Abstract
We recently developed a cell culture system for hepatitis E virus (HEV) in PLC/PRF/5 and A549 cells, using fecal specimens from HEV-infected patients. Since transfusion-associated hepatitis E has been reported, we examined PLC/PRF/5 and A549 cells for the ability to support replication of HEV in various serum samples obtained from 23 patients with genotype 1, 3, or 4 HEV. HEV progenies emerged in culture media of PLC/PRF/5 cells, regardless of the coexistence of HEV antibodies in serum but dependent on the load of HEV inoculated (31% at 2.0 × 104 copies per well and 100% at ≥3.5 × 104 copies per well), and were successfully passaged in A549 cells. HEV particles in serum, with or without HEV antibodies, banded at a sucrose density of 1.15 to 1.16 g/ml, which was markedly lower than that for HEV particles in feces, at 1.27 to 1.28 g/ml, and were nonneutralizable by immune sera in this cell culture system. An immuno-capture PCR assay of HEV virions treated with or without detergent indicated that HEV particles in serum are associated with lipids and HEV ORF3 protein, similar to those in culture supernatant. By immunoprecipitation, it was found that >90% of HEV particles in the circulation exist as free virions not complexed with immunoglobulins, despite the coexistence of HEV antibodies. These results suggest that our in vitro cell culture system can be used for propagation of a wide variety of HEV strains in sera from various infected patients, allowing extended studies on viral replication specific to different HEV strains.
Hepatitis E, an acute viral hepatitis caused by infection with hepatitis E virus (HEV), is a globally distributed human disease. In developing countries of Asia, Africa, and Latin America, where sanitation conditions are not well maintained, HEV infection is transmitted via the fecal-oral route through virus-contaminated water or food, with substantial mortality in pregnant women (7, 33). In industrialized countries, autochthonous hepatitis E is far more common than previously recognized and has a predilection for older men, in whom it causes substantial morbidity and mortality (5, 13, 31, 36, 44). HEV is the sole member of the genus Hepevirus within the family Hepeviridae (6). It is a single-stranded, positive-sense, polyadenylated RNA molecule of approximately 7.2 kb in size, with short 5′- and 3′-untranslated regions (53). The genomic RNA contains three open reading frames (ORFs). ORF1 encodes nonstructural proteins involved in virus replication and virus protein processing. ORF2 and ORF3 overlap, and the ORF2 and ORF3 proteins are translated from a single bicistronic subgenomic RNA (11, 16). ORF2 encodes a 660-amino-acid (aa) capsid protein. ORF3 encodes a small phosphorylated protein (113 or 114 aa) that is essential for viral infectivity in vivo and for virion release (10, 16, 60, 63). The viral capsid protein induces neutralizing antibodies after immunization (8, 14, 26, 51) or during the course of infection (40, 41).
Four major genotypes (genotypes 1 to 4) of HEV have been identified in mammalian species. The viruses in genotypes 1 and 2 are maintained among humans only and are responsible for waterborne epidemics of HEV infection in developing countries. Genotype 3 HEV has been found worldwide, and genotype 4 HEV was isolated in Asia (23, 32, 39). Recent comprehensive molecular and serological studies have led to the consensus that hepatitis E is a zoonosis with a reservoir in pigs and, possibly, a range of other mammals (27, 43, 47, 48, 57, 65). Genotype 3 and 4 HEVs are considered to undergo zoonotic transmission (23, 32). Recently, HEV was found in farmed rabbits in China, possibly representing a novel genotype (66). These rabbits are reared for their fur, and there is, as yet, no evidence of transmission to humans.
Recently, using inocula comprised of fecal suspensions with high HEV loads, originally obtained from Japanese patients who contracted domestic infection of genotype 3 HEV (the JE03-1760F strain) or genotype 4 HEV (the HE-JF5/15F strain), we developed an efficient cell culture system for HEV in PLC/PRF/5 and A549 cells, which yielded the highest HEV load of 108 copies/ml in the culture supernatant, and we successfully propagated six or more generations in serial passages of culture supernatant (22, 55, 56). Infection of humans with HEV via blood transfusion has been reported not only in developing countries (genotype 1) (1, 18) but also in industrialized countries, including Japan (genotypes 3 and 4) (2, 24, 25, 28, 54), suggesting that HEV in serum samples can also be propagated in cultured cells. Therefore, in the present study, we examined whether HEV strains in serum samples obtained from various patients with sporadic acute hepatitis E can replicate in PLC/PRF/5 and A549 cells and produce infectious progenies in culture media, in relation to HEV load, genotype, and coexistence of HEV antibodies. In addition, in an attempt to clarify why HEV strains in sera are infectious in cultured cells despite the presence of HEV antibodies, HEV particles in serum samples were characterized and compared with those in culture supernatant and feces.
MATERIALS AND METHODS
HEV.
With informed consent from patients, 24 serum samples (S1 to S3, S4a, S4b, and S5 to S23) with HEV loads of 1.7 × 105 to 4.1 × 107 copies/ml were collected at the acute phase from 23 patients who contracted domestic or imported infection with a genotype 1, 3, or 4 HEV strain and developed acute hepatitis E during 2001 to 2008 in Japan, and these were used as inocula (Table 1). A fecal suspension containing a high load of wild-type HEV (JE03-1760F strain; 2.0 × 107 copies/ml) (55) and a culture supernatant containing a high load of cell culture-generated JE03-1760F variant (1.2 × 108 copies/ml) (22) were used as the references in this study.
TABLE 1.
Serum samples obtained from hepatitis E patients and inoculated onto cultured cells
| Patient no. | Age (yr) and sexb | HEV genotype | Sample no. | Days after onset | ALT level (IU/liter) | Relative titer of anti-HEV ORF2/relative titer of anti-HEV ORF3a |
HEV RNA titer (copies/ml) | ||
|---|---|---|---|---|---|---|---|---|---|
| IgG class | IgM class | IgA class | |||||||
| 1 | 60 M | 4 | S1 | 2 | 1,036 | −/− | −/− | −/− | 2.3 × 107 |
| 2 | 47 M | 3 | S2 | Unknown | 178 | −/− | −/− | −/− | 1.1 × 106 |
| 3 | 35 M | 4 | S3 | 3 | 829 | −/− | −/− | −/− | 1.8 × 106 |
| 4 | 67 M | 3 | S4a | 15 | 285 | 1:9,500/1:200 | 1:3,300/1:1,000 | 1:3,100/1:190 | 3.6 × 105 |
| 67 M | 3 | S4b | 22 | 15 | 1:13,000/1:340 | 1:3,200/1:300 | 1:1,600/1:130 | 1.7 × 105 | |
| 5 | 23 M | 1 | S5 | 2 | 4,538 | 1:4,200/1:110 | 1:1,600/1:650 | 1:1,700/1:480 | 2.8 × 106 |
| 6 | 78 F | 4 | S6 | 10 | 1,763 | 1:340,000/1:210,000 | 1:7,000/1:260 | 1:11,000/1:8,900 | 7.6 × 106 |
| 7 | 42 M | 3 | S7 | Unknown | 1,364 | 1:1,500/1:120 | 1:820/1:170 | 1:2,000/1:870 | 9.6 × 106 |
| 8 | 44 M | 3 | S8 | 5 | 1,082 | 1:7,100/1:3,400 | 1:30,000/1:1,900 | 1:5,000/1:2,800 | 1.1 × 107 |
| 9 | 46 M | 4 | S9 | 7 | 4,100 | 1:88,000/1:6,300 | 1:550/1:7,900 | 1:280/1:9,800 | 5.5 × 105 |
| 10 | 47 M | 4 | S10 | 7 | 2,492 | 1:6,300/1:4,400 | 1:5,500/1:3,700 | 1:5,000/1:910 | 1.7 × 107 |
| 11 | 61 M | 3 | S11 | 7 | 1,287 | 1:37,000/1:7,900 | 1:13,000/1:110 | 1:40,000/1:5,000 | 8.8 × 106 |
| 12 | 59 M | 4 | S12 | 3 | 3,660 | 1:10,000/1:990 | 1:2,000/1:7,400 | 1:32,000/1:9,500 | 3.7 × 106 |
| 13 | 52 M | 3 | S13 | 2 | 2,965 | −/1:6,300 | 1:20,000/1:5,600 | 1:170/1:63,000 | 1.2 × 107 |
| 14 | 61 M | 4 | S14 | 0 | 4,471 | 1:130/1:1,900 | 1:200/1:5,000 | 1:760/1:3,500 | 3.4 × 106 |
| 15 | 63 M | 4 | S15 | 7 | 1,140 | 1:230,000/1:64,000 | 1:430/1:5,000 | 1:3,700/1:7,900 | 8.1 × 105 |
| 16 | 60 M | 4 | S16 | 7 | 1,412 | 1:22,000/1:4,000 | 1:28,000/1:10,000 | 1:7,900/1:5,600 | 7.0 × 105 |
| 17 | 66 M | 3 | S17 | 7 | 1,813 | 1:14,000/1:1,000 | 1:3,400/1:1,100 | 1:7,100/1:1,100 | 2.2 × 106 |
| 18 | 42 M | 3 | S18 | 5 | 1,434 | 1:400/1:6,600 | 1:12,000/1:10,000 | 1:530/1:3,400 | 7.8 × 106 |
| 19 | 46 M | 3 | S19 | 7 | 3,047 | 1:6,900/1:140 | 1:3,800/1:610 | 1:5,000/1:790 | 1.8 × 107 |
| 20 | 56 F | 3 | S20 | 4 | 427 | 1:6,000/1:330 | 1:4,000/1:750 | 1:2,300/1:180 | 2.6 × 106 |
| 21 | 46 F | 4 | S21 | 7 | 1,548 | 1:8,800/1:3,500 | 1:6,300/1:20,000 | 1:40,000/1:2,100 | 4.1 × 107 |
| 22 | 54 M | 4 | S22 | 6 | 3,035 | 1:3,800/1:120 | 1:4,500/1:1,100 | 1:20,000/1:1,000 | 4.8 × 106 |
| 23 | 48 M | 4 | S23 | 8 | 6,102 | 1:15,000/1:14,000 | 1:13,000/1:4,300 | 1:7,900/1:11,000 | 3.2 × 106 |
Relative titers of anti-HEV ORF2 and anti-HEV ORF3 were determined by the methods described in Materials and Methods. −, negative (<1:100) for the indicated class of antibody.
