Abstract
We evaluated the performance of the prototype Cobas AmpliPrep/Cobas TaqMan HIV-1 test, version 2.0, using prospective and archived clinical samples initially underquantitated by the Cobas AmpliPrep/Cobas TaqMan HIV-1 test. The performance of the new test was significantly improved, and the majority of the underquantitation observed with the first-version test was eliminated.
Plasma HIV viral load quantification is a validated tool for monitoring human immunodeficiency virus type 1 (HIV-1) infection (7). In 2005, Abbott Molecular (Rungis, France) and Roche Diagnostics GmbH (Mannheim, Germany) launched CE-marked (European Community) tests with the automated extraction of nucleic acids coupled to real-time PCR, giving a broader linear range of quantification and fewer time-consuming manipulations (2, 6, 9). For accurate viral load quantitation by PCR, HIV genetic diversity poses a major difficulty, especially in patients infected by non-B subtypes (1, 4, 6, 10, 11). In France, the frequency of naïve chronically infected patients with non-B subtypes reached 42% in 2006 to 2007 (D. Descamps, M. L. Chaix, A. Storto, F. Barin, S. Pakianather, A. G. Marcellin, M. Wirden, B. Masquelier, F. Brun-Vezinet, D. Costagliola, and the ANRS AC11 Resistance Group, presented at the XVI Conference on Retroviruses and Opportunistic Infections, Montreal, Canada, 8 to 11 February 2009).
Recently, we reported significant HIV-1 viral load discrepancies between the Cobas AmpliPrep/Cobas TaqMan HIV-1 test (TaqMan 1; also termed CAP/CTM v1.0) and the Cobas Amplicor HIV-1 Monitor test, version 1.5 (reference assay; also termed CA/HIM v1.5). TaqMan 1 underquantified not only divergent subtypes like CRF-02 but also subtype B isolates (5). Reports on underquantitation spurred the development of a second-generation real-time PCR assay, the Cobas AmpliPrep/Cobas TaqMan HIV-1 test, version 2.0 (TaqMan 2; also termed CA/CTM v2.0), which simultaneously amplifies and detects two targets of the HIV-1 genome.
The aim of this study was to evaluate the performance of this new prototype side by side with TaqMan 1. Results for both tests were compared to data for the reference assay.
Two ANRS (Agence Nationale de Recherche sur le SIDA) member virology laboratories in Paris, France (Bichat-Claude Bernard and Necker Hospitals), performed the evaluation using a panel of archived samples and a prospective panel of routine clinical plasma samples from the two ANRS virology laboratories. The archived panel included 25 plasma samples, stored at −80°C, for which the TaqMan 1 results previously had deviated from the reference assay results by ≥0.5 log10 (5). The prospective panel consisted of 263 routine plasma samples with detectable HIV-1 viral load. All specimens were diluted in HIV-1-negative plasma to generate about 5 ml, were aliquoted, and were stored frozen until single use. Reagents were provided by Roche Molecular Diagnostics.
Analyses for the TaqMan tests were performed in the two French laboratories, and the reference assay results were provided by Roche Molecular Diagnostics. Agreement between the three assays was evaluated by Bland-Altman plots (3). Underquantitation was defined as a greater-than −0.5 log10-titer deviation from the reference test.
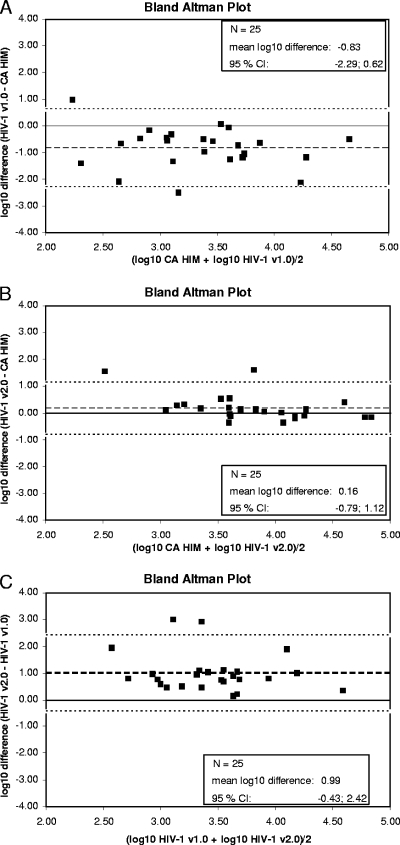
Among the 25 archived samples, the HIV-1 subtype distribution was the following: A (n = 3), B (n = 9), F (n = 1), G (n = 1), CRF02 (n = 10), and CRF06 (n = 1). The mean viral load was 2.94, 3.94, and 3.77 log10 copies/ml for TaqMan 1, TaqMan 2, and the reference assay, respectively (Fig. 1A, B, C, and Table 1). Seventeen of the 25 specimens were confirmed as underquantitated by more than 0.5 log10 in TaqMan 1 compared to results for the reference test (>1 log10, n = 10; −0.5 to −1 log10, n = 7). Among these samples, all 17 differed from the reference by less than −0.5 log10 in the new test. Five out the 25 samples were underquantitated in a range of −0.3 to −0.5 log10, and the underquantitation previously described was not reproducible in this study for three samples (5).
