad. Sci. USA 103:8305-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajan, S., and L. Saiman. 2002. Pulmonary infections in patients with cystic fibrosis. Semin. Respir. Infect. 17:47-56. [DOI] [PubMed] [Google Scholar]
- 19.Saiman, L., and J. Siegel. 2004. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spicuzza, L., C. Sciuto, G. Vitaliti, G. Di Dio, S. Leonardi, and M. La Rosa. 2009. Emerging pathogens in cystic fibrosis: ten years of follow-up in a cohort of patients. Eur. J. Clin. Microbiol. Infect. Dis. 28:191-195. [DOI] [PubMed] [Google Scholar]
- 21.Steinkamp, G., B. Wiedemann, E. Rietschel, A. Krahl, J. Gielen, H. Bärmeier, and F. Ratjen. 2005. Prospective evaluation of emerging bacteria in cystic fibrosis. J. Cyst. Fibros. 4:41-48. [DOI] [PubMed] [Google Scholar]
- 22.Tan, K., S. P. Conway, K. G. Brownlee, C. Etherington, and D. G. Peckham. 2002. Alcaligenes infection in cystic fibrosis. Pediatr. Pulmonol. 34:101-104. [DOI] [PubMed] [Google Scholar]
- 23.Tena, D., R. Carranza, J. R. Barberá, S. Valdezate, J. M. Garrancho, M. Arranz, and J. A. Sáez-Nieto. 2005. Outbreak of long-term intravascular catheter-related bacteremia due to Achromobacter xylosoxidans subspecies xylosoxidans in a haemodialysis unit. Eur. J. Clin. Microbiol. Infect. Dis. 24:727-732. [DOI] [PubMed] [Google Scholar]
- 24.Vonberg, R. P., and P. Gastmeier. 2005. Isolation of infectious cystic fibrosis patients: results of a systematic review. Infect. Control Hosp. Epidemiol. 26:401-409. [DOI] [PubMed] [Google Scholar]
- 25.Weitkamp, J. H., Y. W. Tang, D. W. Haas, N. K. Midha, and J. E. Crowe, Jr. 2000. Recurrent Achromobacter xylosoxidans bacteremia associated with persistent lymph node infection in a patient with hyper-immunoglobulin M syndrome. Clin. Infect. Dis. 31:1183-1187. [DOI] [PubMed] [Google Scholar]