F, female; M, male.
Detection of antibodies to HEV.
To detect anti-HEV ORF2 IgG, anti-HEV ORF2 IgM, and anti-HEV ORF2 IgA in serum samples, enzyme-linked immunosorbent assays (ELISAs) were performed using purified recombinant ORF2 protein that had been expressed in silkworm pupae (29), as described previously (49). The optical density (OD) of each sample was read at 450 nm. The cutoff values used for the anti-HEV ORF2 IgG, anti-HEV ORF2 IgM, and anti-HEV ORF2 IgA assays were 0.175, 0.440, and 0.642, respectively (49). Samples with OD values for anti-HEV ORF2 IgG, IgM, or IgA equal to or greater than the respective cutoff value were considered to be positive for anti-HEV ORF2 IgG, IgM, or IgA, respectively. The specificity of the anti-HEV ORF2 assays was verified by absorption with the same recombinant ORF2 protein that was used as the antigen probe, as described previously (49). The relative titers of anti-HEV ORF2 IgG, IgM, and IgA antibodies were determined by end-point ELISA, i.e., the serum dilution that gave the OD value of each cutoff point was estimated by testing multiple dilutions of the serum.
In addition, anti-HEV ORF3 IgG, IgM, and IgA antibodies were detected by an ELISA using the synthetic peptide spORF3-C24, with the sequence CLG ATS PSA PPL PPV VDL PQL GLR R, covering aa 90 to 113 at the C terminus of the ORF3 protein of the JE03-1760F strain (52), and the cutoff value was provisionally set at 0.2 for each class of ORF3 antibodies. The relative titers of anti-HEV ORF3 IgG, IgM, and IgA antibodies were also determined by end-point ELISA, i.e., the serum dilution that gave an OD value of 0.2 was estimated by testing multiple dilutions of the serum.
Detection and sequence analysis of HEV RNA.
For detection of HEV RNA, total RNA was extracted from 10 to 100 μl of serum sample or culture supernatant by use of TRIzol-LS reagent (Invitrogen, Carlsbad, CA) and subjected to a nested reverse transcription-PCR (RT-PCR) that amplifies a 457-nucleotide (nt) sequence in ORF2, as described previously (29). The amplification product was sequenced directly on both strands, using a BigDye Terminator v3.1 cycle sequencing kit on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Sequence analysis was performed using Genetyx-Mac 12.2.7 (Genetyx Corp., Tokyo, Japan) and ODEN, version 1.1.1, from the DNA Data Bank of Japan (DDBJ; National Institute of Genetics, Mishima, Japan) (17). A phylogenetic tree was constructed by the neighbor-joining method (38), based on the 412-nt ORF2 sequence. Bootstrap values were determined for 1,000 resamplings of the data sets (9).
Quantitation of HEV RNA.
HEV RNA was quantitated by real-time RT-PCR according to a previously described method (55), with slight modifications. In brief, total RNA extracted from 100 μl of diluted serum sample, fecal supernatant, or culture medium was subjected to real-time RT-PCR with a QuantiTect Probe RT-PCR kit (Qiagen, Tokyo, Japan), a sense primer (5′-GGT GGT TTC TGG GGT GAC-3′), an antisense primer (5′-AGG GGT TGG TTG GAT GAA-3′), and a probe consisting of an oligonucleotide with a 5′ reporter dye (6-carboxyfluorescein [FAM]) and a 3′ quencher dye (6-carboxytetramethylrhodamine [TAMRA]) (5′-FAM-TGA TTC TCA GCC CTT CGC-TAMRA-3′) on an Applied Biosystems 7900HT Fast real-time PCR system (Applied Biosystems). The thermal cycler conditions were 50°C for 30 min, 95°C for 15 min, and 50 cycles of 94°C for 15 s, 56°C for 30 s, and 76°C for 30 s.
Cell culture and virus inoculation.
PLC/PRF/5 (ATCC CRL-8024) and A549 (ATCC CCL-185) cells were grown in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS; Invitrogen), 100 U/ml of penicillin G, 100 μg/ml of streptomycin, and 2.5 μg/ml of amphotericin B (growth medium) at 37°C in a humidified 5% CO2 atmosphere as described previously (55). For virus infection, cells (5 × 105 cells) in 2 ml of medium were added to wells (diameter of 3.5 cm) of a six-well plate (Iwaki, Tsukuba, Japan) 2 days before virus infection. Monolayers of confluent cultured cells in a six-well plate were washed three times with 1 ml of phosphate-buffered saline (PBS) without Ca2+ and Mg2+ [PBS(−)] and inoculated with 0.2 ml of serum sample, culture supernatant, or fecal suspension that had been diluted with PBS(−) containing 0.2% (wt/vol) bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, MO) and filtered through a microfilter with a pore size of 0.22 μm (Millex-GV; Millipore Corp., Bedford, MA). In passage, 0.2 ml of culture supernatant that had been filtered through a 0.22-μm microfilter was inoculated on a monolayer of PLC/PRF/5 or A549 cells. After being inoculated at room temperature for 1 h, the solution was removed and 2 ml of maintenance medium was added. The maintenance medium used for virus culture was a 1:1 mixture of DMEM and medium 199 (Invitrogen) containing 2% (vol/vol) heat-inactivated FBS and 30 mM MgCl2 (final concentration); other supplements were the same as those in the growth medium. Culture was performed at 35.5°C in a humidified 5% CO2 atmosphere. On the day following inoculation, the inoculated cells were washed five times with 1 ml of PBS(−), and thereafter, 2 ml of maintenance medium was added. Then, every other day from day 2 after inoculation, one-half (1 ml) of the culture medium was replaced with fresh maintenance medium, and harvested media were stored at −80°C until virus titration. The infected tissue culture microplates were examined daily under an inverted microscope for specific cytopathic effect (CPE).
Neutralization test.
Serum samples that were positive for the IgG, IgM, and IgA classes of anti-HEV antibodies (pool 1 to pool 3 and sample 03-2150) were obtained during the convalescent phase from four patients with sporadic acute hepatitis E (Table 2). One additional serum sample (08-1340), containing only the IgG class of anti-HEV antibody, was obtained 5.7 years after disease onset from a patient with HEV infection, from whom the serum sample 03-2150 and fecal specimen JE03-1760F had been obtained (50). All serum samples used for the neutralization test were negative for HEV RNA.
TABLE 2.
Serum samples used in neutralization testc
| Serum sample | Days (yr) after disease onsetb | Relative titer of anti-HEV ORF2/relative titer of anti-HEV ORF3a |
Genotype of HEV recovered from patient | ||
|---|---|---|---|---|---|
| IgG class | IgM class | IgA class | |||
| Pool 1 | 13-23 | 1:5,200/1:620 | 1:2,400/1:140 | 1:790/1:250 | 3 |
| Pool 2 | 37-54 | 1:7,100/− | 1:610/1:110 | 1:590/− | 1 |
| Pool 3 | 24-36 | 1:98,000/1:4,300 | 1:3,500/1:4,600 | 1:2,000/1:1,900 | 4 |
| 03-2150 | 40 | 1:32,000/1:4,000 | 1:4,200/1:1,200 | 1:2,400/1:630 | 3 |
| 08-1340 | 2,088 (5.7) | 1:3,400/− | −/− | −/− | 3 |
Relative titers of anti-HEV ORF2 and anti-HEV ORF3 were determined by the methods described in Materials and Methods. −, negative (<1:100) for the indicated class of antibody.
Pooled sera were obtained during the convalescent phase, on the indicated days after disease onset, from three patients with hepatitis E.
All samples were negative for HEV RNA.
A solution of 0.2 ml containing the same amount of HEV RNA (serum sample, fecal suspension, or culture supernatant; 2.0 × 104 or 5.0 × 104 copies) and each of various antibody-positive serum samples diluted at 1:2.5, 1:25, or 1:250 with PBS(−) containing 0.2% BSA as well as anti-ORF2 monoclonal antibody (MAb) (H6225) (51), anti-ORF3 MAb (TA0536) (52), or negative-control MAb (905) (46) at a concentration of 2 mg/ml, was incubated at room temperature for 1 h and then inoculated onto monolayers of A549 cells in a six-well microplate. In addition, a solution of 0.2 ml containing the same amount of culture supernatant treated with lipid solvent and/or protease, as well as anti-ORF2 MAb (H6225; 0.2 or 2 mg/ml) (51), anti-ORF3 MAb (TA0536; 0.2 or 2 mg/ml) (52), and antibody-positive serum sample pool 1 diluted 1:25 or 1:250, was incubated at room temperature for 1 h and then inoculated onto A549 cells. After 1 h, the supernatant was removed and 2 ml of maintenance medium was added. The protocol after infection and the maintenance of cell culture were performed as described above.
Equilibrium centrifugation in sucrose density gradient.
A density gradient was prepared in an SW60Ti tube (Beckman Coulter Inc., Fullerton, CA) (0.8 ml of 60% [wt/wt] sucrose, 0.6 ml each of 50%, 40%, 30%, and 20% [wt/wt] sucrose, and 0.3 ml of 10% [wt/wt] sucrose in Tris-HCl buffer [0.01 M, pH 7.5] supplemented with 1 mM EDTA and 150 mM NaCl [TEN]). One hundred microliters each of serum sample, a fecal suspension containing the JE03-1760F strain, and a culture supernatant containing a cell culture-generated JE03-1760F variant, as well as culture supernatants treated with 5% (vol/vol) Tween 20, 10% (vol/vol) chloroform, or a solution containing 0.1% (vol/vol) NP-40, 0.1% (vol/vol) 2-mercaptoethanol (2-ME), and/or 0.1% (wt/vol) pronase E, was layered onto the surface of the gradient and overlaid with 0.4 ml of TEN. The tube was centrifuged at 179,200 × g at 10°C for 48 h, and 150-μl fractions were then recovered from the surface. The density of each fraction was measured by refractometry.