FIG. 1.
Bland-Altman plots for comparisons between CAP/CTM v1.0, CAP/CTM v2.0, and CA/HIM v1.5 in a panel of 25 archived plasma samples. (A) Bland-Altman plot for the comparison between CAP/CTM v1.0 and CA/HIM v1.5 in 25 archived samples. The broken line indicates the mean log10 difference, and the dotted lines represent the borders of the 95% confidence intervals (−2.29 and 0.62). (B) Bland-Altman plot for the comparison between CAP/CTM v2.0 and CA/HIM v1.5 in 25 archived samples. The broken line indicates the mean log10 difference, and the dotted lines represent the borders of the 95% confidence intervals (−0.79 and 1.12). (C) Bland-Altman plot for the comparison between CAP/CTM v1.0 and CAP/CTM v2.0 in 25 archived samples. The broken line indicates the mean log10 difference, and the dotted lines represent the borders of the 95% confidence intervals (−0.43 and 2.42).
TABLE 1.
Deming regression and Bland-Altman plots for measurement of agreement between CAP/CTM v1.0, CAP/CT v2.0, and CA/HIM v1.5a
| Samples analyzed | Deming regression |
Bland-Altman difference plot |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Equation | Confidence interval of intercept | R2b | Bias (log10 difference) | SD | Limits of agreement (90% IC) | Significant | No. (%) of outliers of |
||
| >0.5 log copies/ml | >1 log copies/ml | ||||||||
| Archived samples | |||||||||
| CAP/CTM v1.0-CA/HIM v1.5 | y = 0.70* x + 0.32 | −0.92, 1.56 | 0.198 | −0.83 | 0.742 | −1.085, −0.577 | Yes | 17 (68.0) | 10 (40.0) |
| CAP/CTM v2.0-CA/HIM v1.5 | y = 0.63* x + 1.55 | 0.81, 2.26 | 0.586 | 0.16 | 0.489 | −0.005, 0.330 | No | 4 (16.0) | 2 (8.0) |
| CAP/CTM v2.0-CAP/CTM v1.0 | y = 0.49* x + 2.50 | 1.48, 3.52 | 0.060 | 0.99 | 0.724 | 0.746, 1.242 | Yes | 21 (84.0) | 9 (36.0) |
| Prospective samples | |||||||||
| CAP/CTM v1.0-CA/HIM v1.5 | y = 1.04* x − 0.32 | −0.58, −0.06 | 0.738 | −0.15 | 0.393 | −0.186, −0.106 | Yes | 34 (13.1) | 12 (4.6) |
| CAP/CTM v2.0-CA/HIM v1.5 | y = 0.95* x + 0.26 | 0.07, 0.45 | 0.839 | 0.08 | 0.299 | 0.049, 0.110 | Yes | 24 (9.3) | 3 (1.2) |
| CAP/CTM v2.0-CAP/CTM v1.0 | y = 0.93* x + 0.48 | 0.28, 0.68 | 0.797 | 0.22 | 0.346 | 0.186, 0.257 | Yes | 30 (11.5) | 9 (3.5) |
The reference method is always the second method.
R2, coefficient of determination.