Immuno-capture RT-PCR assay.
The immuno-capture RT-PCR assay was performed by a previously described method (51). Briefly, anti-ORF2 MAb (H6225), anti-ORF3 MAb (TA0536), anti-human IgG MAb (G19) (Institute of Immunology Co. Ltd., Tokyo, Japan), anti-human IgM MAb (M49) (58), anti-human IgA MAb (A13) (30), and MAb 905, raised against hepatitis B virus (HBV) e antigen (46) and used as a negative control, were used in this assay. These MAbs were biotinylated and purified by a previously described method (51). The wells of a Reacti-Bind high-binding-capacity streptavidin-coated plate (Thermo Fisher Scientific Inc., Pittsburgh, PA) were washed with saline, and 100 μl of 1 μg/ml biotinylated MAb in PBS containing 0.1% BSA was added to each well. The wells were incubated with gentle shaking at room temperature for 1 h and were then washed with saline. One hundred microliters of each virus sample was added to each well, incubated with gentle shaking at room temperature for 2 h, and then left standing at 4°C overnight. The solution in each well was removed, and the wells were washed with saline. The RNA in each well was then extracted with TRIzol-LS reagent and subjected to quantitative detection of HEV RNA. In addition, to examine whether prior treatment with detergents and/or protease affects the binding efficiency of MAbs and HEV particles, 5 μl of serum sample diluted 1:50 with TEN, of culture supernatant, or of fecal suspension was mixed with 50 μl of Tween 20 (0.22% or 5.5%) or a solution containing 0.11% NP-40, 0.11% 2-ME, and/or 0.11% pronase E, incubated at 37°C for 2 h, diluted 1:100 with PBS containing 0.1% BSA, and then subjected to the immuno-capture RT-PCR assay as described above.
Immunoprecipitation of HEV particles.
HEV particles complexed with immunoglobulins were detected as follows. A 50-μl aliquot of test serum diluted 1:200 with saline was added to 50 μl of goat antiserum to human IgG/IgM/IgA (whole molecule) (ICN Pharmaceuticals Inc., Aurora, OH) or to normal goat serum (Chemicon International Inc., Temecula, CA). As a control, fecal supernatant mixed with an immune serum (03-2150) was added to goat antiserum to human IgG/IgM/IgA or to normal goat serum. Each mixture was incubated at 37°C for 1 h and at 4°C overnight and then separated into precipitate and supernatant fractions by centrifugation (at 1,600 × g at 4°C for 5 min). HEV RNA in the precipitate fraction was quantitated.
Nucleotide sequence accession numbers.
The nucleotide sequences of HEV isolates determined in the present study have been deposited in the GenBank/EMBL/DDBJ databases under the following accession numbers: AB525039 to AB525077.
RESULTS
Inoculation of serum HEV onto PLC/PRF/5 or A549 cells.
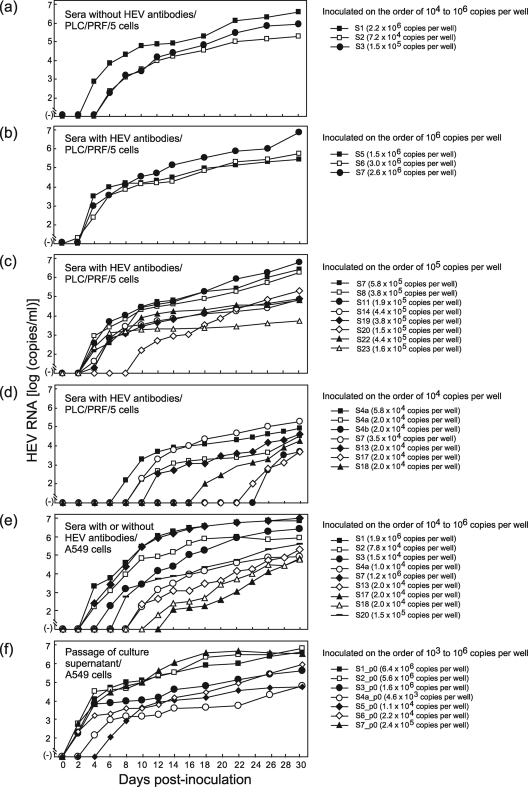
Three serum samples (S1 to S3) containing an HEV strain of genotype 3 or 4, but without HEV antibodies, were inoculated onto fresh monolayers of PLC/PRF/5 cells in a six-well microplate at a viral load of 7.2 × 104 to 2.2 × 106 copies per well (Table 3). In the primary propagation (passage 0), HEV RNA was first detected in the culture medium of PLC/PRF/5 cells on the 4th to 6th day postinoculation (dpi), with a viral load of 1.8 × 102 to 7.2 × 103 copies/ml, and continued to increase thereafter, with a maximum load of 1.8 × 105 to 3.7 × 106 copies/ml on 30 dpi (Fig. 1a). When 21 other HEV RNA-positive serum samples (S4a, S4b, and S5 to S23) that tested simultaneously positive for IgG, IgM, and IgA classes of HEV ORF2 and ORF3 antibodies were inoculated onto PLC/PRF/5 cells (7 samples were inoculated at two or three distinct viral loads), HEV progenies were released into the culture medium in all 13 samples upon inoculation at an HEV load of 3.5 × 104 to 3.0 × 106 copies per well and in 5 (31%) of 16 samples upon inoculation at an HEV load of 2.0 × 104 copies per well. At 30 dpi, the HEV load in the culture supernatant was high, at 2.9 × 105 to 7.2 × 106 (median, 5.8 × 105) copies/ml, upon inoculation of HEV on the order of 106 copies per well (Fig. 1b), while it was 5.3 × 103 to 6.0 × 106 (median, 1.3 × 105) copies/ml upon inoculation of HEV on the order of 105 copies per well (Fig. 1c). When inoculated at 2.0 × 104 to 5.8 × 104 copies per well, the viral load in the culture supernatant reached only 4.7 × 103 to 2.0 × 105 (median, 3.4 × 104) copies/ml on 30 dpi (Fig. 1d).
TABLE 3.
Detection of HEV RNA in culture supernatants of PLC/PRF/5 cells inoculated with serum samples at the indicated HEV RNA titers
| Sample no. | HEV RNA titer in inoculum (copies per well) | Detectability of HEV RNAa | HEV RNA titer at 30 dpi (copies/ml)b | HEV isolate in culture medium |
|---|---|---|---|---|
| S1 | 2.2 × 106 | + | 3.7 × 106 | S1_p0 |
| S2 | 7.2 × 104 | + | 1.8 × 105 | S2_p0 |
| S3 | 1.5 × 105 | + | 8.9 × 105 | S3_p0 |
| S4a | 5.8 × 104 | + | 8.3 × 104 | S4a-1_p0 |
| 2.0 × 104 | + | 3.4 × 104 | S4a-2_p0 | |
| S4b | 2.0 × 104 | + | 5.0 × 103 | S4b_p0 |
| S5 | 1.5 × 106 | + | 2.9 × 105 | S5_p0 |
| S6 | 3.0 × 106 | + | 5.8 × 105 | S6_p0 |
| S7 | 2.6 × 106 | + | 7.2 × 106 | S7-1_p0 |
| 5.8 × 105 | + | 2.6 × 106 | S7-2_p0 | |
| 3.5 × 104 | + | 2.0 × 105 | S7-3_p0 | |
| S8 | 3.8 × 105 | + | 1.7 × 106 | S8_p0 |
| 2.0 × 104 | − | NA | ||
| S9 | 2.0 × 104 | − | NA | |
| S10 | 2.0 × 104 | − | NA | |
| S11 | 1.9 × 105 | + | 6.0 × 106 | S11_p0 |
| 2.0 × 104 | − | NA | ||
| S12 | 2.0 × 104 | − | NA | |
| S13 | 2.0 × 104 | + | 4.3 × 104 | S13_p0 |
| S14 | 4.4 × 105 | + | 6.7 × 104 | S14_p0 |
| 2.0 × 104 | − | NA | ||
| S15 | 2.0 × 104 | − | NA | |
| S16 | 2.0 × 104 | − | NA | |
| S17 | 2.0 × 104 | + | 4.7 × 103 | S17_p0 |
| S18 | 2.0 × 104 | + | 1.7 × 104 | S18_p0 |
| S19 | 3.8 × 105 | + | 7.4 × 104 | S19_p0 |
| 2.0 × 104 | − | NA | ||
| S20 | 1.5 × 105 | + | 1.9 × 105 | S20_p0 |
| 2.0 × 104 | − | NA | ||
| S21 | 2.0 × 104 | − | NA | |
| S22 | 4.4 × 105 | + | 6.7 × 104 | S22_p0 |
| S23 | 1.6 × 105 | + | 5.3 × 103 | S23_p0 |
−, negative for HEV RNA during the observation period (0 to 30 dpi).
NA, not applicable.
FIG. 1.
Quantitation of HEV RNA in culture supernatants of PLC/PRF/5 or A549 cells inoculated with serum samples, with or without HEV antibodies (passage 0) and at various HEV loads (104 to 106 copies per well) (a to e), or in culture medium of passage 0 (f). (f) The harvested culture supernatant of passage 0 was purified by passage through a microfilter with a pore size of 0.22 μm (see Materials and Methods) and then inoculated onto fresh A549 cells.
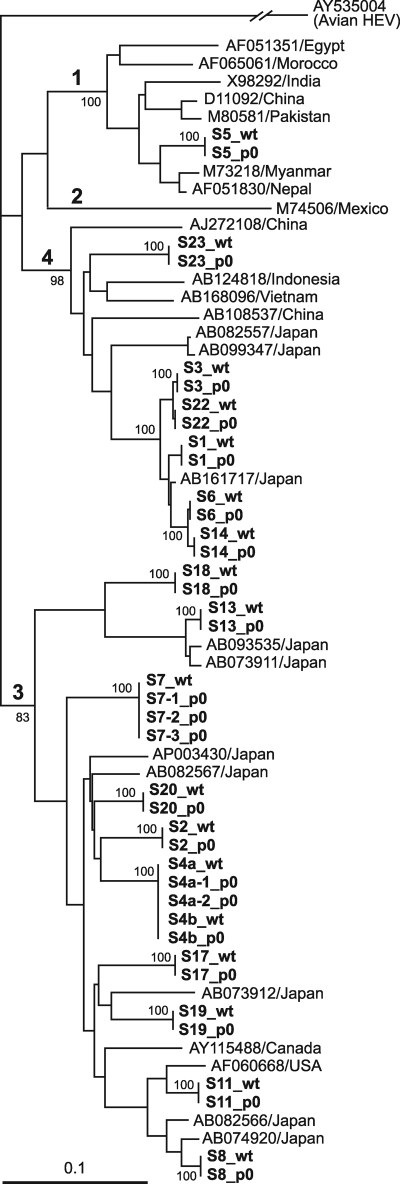
The serum-derived wild-type HEV and its cell culture-generated variant were 100% identical in nucleotide sequence for 412 nt within the ORF2 gene for all 21 pairs (Fig. 1a to d). The phylogenetic tree constructed based on the 412-nt ORF2 sequence depicted that each pair of the wild type and its cell culture-produced variant segregated into a cluster within genotype 1, 3, or 4, with a bootstrap value of 100% (Fig. 2), confirming that various HEV strains of serum origin were successfully propagated in cultured cells.
FIG. 2.
Phylogenetic tree constructed by the neighbor-joining method, based on the partial nucleotide sequence of the ORF2 region (412 nt) of 64 HEV isolates, using an avian HEV (GenBank accession no. AY535004) as an outgroup. In addition to 39 HEV isolates obtained in the present study, which are indicated in bold, 24 representative HEV isolates of genotypes 1 to 4 whose common 412-nt sequence is known were included for comparison, and their accession numbers, followed by the names of the countries where they were isolated, are indicated. “wt” denotes an HEV isolate recovered from a serum sample that was used to inoculate cells, and “p0” represents an HEV isolate obtained from culture supernatant harvested 30 days after inoculation of the corresponding serum sample. Bootstrap values are indicated for the major nodes as percentages of the data obtained from 1,000 resamplings. Bar, 0.1 nucleotide substitution per site.
When representative serum samples, including 3 antibody-negative (S1 to S3) and 6 antibody-positive (S4a, S7, S13, S17, S18, and S20) samples, were inoculated onto A549 cells at an HEV load ranging from 1.0 × 104 copies per well to 1.9 × 106 copies per well, HEV progenies emerged in the culture medium in all samples tested, regardless of the presence or absence of HEV antibodies in the serum samples (Fig. 1e). It was notable that during primary propagation, CPE was not observed in the PLC/PRF/5 and A549 cells, despite differences in the titer of the inoculum and HEV load in each inoculation.
Supernatant passage of HEV in A549 cells.
When HEV progenies in randomly selected culture supernatants of primary propagation (passage 0) were inoculated onto PLC/PRF/5 and A549 cells at a load of 1.0 × 104 copies per well, A549 cells supported more efficient multiplication of HEV than did PLC/PRF/5 cells. Therefore, in passage, A549 cells were used as host cells for virus culture in place of PLC/PRF/5 cells. All HEV progenies (S1_p0 to S3_p0, S4a_p0, S4b_p0, and S5_p0 to S23_p0) released into the culture medium of PLC/PRF/5 cells that had been inoculated with serum samples (S1 to S3, S4a, S4b, and S5 to S23, respectively) (Table 3) were successfully passaged in A549 cells, as depicted in Fig. 1f for representative progenies. Although inoculated at various HEV loads, ranging from 4.6 × 103 copies per well to 6.4 × 106 copies per well, HEV RNA in the passage appeared at 2 to 6 dpi and reached a maximum load of 5.3 × 104 to 6.6 × 106 copies/ml at 30 dpi in the culture medium of A549 cells (Fig. 1f). During the passage, CPE was not observed in the A549 cells, despite differences in the changing profile of HEV load in each passage.
Neutralization of serum HEV by anti-HEV immune sera or MAbs.
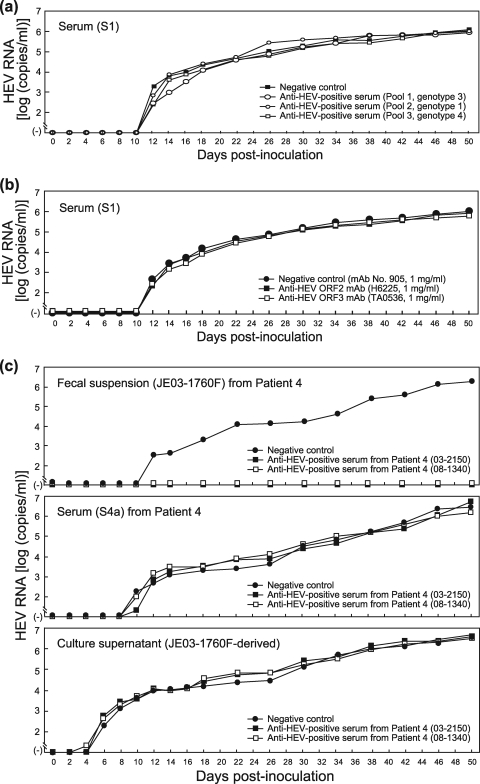
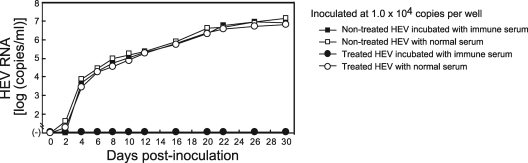
Three serum samples (pool 1 to pool 3) (Table 2), containing IgG, IgM, and IgA classes of anti-HEV antibodies obtained from patients who had contracted infection with HEV genotypes 3, 1, and 4, respectively, were tested for the ability to neutralize a serum-derived HEV strain of genotype 3 without concurrent HEV antibodies (S1) in the present cell culture system. One hundred microliters of each serum pool (pool 1 to pool 3), diluted 1:2.5 with PBS(−), was mixed with an equal volume of the diluted serum (S1) containing 5.0 × 104 copies of HEV, kept at room temperature for 1 h, and then inoculated onto monolayers of A549 cells in a six-well microplate. In each well containing a sample from pool 1, pool 2, or pool 3, HEV RNA was first detectable at 12 dpi and continued to increase, reaching 106 copies/ml at 50 dpi, as in wells with the control serum (anti-HEV-negative serum) (Fig. 3a). Of note, HEV particles in feces were neutralized by each serum pool (pool 1 to pool 3), even diluted 1:250, indicating that HEV particles in serum are nonneutralizable by immune sera with a 100-fold-higher concentration of anti-HEV antibodies in this cell culture system. In addition, HEV particles in the serum samples (S1) were not neutralized by anti-ORF2 MAb (H6225) or anti-ORF3 MAb (TA0536), even at a final concentration of 1 mg/ml (Fig. 3b).
FIG. 3.
(a) Quantitation of HEV RNA in culture supernatants of A549 cells inoculated with a serum sample (S1) with a viral load of 5.0 × 105 copies/ml that had been mixed with a pooled serum sample obtained during the early convalescent phase (pool 1, 2, or 3) and positive for IgM-, IgA-, and IgG-class anti-HEV antibodies or with an antibody-negative serum and cultured for 50 days. (b) Quantitation of HEV RNA in culture supernatants of A549 cells inoculated with a serum sample (S1) with a viral load of 5.0 × 105 copies/ml that had been mixed with an anti-ORF2 MAb (H6225) (51), anti-ORF3 MAb (TA0536) (52), or negative-control MAb (905) (46) at a final concentration of 1 mg/ml and cultured for 50 days. (c) Quantitation of HEV RNA in culture supernatants of A549 cells inoculated with a fecal suspension (JE03-1760F), a serum sample (S4a), or a culture supernatant of JE03-1760F (patient 4) origin with an HEV load of 2.0 × 105 copies/ml that had been mixed with a serum sample (03-2150) positive for IgM-, IgA-, and IgG-class anti-HEV antibodies but negative for HEV RNA, another serum sample (08-1340) positive only for IgG anti-HEV, or an antibody-negative serum and cultured for the indicated number of days. The fecal suspension, serum sample (S4a), and culture supernatant contained HEV of the same strain (JE03-1760F), and serum samples (S4a, 03-2150, and 08-1340) were obtained from the JE03-1760F patient (patient 4 in this study) 15, 40, and 2,088 days, respectively, after disease onset.
To confirm this observation, 100 μl each of serum samples containing IgG, IgM, and IgA classes of anti-HEV antibodies (03-2150 in Table 2) or IgG-class anti-HEV only (08-1340), diluted 1:2.5, both of which were obtained from the JE03-1760F patient (patient 4), 40 days and 5.7 years, respectively, after disease onset, was mixed with an equal volume of diluted inoculum of JE03-1760F origin (2.0 × 104 copies per well) and kept at room temperature for 1 h. When a fecal suspension containing the JE03-1760F strain was used as an inoculum and mixed with serum 03-2150 or 08-1340, the harvested culture supernatants were negative for HEV RNA throughout the observation period of 50 days, although HEV RNA appeared at 12 dpi and increased thereafter (1.8 × 106 copies/ml at 50 dpi) when mixed with the control serum (anti-HEV-negative serum), indicating that sera 03-2150 and 08-1340 have the ability to neutralize fecal HEV in this cell culture system (Fig. 3c). However, when serum S4a, obtained from patient 4 15 days after disease onset, was used as an inoculum, HEV RNA became detectable in the culture supernatant on the 10th day and thereafter, even with prior incubation with serum 03-2150 or 08-1340. Similarly, when the cell culture-generated JE03-1760F variant was used as an inoculum, the harvested culture supernatants were positive for HEV RNA on the 6th day after inoculation (Fig. 3c). These results indicate that immune sera have little or no ability to neutralize even HEV particles of the same strain in serum and culture supernatant.