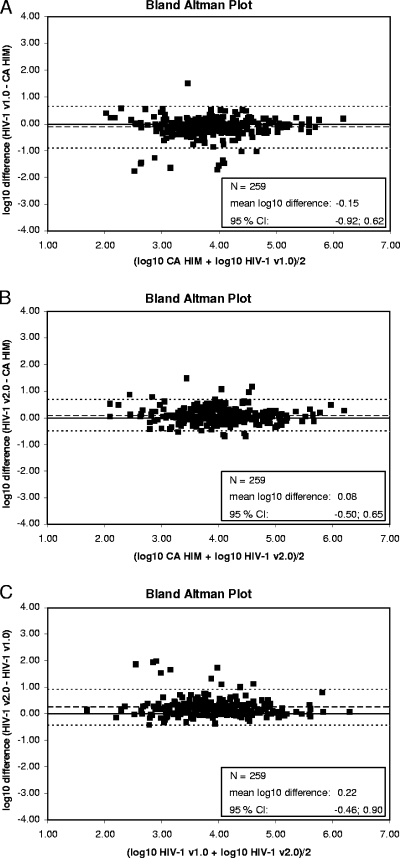
Among the 263 prospective samples, HIV-1 subtypes were available for 159 patients, and the distribution was A (n = 10), B (n = 66), C (n = 4), D (n = 4), F (n = 3), G (n = 5), H (n = 1), J (n = 1), CRF01 (n = 6), CRF02 (n = 50), CRF04 (n = 1), CRF05 (n = 1), CRF06 (n = 3), CRF12 (n = 1), CRF18 (n = 2), and CRF36 (n = 1). Valid positive results were obtained for 259 samples in all three tests (Fig. 2A, B, C, and Table 1). The mean viral load was 3.95, 4.16, and 4.10 log10 copies/ml for TaqMan 1, TaqMan 2, and the reference assay, respectively. Twenty-seven (10.4%) of the 259 samples were underquantitated with TaqMan 1 by more than 0.5 log10 (>1 log10, n = 11; −0.5 to −1 log10, n = 16) (Table 1). Nine of the 11 samples underquantitated by >1 log10 fell in a range of less than −0.5 log10 difference, and 2/11 fell in a range of −0.7 to −0.5 log10 difference in the new test compared to results for the reference. Among the 16 samples underquantitated between −0.5 and −1.0 log10, 15 were within less than −0.5 log10 difference in the new test. On the contrary, 7 of the 259 samples (2.7%) were underquantitated with the reference test by more than 0.5 log10 (>1 log10, n = 1; 0.5 to 1 log10, n = 6) compared to the results of TaqMan 1. Two samples not underquantitated by TaqMan 1 showed titers of −0.55 and −0.66 log10 in the new test.
FIG. 2.
Bland-Altman plots for comparisons between CAP/CTM v1.0, CAP/CTM v2.0, and CA/HIM v1.5 in a panel of prospectively collected plasma samples. (A) Bland-Altman plot for the comparison between CAP/CTM v1.0 and CA/HIM v1.5 in 259 valid result pairs for a panel of prospective routine clinical samples. The broken line indicates the mean log10 difference, and the dotted lines represent the borders of the 95% confidence intervals (−0.92 and 0.62). (B) Bland-Altman plot for the comparison between CAP/CTM v2.0 and CA/HIM v1.5 in 259 valid result pairs for a panel of prospective routine clinical samples. The broken line indicates the mean log10 difference, and the dotted lines represent the borders of the 95% confidence intervals (−0.50 and 0.65). (C) Bland-Altman plot for the comparison between CAP/CTM v1.0 and CAP/CTM v2.0 (HIV-1 v2.0) in 259 valid result pairs for a panel of prospective routine clinical samples. The broken line indicates the mean log10 difference, and the dotted lines represent the borders of the 95% confidence intervals (−0.46 and 0.90).
This study evaluated a novel real-time PCR test for HIV-1 viral load quantification, TaqMan 2, side by side with the previous version of the test and compared them to the reference assay. The new test includes primers and a probe for a second highly conserved region of the HIV-1 genome (the 5′ long terminal repeat [5′LTR]) in addition to the gag primers and probe of TaqMan 1 to address observed underquantitation.
The performance of the new test was significantly improved by resolving the majority of the underquantitation seen in the previous test version. Underquantitation at low frequency also occurred in the reference assay and is mitigated in the new test as well. Sequencing was achieved for four of five samples underquantitated by >1 log10 in the reference test compared results for the new test, and it confirmed multiple mismatches in gag primers and probe-binding regions of the reference test. As HIV-1 diversity and viral recombination increase, these results underline the importance of primer and probe design for viral load quantification assays combined with novel concepts as the simultaneous amplification and detection of two HIV-1 targets. A previous report already has shown that real-time PCR with amplification within the 5′LTR region might bring better quantification of some African viral strains than the reference assay (8). In addition, it emphasizes the need for the surveillance of commercialized assays to ensure the accurate viral load monitoring of HIV-1-infected patients.
Footnotes
Published ahead of print on 3 February 2010.
REFERENCES
- 1.Alaeus, A., K. Lidman, A. Sonnerborg, and J. Albert. 1997. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS 11:859-865. [DOI] [PubMed] [Google Scholar]
- 2.Berger, A., L. Scherzed, M. Sturmer, W. Preiser, H. W. Doerr, and H. F. Rabenau. 2002. Evaluation of the Cobas AmpliPrep/Cobas Amplicor HIV-1 Monitor Ultrasensitive Test: comparison with the Cobas Amplicor HIV-1 Monitor test (manual specimen preparation). J. Clin. Virol. 25(Suppl. 3):S103-S107. [DOI] [PubMed] [Google Scholar]
- 3.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed] [Google Scholar]
- 4.Damond, F., C. Apetrei, D. Descamps, F. Brun-Vezinet, and F. Simon. 1999. HIV-1 subtypes and plasma RNA quantification. AIDS 13:286-288. [DOI] [PubMed] [Google Scholar]
- 5.Damond, F., B. Roquebert, A. Benard, G. Collin, M. Miceli, P. Yeni, F. Brun-Vezinet, and D. Descamps. 2007. Human immunodeficiency virus type 1 (HIV-1) plasma load discrepancies between the Roche Cobas AMPLICOR HIV-1 MONITOR Version 1.5 and the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 assays. J. Clin. Microbiol. 45:3436-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gueudin, M., J. C. Plantier, V. Lemee, M. P. Schmitt, L. Chartier, T. Bourlet, A. Ruffault, F. Damond, M. Vray, and F. Simon. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime Extraction-Quantification Systems for HIV-1 Subtypes. J. Acquir Immune Defic Syndr. [DOI] [PubMed]
- 7.Mellors, J. W., A. Munoz, J. V. Giorgi, J. B. Margolick, C. J. Tassoni, P. Gupta, L. A. Kingsley, J. A. Todd, A. J. Saah, R. Detels, J. P. Phair, and C. R. Rinaldo, Jr. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 8.Rouet, F., D. K. Ekouevi, M. L. Chaix, M. Burgard, A. Inwoley, T. D. Tony, C. Danel, X. Anglaret, V. Leroy, P. Msellati, F. Dabis, and C. Rouzioux. 2005. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. J. Clin. Microbiol. 43:2709-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schumacher, W., E. Frick, M. Kauselmann, V. Maier-Hoyle, R. van der Vliet, and R. Babiel. 2007. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the Cobas AmpliPrep/Cobas TaqMan system. J. Clin. Virol. 38:304-312. [DOI] [PubMed] [Google Scholar]
- 10.Scott, L. E., L. D. Noble, J. Moloi, L. Erasmus, W. D. Venter, and W. Stevens. 2009. Evaluation of the Abbott m2000 realtime human immunodeficiency virus type 1 (HIV-1) assay for HIV load monitoring in South Africa compared to the Roche Cobas AmpliPrep-Cobas Amplicor, Roche Cobas AmpliPrep-Cobas TaqMan HIV-1, and BioMerieux NucliSENS EasyQ HIV-1 assays. J. Clin. Microbiol. 47:2209-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirden, M., R. Tubiana, F. Marguet, I. Leroy, A. Simon, M. Bonmarchand, Z. Ait-Arkoub, R. Murphy, A. G. Marcelin, C. Katlama, and V. Calvez. 2009. Impact of discrepancies between the Abbott realtime and Cobas TaqMan assays for quantification of human immunodeficiency virus type 1 group M non-B subtypes. J. Clin. Microbiol. 47:1543-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]