Buoyant density of serum HEV.
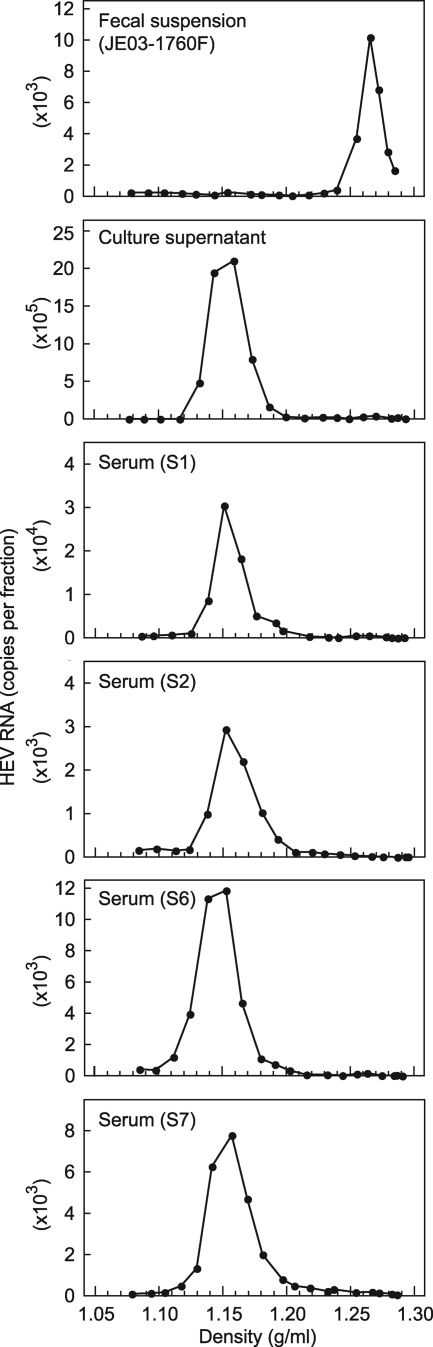
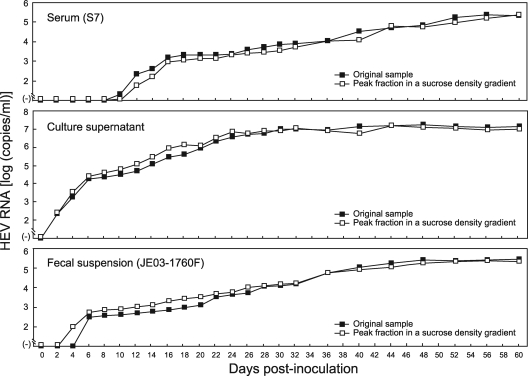
In an attempt to clarify why HEV in serum was nonneutralizable, HEV RNA-positive serum samples, a fecal suspension containing the JE03-1760F strain, and a culture supernatant containing the cell culture-produced JE03-1760F variant were subjected to ultracentrifugation in a sucrose density gradient. Like the case for the HEV progenies in culture medium, HEV particles in serum samples, with or without HEV antibodies, banded at a density of 1.15 to 1.16 g/ml, which was markedly lower than that for HEV particles in feces, at 1.27 to 1.28 g/ml (Fig. 4), suggesting that HEV particles in serum and culture supernatant are associated with a very-low-density material, possibly lipids.
FIG. 4.
Sucrose density gradient fractionation of HEV in a fecal specimen (JE03-1760F) and in a culture supernatant containing cell culture-produced HEV, as controls, and in serum samples with (S6 and S7) and without (S1 and S2) HEV antibodies.
When HEV particles in the peak fraction from the ultracentrifugation procedure that were derived from a serum sample (S7), a culture supernatant, and a fecal suspension (JE03-1760F) were inoculated onto A549 cells at HEV loads of 1.0 × 104, 1.0 × 105, and 1.2 × 104 copies per well, respectively, the appearance and increase of the HEV progenies in the culture supernatant were nearly identical to those of the respective original samples (Fig. 5), indicating that HEV particles in serum and culture supernatant banding at a density of 1.15 to 1.16 g/ml are infectious in cell culture and that the peak fraction from ultracentrifugation contains infectious particles corresponding to those in the original, unfractionated sample, like the case for those in feces, banding at 1.27 to 1.28 g/ml.
FIG. 5.
Comparison of HEV RNA titers in culture supernatants of cultured cells obtained during the observation period of 60 days after inoculation between an original serum sample (S7) and its peak fraction in a sucrose density gradient, between an original culture supernatant and its peak fraction in a density gradient, and between an original fecal suspension (JE03-1760F) and its peak fraction in a density gradient (Fig. 4). The loads of HEV inoculated were 1.0 × 104 copies per well for the serum sample and its peak fraction, 1.0 × 105 copies per well for the culture supernatant and its peak fraction, and 1.2 × 104 copies per well for the fecal specimen and its peak fraction.
Association of serum HEV with human immunoglobulins.
To characterize the association of HEV particles with human immunoglobulins in the sera of HEV-infected patients, the HEV virions in the serum samples (S1 to S3, S6, S7, S19, and S21), with or without circulating HEV antibodies, were subjected to immunoprecipitation with goat anti-human IgG/IgM/IgA, and HEV RNA was quantitated in the precipitate fraction. Only ≤8.1% of the PCR signals were recovered from the precipitate fraction (Table 4), suggesting that HEV exists in the circulation predominantly as free virions, even in the presence of high-titer HEV antibodies. To confirm this finding, immuno-capture RT-PCR was performed to detect HEV virions in the peak fraction from sucrose density gradient ultracentrifugation, using anti-human IgG/IgM/IgA MAbs. Only ≤8.5% of viral particles in serum were captured by anti-human IgG/IgM/IgA MAbs, despite the presence of HEV antibodies (Table 5), indicating that the great majority of HEV particles in the circulation exist as free virions not complexed with IgG, IgM, or IgA.
TABLE 4.
Immunoprecipitation of HEV particles in serum with goat serum against human IgG/IgM/IgA
| Sample with HEV RNA (copies/test) | Immunoprecipitation (%) with: |
|
|---|---|---|
| Goat anti-human IgG/IgM/IgA | Normal goat serum | |
| HEV RNA-positive seraa | ||
| Anti-HEV-negative sera | ||
| S1 (5.8 × 103) | 3.5 | 1.7 |
| S2 (2.8 × 102) | 6.0 | 0.0 |
| S3 (4.5 × 102) | 0.9 | 0.0 |
| Anti-HEV-positive sera | ||
| S6 (1.9 × 103) | 5.4 | 1.8 |
| S7 (2.4 × 103) | 5.4 | 0.4 |
| S19 (4.5 × 103) | 8.1 | 0.4 |
| S21 (1.0 × 104) | 2.4 | 1.0 |
| Fecal suspension (7.0 × 104) mixed with serum sample 03-2150b | 98.2 | 0.0 |
TABLE 5.
Bound HEV after immuno-capture by anti-human IgG, IgM, and IgA MAbs
| Serum or fecal sample | Total copy no. of HEV applied to each well | % of captured HEV in total HEV per well |
|
|---|---|---|---|
| Anti-human IgG/IgM/IgA MAbs (G19/M49/A13) | Negative control (MAb 905) | ||
| HEV RNA-positive seraa | |||
| Anti-HEV-negative sera | |||
| S1 | 2.5 × 103 | 1.7 | 0.0 |
| S2 | 2.7 × 102 | 2.1 | 0.0 |
| S3 | 3.5 × 102 | 1.8 | 0.0 |
| Anti-HEV-positive sera | |||
| S4a | 2.9 × 102 | 0.9 | 0.0 |
| S6 | 6.5 × 103 | 8.5 | 0.2 |
| S7 | 3.7 × 103 | 6.6 | 0.5 |
| Fecal suspension mixed with serum sample 03-2150b | 7.8 × 103 | 84.4 | 0.1 |
Association of serum HEV with lipids and ORF3 protein.
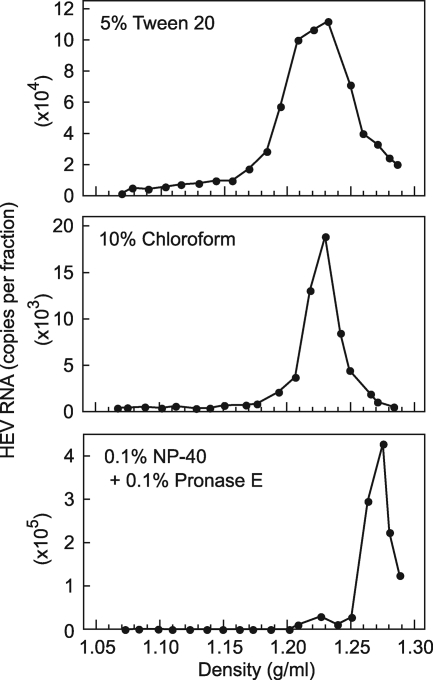
To examine whether HEV particles in serum are associated with lipids and HEV ORF3 protein, similar to those in culture supernatant (63), the immuno-capture RT-PCR assay was performed using anti-ORF2 MAb (H6225) and anti-ORF3 MAb (TA0536) (Table 6). Like the case for fecal HEV, although 92.2 to 93.1% of viral particles were captured by anti-ORF2 MAb, no virus was captured by anti-ORF3 MAb, regardless of the presence or absence of 0.2% Tween 20. On the other hand, when viral particles in a viremic serum (S1) and a culture supernatant of HEV-infected PLC/PRF/5 cells were tested, they were both captured by anti-ORF2 and anti-ORF3 MAbs, although only partially (16.5 to 38.5%) in the presence of 0.2% Tween 20. Of note, upon treatment with 5% Tween 20, the efficiency of binding of HEV particles in serum and culture supernatant by anti-ORF2 MAb increased to 55.7% and 69.0%, respectively, and that by anti-ORF3 MAb increased to 90.4% and 76.8%, respectively. However, the buoyant density of HEV particles treated with 5% Tween 20, even after treatment with 10% chloroform, did not shift to 1.27 to 1.28 g/ml, corresponding to that of fecal HEV: the Tween 20-treated HEV particles were distributed at 1.17 to 1.29 g/ml, peaking at 1.23 g/ml, and the chloroform-treated HEV particles were found at 1.19 to 1.27 g/ml, with a peak density of 1.23 g/ml (Fig. 6). Next, 5 μl of the HEV RNA-positive serum sample (S1), diluted 1:50 with TEN, was mixed with 50 μl of TEN buffer containing 0.1% NP-40, 0.1% 2-ME, and 0.1% pronase E and incubated at 37°C for 2 h, a method reported to liberate core particles from HBV virions (45). The efficiency of binding of HEV particles in serum by anti-ORF2 MAb increased substantially, to 86.3%, and that by anti-ORF3 MAb decreased to 0%. The buoyant density of the treated HEV particles in serum shifted to 1.27 g/ml, similar to that of fecal HEV particles as well as cell culture-generated HEV particles that had been treated with NP-40, 2-ME, and pronase E. Upon treatment with NP-40 and pronase E (without the addition of 2-ME), a similar finding was obtained (Fig. 6). These results suggest that serum-derived HEV particles are associated with lipids and ORF3 protein, similar to those in culture supernatant.
TABLE 6.
Reactivities of anti-ORF2 MAb and anti-ORF3 MAb with HEV particles treated with various reagents, evaluated by immuno-capture PCR
| Treatment | % of captured HEV in total HEV per well |
|||||
|---|---|---|---|---|---|---|
| Serum |
Culture supernatant |
Feces |
||||
| Anti-ORF2 MAb (H6225) | Anti-ORF3 MAb (TA0536) | Anti-ORF2 MAb (H6225) | Anti-ORF3 MAb (TA0536) | Anti-ORF2 MAb (H6225) | Anti-ORF3 MAb (TA0536) | |
| Tween 20 | ||||||
| 0.2% | 38.5 | 37.6 | 22.1 | 16.5 | 92.2 | 0.0 |
| 5% | 55.7 | 90.4 | 69.0 | 76.8 | 95.7 | 0.4 |
| 10% Chloroform | ||||||
| 25°C, 10 min | 28.4 | 64.7 | 87.3 | 54.4 | 92.0 | 0.1 |
| 37°C, 30 min | 35.7 | 21.5 | 62.9 | 9.5 | 99.2 | 0.0 |
| NP-40 + 2-ME + pronase Ea | 86.3 | 0.0 | 98.7 | 0.2 | 99.9 | 1.3 |
| NP-40 + 2-MEa | 78.3 | 84.2 | 97.2 | 77.9 | 99.8 | 0.4 |
| NP-40 + pronase Ea | 90.4 | 0.6 | 99.0 | 0.2 | 99.4 | 0.0 |
| None | 1.0 | 1.5 | 6.3 | 5.7 | 93.1 | 0.0 |
Treated with 0.1% NP-40, 0.1% 2-ME, and/or 0.1% pronase E.
FIG. 6.
Sucrose density gradient fractionation of HEV in culture supernatants treated with 5% Tween 20, 10% chloroform, or a solution containing 0.1% NP-40 and 0.1% pronase E.
Neutralization of HEV particles in cell culture treated with lipid solvents and/or protease by an anti-HEV immune serum or MAb (anti-ORF2 or anti-ORF3).
To examine whether the HEV particles in a cell culture treated with lipid solvent can be neutralized by an anti-HEV immune serum or MAb, 100 μl of a serum pool (pool 1; diluted 1:25 or 1:250), anti-ORF2 MAb (H6225; 0.2 or 2 mg/ml), or anti-ORF3 MAb (TA0536; 0.2 or 2 mg/ml) was mixed with an equal volume of the diluted culture supernatant (treated with 5% Tween 20 and containing 2.4 × 104 copies of HEV), kept at room temperature for 1 h, and then inoculated onto A549 cells in a six-well microplate. The treated HEV particles replicated as efficiently as those without detergent treatment and were neutralized partially by anti-HEV immune serum, anti-ORF2 MAb, and anti-ORF3 MAb, in an antibody dose-dependent manner (Table 7).
TABLE 7.
Neutralization of cell culture-generated HEV particles treated with 5% Tween 20 by an anti-HEV immune serum or MAb (anti-ORF2 or anti-ORF3)
| Days after inoculation | HEV RNA titer (copies/ml) in culture supernatanta |
|||||||
|---|---|---|---|---|---|---|---|---|
| Nontreated HEV (without antibody)b | 5% Tween 20-treated HEV |
|||||||
| Without antibodyb | Immune serum (pool 1) |
Anti-ORF2 MAb (H6225) |
Anti-ORF3 MAb (TA0536) |
|||||
| 1:50 | 1:500 | 1 mg/mlc | 0.1 mg/mlc | 1 mg/mlc | 0.1 mg/mlc | |||
| 0 | − | − | − | − | − | − | − | − |
| 2 | 20 | 40 | − | − | − | − | − | − |
| 4 | 9.9 × 103 | 7.5 × 103 | − | − | − | − | − | 160 |
| 6 | 5.7 × 104 | 3.1 × 104 | − | 60 | − | 160 | 120 | 3.3 × 104 |
| 8 | 2.5 × 105 | 6.1 × 104 | 40 | 160 | − | 760 | 3.6 × 103 | 8.8 × 104 |
| 10 | 9.9 × 105 | 7.5 × 105 | 180 | 720 | − | 1.0 × 103 | 1.7 × 104 | 3.4 × 105 |
| 12 | 2.8 × 106 | 2.2 × 106 | 660 | 3.4 × 103 | 140 | 1.1 × 104 | 2.0 × 104 | 9.4 × 105 |
| 14 | 5.9 × 106 | 5.5 × 106 | 1.1 × 103 | 2.1 × 104 | 340 | 7.2 × 104 | 4.5 × 104 | 4.0 × 106 |
−, negative for HEV RNA.
No prior incubation with anti-HEV antibody.
Final concentration of anti-HEV antibody in each inoculum.
To further explore whether cell culture-produced HEV particles treated with detergent and protease, with a buoyant density of 1.27 to 1.28 g/ml in sucrose, can be neutralized by an anti-HEV immune serum, 100 μl of immune serum (pool 1; diluted 1:25) was mixed with an equal volume of the culture supernatant (containing 1.0 × 104 copies of HEV that had been treated with 0.1% NP-40 and 0.1% pronase E and diluted 1:100), kept at room temperature for 1 h, and then inoculated onto A549 cells. When cells were inoculated with the treated HEV particles, HEV RNA was first detected in the culture medium at 4 dpi and continued to be detectable up to the end of the observation period of 30 days, reaching a maximum load of 6.3 × 106 copies/ml, similar to the case for particles without treatment (Fig. 7). In contrast, in wells inoculated with the treated HEV particles that had been incubated with an immune serum, HEV RNA was not detectable until 30 days, indicating that virions treated with lipid solvent and protease, possessing the same character as virions from feces, are neutralizable by anti-HEV immune sera.
FIG. 7.
Quantitation of HEV RNA in culture supernatants of A549 cells inoculated with a culture supernatant with a viral load of 1.0 × 105 copies/ml that had been treated with or without 0.1% NP-40 and 0.1% pronase E, mixed with a normal serum or pooled serum sample obtained at the early convalescent phase (pool 1) and positive for IgM-, IgA-, and IgG-class anti-HEV ORF2 and ORF3 antibodies, and cultured for 30 days.
DISCUSSION
Only a limited number of HEV strains of genotype 3 (JE03-1760F) and genotype 4 (HE-JF5/15F) in fecal specimens have been propagated efficiently in PLC/PRF/5 and A549 cells (55, 56). However, the present study revealed that various HEV strains of genotype 1, 3, or 4 in serum samples obtained from patients with domestic or imported hepatitis E can also infect and replicate efficiently in PLC/PRF/5 and A549 cells. Of note, when inoculated at an HEV load of ≥3.5 × 104 copies per well in a six-well microplate, HEV strains in all serum samples tested were successfully propagated in cultured cells, while upon inoculation at 2.0 × 104 copies per well, HEV strains in only 30% of serum samples grew in cultured cells. In other words, if the HEV load in serum is 3.5 × 105 copies/ml or higher, PLC/PRF/5 and A549 cells can support multiplication of HEV strains in all serum samples, with or without concurrent HEV antibodies, irrespective of HEV genotype. Progenies of serum-derived HEV strains in culture supernatant were successfully passaged in A549 cells, indicating that HEV progenies of serum origin released from cultured cells are infectious, similar to those of feces origin.
It has been reported that fecal samples contain various amounts of metabolic and phenolic compounds and polysaccharides and have a very heterogeneous composition (62). Like the case for the HE-JF5/15F strain which was used for the establishment of a genotype 4 HEV cell culture system (56), when the fecal specimen was inoculated onto monolayers of PLC/PRF/5 cells at a dilution of 1:5, as was done for the JE03-1760F specimen (55), nearly all cells detached from the bottoms of all six wells inoculated, probably due to cell toxicity of the fecal sample. Approximately 30% of cells detached from the bottoms of the wells just after inoculation of the 10-fold-diluted fecal suspension, while such cell damage was not observed in any of the six wells tested when the 25-fold-diluted fecal specimen was used for inoculation of HEV onto PLC/PRF/5 or A549 cells, indicating that at least 5- to 25-fold dilution may be required when a fecal specimen is used as the inoculum in our cell culture system for HEV. In contrast, serum samples were successfully inoculated at a dilution of only 1:2, without apparent cell detachment, in the present study. In general, serum samples are routinely collected at the first examination from patients with a clinical diagnosis of acute hepatitis and are used for serological and molecular tests of hepatitis virus infection, while fecal specimens are rarely collected from patients for such purposes, although they may be collected after the diagnosis of hepatitis A or E is established. Therefore, serum samples are available earlier after disease onset than fecal specimens for all patients diagnosed as having hepatitis E and most likely have higher HEV loads (50), which will greatly increase the ease with which inoculation of clinical samples with HEV onto cultured cells can be carried out.
Since transmission of HEV and subsequent development of hepatitis E via transfusion of viremic blood have been reported (1, 2, 18, 24, 25, 28, 54), our present observation that various HEV strains in serum samples were able to replicate in cultured cells and produce infectious progenies in the culture supernatant may be conceivable. Unexpectedly, however, HEV particles in serum samples grew in cultured cells despite the presence of HEV antibodies. The present studies indicated that over 90% of HEV particles in the circulation are free of immunoglobulins, even in the presence of IgM HEV antibodies. Similar to cell culture-generated HEV particles, HEV particles in serum were nonneutralizable by immune sera that can definitely neutralize the infection of HEV in feces in the cell culture system, and they banded at a sucrose density of 1.15 to 1.16 g/ml, which was identical to that in culture supernatant but markedly lower than that in feces (1.27 to 1.28 g/ml). Without prior treatment with detergent, few or no virus particles in both serum and cell culture were captured by anti-ORF2 MAb and anti-ORF3 MAb. However, after treatment with 5% Tween 20, the efficiencies of binding of HEV particles in serum by anti-ORF2 MAb and anti-ORF3 MAb increased markedly, to 55.7% and 90.4%, respectively, showing a similar pattern to that for cell culture-produced HEV particles (Table 6). These results suggest that HEV virions in serum possess the ORF3 protein on their surfaces, in association with lipids, similar to those in the supernatant of cultured cells (63), and would reconcile the apparently contradictory observation that IgM and IgA antibodies, which are capable of viral neutralization, and infectious virus coexist in acute-phase serum specimens, as reported for hepatitis A virus (HAV) (19, 35).
Provost et al. (34) reported that the infectious HAV present in acute-phase marmoset sera banded at two densities in CsCl (1.34 g/ml and 1.15 g/ml), suggesting that the less-dense particles represent lipid-associated virions. A similar banding pattern was observed by Lemon and Binn (20), who reported that the HAV released from infected BS-C-1 cells was incompletely neutralized when it was incubated with a variety of convalescent-phase sera (nonneutralizable fraction of 17 to 32%) and that chloroform extraction of virus resulted in a substantial reduction of the nonneutralizable fraction (to <1%). Nonneutralizable HAV recovered from untreated cell culture supernatant fluids sedimented heterogeneously and less rapidly than normal virus in rate-zonal sucrose gradients and also banded at a lower density in CsCl (1.14 to 1.18 g/ml) than did normal, neutralizable virus (1.32 g/ml). It seems that HEV particles in serum and culture supernatant are similar to the nonneutralizable fraction of HAV in cell culture in that they are resistant to antibody-mediated neutralization and are likely to be associated with lipids. However, in contrast to HAV, HEV particles in serum samples and culture supernatant showed a monophasic pattern in sucrose gradients, and essentially all HEV particles in these samples appeared to be nonneutralizable. In addition, chloroform treatment did not result in a shift of HEV particles in serum and culture medium to those with a density of 1.27 to 1.28 g/ml, corresponding to that of fecal HEV. Treatment with TEN buffer containing 0.1% NP-40 and 0.1% pronase E caused a shift of HEV particles in serum and cell culture to peak at a sucrose density of 1.27 g/ml. Of practical importance, these observations suggest that HEV exists as an “enveloped” virus in the circulation, similar to cell culture-produced HEV (63).
The inability of serum anti-HEV antibodies to neutralize infection of serum-derived virions appears to be at odds with the epidemiology of HEV infection (7, 36) and with the results of preclinical and clinical trials of a recombinant ORF2-based vaccine (42). The titers of serum antibodies (at the dilution used) affect the efficiency of virus neutralization. Therefore, we cannot rule out the possibility that the titer of anti-HEV antibodies that can neutralize feces-derived virions was too low to neutralize serum-derived HEV given the different membrane compositions. However, HEV in serum was not neutralized by each immune serum (pool 1 to pool 3 or 03-2150) diluted 1:5 (final concentration), although HEV in feces was neutralized by the immune sera diluted 1:500, suggesting that HEV in serum is nonneutralizable even by immune sera with a 100-fold-higher titer of anti-HEV antibodies in our cell culture system for HEV. In support of this finding, serum-derived HEV was not neutralized by anti-ORF2 MAb (H6225) or anti-ORF3 MAb (TA0536), even at a final concentration of 1 mg/ml, although the anti-ORF2 MAb could protect efficiently against infection of fecal HEV at an antibody concentration of 10 μg/ml in our previous study (51). Natural infection with HEV usually follows ingestion of virus from water or food contaminated with HEV. Although the sequence of events that begins with entry via the gastrointestinal tract and ultimately results in hepatitis has not been resolved completely (7), and although extrahepatic sites of HEV, such as lymph node, small intestine, and colon tissues, have been identified in pigs infected with HEV (61), it is tempting to speculate that the primary site of replication for HEV is the hepatocyte, similar to that for HAV (15), and that HEV particles entering through the gastrointestinal tract, unaccompanied by lipids, can be neutralized by immune sera before reaching the liver via the portal vein. This speculation may help to explain how humans who were immunized with a recombinant ORF2-based vaccine can be protected against infection with HEV via the fecal-oral route.
It was previously reported that the ORF3 protein binds to the ORF2 capsid protein (59) and localizes to endocytic membranes (3). The apparent association of HEV virions with lipids suggests that cellular membranes play an important role in the assembly and release of HEV from noncytopathically infected cells. The immuno-capture RT-PCR assay in the present study indicated the presence of the ORF3 protein on the surfaces of not only HEV particles in cell culture but also those in serum samples, despite the absence of ORF3 protein on fecal HEV. In contrast to the HEV particles in cell culture treated with detergent and protease, those treated with detergent only (5% Tween 20) were neutralized only partially by immune serum containing anti-HEV ORF2 and ORF3 antibodies, an anti-ORF2 MAb (H6225) that can neutralize fecal HEV (51), or an anti-ORF3 MAb (TA0536) directed against a C-terminal immunodominant epitope (52) (Table 7 and Fig. 7). The ORF3 protein was accessible to anti-ORF3 MAb (TA0536) following detergent or chloroform treatment. These findings suggest that the C-terminal portion of the ORF3 protein is not exposed on the surfaces of HEV particles in serum and culture supernatant and may be covered with lipid membranes. The partial neutralization of the detergent-treated HEV virions in cell culture by anti-HEV antibodies may be ascribable to incomplete exposure of ORF2 and ORF3 proteins after treatment with detergent only. In support of this speculation, 10 to 44% of the 5% Tween 20-treated HEV particles in cell culture and serum were not captured by anti-ORF2 and anti-ORF3 MAbs in the immuno-capture studies (Table 6). The ORF3 protein is likely to act as an adapter to link various intracellular transduction pathways, creating a host cell environment favorable for HEV replication and assembly (4). Our previous study using a wild-type JE03-1760F cDNA clone and its ORF3-null mutant demonstrated that the ORF3 protein is required for virion release from cultured cells (63). Taken together, the data indicate that it is very likely that HEV particles are released from both infected cultured cells (in vitro) and infected hepatocytes (in vivo) as lipid-associated virions accompanied by ORF3 protein and that the ORF3 protein and lipids are dissociated from the virion after it is shed into the bile duct and then the duodenum, which contain detergent (deoxycholic acid) and protease (trypsin), respectively.
The crystal structure of HEV-like particles was recently elucidated (12, 64). The outer surface of the particle, which is a target for antibodies, is mainly formed by the middle and protruding domains. It was also recently reported that the dimerization domain of the recombinant capsid protein (ORF2 aa 455 to 602), most probably the surface protrusion, is the neutralizing antibody recognition site of HEV and is essential for HEV-host interactions (21). However, a putative receptor for HEV has not yet been isolated, and virtually nothing is known about the mechanism by which HEV enters susceptible cells. Our previous study on virus attachment determined that cloned pJE03-1760F/wt virus in culture medium adsorbed to A549 cells with lower efficiency than that in cell lysate (approximately 3% and 14%, respectively, of the input virus within 1 h), peaking at a sucrose density of 1.27 to 1.28 g/ml (63), suggesting that nonneutralizable HEV particles in serum samples and culture supernatant, which are associated with lipids and band at 1.15 to 1.16 g/ml in sucrose gradients, can bind (though inefficiently) to cultured cells. Our present study indicated that even nonneutralizable HEV particles in serum samples and culture supernatant can be propagated in PLC/PRF/5 and A549 cells. It is interesting that a lipid solvent did not abolish or increase the infectivity of the newly released virions in the infected cell culture, and both cell culture-generated HEV particles and those that were treated with detergent and/or protease were propagated in cultured cells, with nearly identical efficiencies (Table 7 and Fig. 7), regardless of the presence or absence of the ORF3 protein and lipids on the surface. Taken altogether, it seems unlikely that only a fraction of HEV particles in serum samples and culture supernatant, partially unaccompanied by lipids, can bind to the cell surface receptor and enter susceptible cells. In an attempt to provide a plausible explanation(s) for this unexpected and intriguing observed phenomenon, it should be clarified in future studies whether and how nonneutralizable HEV particles in serum and culture supernatant can enter susceptible cells.
In conclusion, the present study indicated that PLC/PRF/5 and A549 cells can support efficient multiplication and passage of a wide variety of HEV strains (genotype 1, 3, or 4) in serum samples obtained from patients with imported or domestic hepatitis E and that HEV particles in serum samples are associated with lipids and ORF3 protein, similar to those in culture supernatant, most likely contributing to the assembly and release of HEV from noncytopathically infected cells in vivo and in vitro. Although lipid-associated virions of other nonenveloped viruses, such as HAV, have been reported (20, 34, 37), the underlying mechanism by which HEV particles in the circulation can be propagated in cultured cells, similar to those in culture supernatant, despite the fact that they are seemingly “enveloped” and nonneutralizable by immune sera, needs to be clarified in future studies.
Acknowledgments
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and from the Ministry of Health, Labor, and Welfare of Japan.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Arankalle, V. A., and L. P. Chobe. 2000. Retrospective analysis of blood transfusion recipients: evidence for post-transfusion hepatitis E. Vox Sang. 79:72-74. [DOI] [PubMed] [Google Scholar]
- 2.Boxall, E., A. Herborn, G. Kochethu, G. Pratt, D. Adams, S. Ijaz, and C. G. Teo. 2006. Transfusion-transmitted hepatitis E in a ‘nonhyperendemic’ country. Transfus. Med. 16:79-83. [DOI] [PubMed] [Google Scholar]
- 3.Chandra, V., A. Kar-Roy, S. Kumari, S. Mayor, and S. Jameel. 2008. The hepatitis E virus ORF3 protein modulates epidermal growth factor receptor trafficking, STAT3 translocation, and the acute-phase response. J. Virol. 82:7100-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandra, V., S. Taneja, M. Kalia, and S. Jameel. 2008. Molecular biology and pathogenesis of hepatitis E virus. J. Biosci. 33:451-464. [DOI] [PubMed] [Google Scholar]
- 5.Dalton, H. R., R. Bendall, S. Ijaz, and M. Banks. 2008. Hepatitis E: an emerging infection in developed countries. Lancet Infect. Dis. 8:698-709. [DOI] [PubMed] [Google Scholar]
- 6.Emerson, S. U., D. Anderson, A. Arankalle, X. J. Meng, M. Purdy, G. G. Schlauder, and S. A. Tsarev. 2004. Hepevirus, p. 853-855. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: VIIIth report of the ICTV. Elsevier/Academic Press, London, United Kingdom.
- 7.Emerson, S. U., and R. H. Purcell. 2006. Hepatitis E virus, p. 3047-3058. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 8.Emerson, S. U., P. Clemente-Casares, N. Moiduddin, V. A. Arankalle, U. Torian, and R. H. Purcell. 2006. Putative neutralization epitopes and broad cross-genotype neutralization of hepatitis E virus confirmed by a quantitative cell-culture assay. J. Gen. Virol. 87:697-704. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 10.Graff, J., H. Nguyen, C. Yu, W. R. Elkins, M. St. Claire, R. H. Purcell, and S. U. Emerson. 2005. The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J. Virol. 79:6680-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graff, J., U. Torian, H. Nguyen, and S. U. Emerson. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 80:5919-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guu, T. S., Z. Liu, Q. Ye, D. A. Mata, K. Li, C. Yin, J. Zhang, and Y. J. Tao. 2009. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. USA 106:12992-12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison, T. J. 1999. Hepatitis E virus—an update. Liver 19:171-176. [DOI] [PubMed] [Google Scholar]
- 14.He, S., J. Miao, Z. Zheng, T. Wu, M. Xie, M. Tang, J. Zhang, M. H. Ng, and N. Xia. 2008. Putative receptor-binding sites of hepatitis E virus. J. Gen. Virol. 89:245-249. [DOI] [PubMed] [Google Scholar]
- 15.Holinger, F. B., and S. U. Emerson. 2006. Hepatitis A virus, p. 911-947. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 16.Huang, Y. W., T. Opriessnig, P. G. Halbur, and X. J. Meng. 2007. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J. Virol. 81:3018-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ina, Y. 1994. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput. Appl. Biosci. 10:11-12. [DOI] [PubMed] [Google Scholar]
- 18.Khuroo, M. S., S. Kamili, and G. N. Yattoo. 2004. Hepatitis E virus infection may be transmitted through blood transfusions in an endemic area. J. Gastroenterol. Hepatol. 19:778-784. [DOI] [PubMed] [Google Scholar]
- 19.Lemon, S. M., L. N. Binn, and R. H. Marchwicki. 1983. Radioimmunofocus assay for quantitation of hepatitis A virus in cell cultures. J. Clin. Microbiol. 17:834-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemon, S. M., and L. N. Binn. 1985. Incomplete neutralization of hepatitis A virus in vitro due to lipid-associated virions. J. Gen. Virol. 66:2501-2505. [DOI] [PubMed] [Google Scholar]
- 21.Li, S., X. Tang, J. Seetharaman, C. Yang, Y. Gu, J. Zhang, H. Du, J. W. Shih, C. L. Hew, J. Sivaraman, and N. Xia. 2009. Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction. PLoS Pathog. 5:e1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenzo, F. R., T. Tanaka, H. Takahashi, K. Ichiyama, Y. Hoshino, K. Yamada, J. Inoue, M. Takahashi, and H. Okamoto. 2008. Mutational events during the primary propagation and consecutive passages of hepatitis E virus strain JE03-1760F in cell culture. Virus Res. 137:86-96. [DOI] [PubMed] [Google Scholar]
- 23.Lu, L., C. Li, and C. H. Hagedorn. 2006. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 16:5-36. [DOI] [PubMed] [Google Scholar]
- 24.Matsubayashi, K., Y. Nagaoka, H. Sakata, S. Sato, K. Fukai, T. Kato, K. Takahashi, S. Mishiro, M. Imai, N. Takeda, and H. Ikeda. 2004. Transfusion-transmitted hepatitis E caused by apparently indigenous hepatitis E virus strain in Hokkaido, Japan. Transfusion 44:934-940. [DOI] [PubMed] [Google Scholar]
- 25.Matsubayashi, K., J. H. Kang, H. Sakata, K. Takahashi, M. Shindo, M. Kato, S. Sato, T. Kato, H. Nishimori, K. Tsuji, H. Maguchi, J. Yoshida, H. Maekubo, S. Mishiro, and H. Ikeda. 2008. A case of transfusion-transmitted hepatitis E caused by blood from a donor infected with hepatitis E virus via zoonotic food-borne route. Transfusion 48:1368-1375. [DOI] [PubMed] [Google Scholar]
- 26.Meng, J., X. Dai, J. C. Chang, E. Lopareva, J. Pillot, H. A. Fields, and Y. E. Khudyakov. 2001. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 288:203-211. [DOI] [PubMed] [Google Scholar]
- 27.Meng, X. J. 2005. Hepatitis E as a zoonotic disease, p. 611-623. In H. C. Thomas, S. Lemon, and A. J. Zuckerman (ed.), Viral hepatitis, 3rd ed. Blackwell Publishing, Malden, MA.
- 28.Mitsui, T., Y. Tsukamoto, C. Yamazaki, K. Masuko, F. Tsuda, M. Takahashi, T. Nishizawa, and H. Okamoto. 2004. Prevalence of hepatitis E virus infection among hemodialysis patients in Japan: evidence for infection with a genotype 3 HEV by blood transfusion. J. Med. Virol. 74:563-572. [DOI] [PubMed] [Google Scholar]
- 29.Mizuo, H., K. Suzuki, Y. Takikawa, Y. Sugai, H. Tokita, Y. Akahane, K. Itoh, Y. Gotanda, M. Takahashi, T. Nishizawa, and H. Okamoto. 2002. Polyphyletic strains of hepatitis E virus are responsible for sporadic cases of acute hepatitis in Japan. J. Clin. Microbiol. 40:3209-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomura, M., M. Imai, F. Tsuda, S. Furuta, Y. Akahane, K. Tachibana, S. Usuda, Y. Miyakawa, and M. Mayumi. 1985. Immunoglobulin A antibody against hepatitis B core antigen in the acute and persistent infection with hepatitis B virus. Gastroenterology 89:1109-1113. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto, H., M. Takahashi, and T. Nishizawa. 2003. Features of hepatitis E virus infection in Japan. Intern. Med. 42:1065-1071. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto, H. 2007. Genetic variability and evolution of hepatitis E virus. Virus Res. 127:216-228. [DOI] [PubMed] [Google Scholar]
- 33.Panda, S. K., D. Thakral, and S. Rehman. 2007. Hepatitis E virus. Rev. Med. Virol. 17:151-180. [DOI] [PubMed] [Google Scholar]
- 34.Provost, P. J., B. S. Wolanski, W. J. Miller, O. L. Ittensohn, W. J. McAleer, and M. R. Hilleman. 1975. Physical, chemical and morphologic dimensions of human hepatitis A virus strain CR326 (38578). Proc. Soc. Exp. Biol. Med. 148:532-539. [DOI] [PubMed] [Google Scholar]
- 35.Purcell, R. H., S. M. Feinstone, J. R. Ticehurst, R. J. Daemer, and B. M. Baroudy. 1984. Hepatitis A virus, p. 9-22. In G. N. Vyas, J. L. Dienstag, and J. H. Hoofnagle (ed.), Viral hepatitis and liver disease. Grune & Stratton, Orlando, FL.
- 36.Purcell, R. H., and S. U. Emerson. 2008. Hepatitis E: an emerging awareness of an old disease. J. Hepatol. 48:494-503. [DOI] [PubMed] [Google Scholar]
- 37.Rueckert, R. 1976. On the structure and morphogenesis of picornaviruses, p. 131-213. In H. Frankel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 6. Plenum Press, New York, NY. [Google Scholar]
- 38.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 39.Schlauder, G. G., and I. K. Mushahwar. 2001. Genetic heterogeneity of hepatitis E virus. J. Med. Virol. 65:282-292. [DOI] [PubMed] [Google Scholar]
- 40.Schofield, D. J., J. Glamann, S. U. Emerson, and R. H. Purcell. 2000. Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J. Virol. 74:5548-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schofield, D. J., R. H. Purcell, H. T. Nguyen, and S. U. Emerson. 2003. Monoclonal antibodies that neutralize HEV recognize an antigenic site at the carboxyterminus of an ORF2 protein vaccine. Vaccine 22:257-267. [DOI] [PubMed] [Google Scholar]
- 42.Shrestha, M. P., R. M. Scott, D. M. Joshi, M. P. Mammen, Jr., G. B. Thapa, N. Thapa, K. S. Myint, M. Fourneau, R. A. Kuschner, S. K. Shrestha, M. P. David, J. Seriwatana, D. W. Vaughn, A. Safary, T. P. Endy, and B. L. Innis. 2007. Safety and efficacy of a recombinant hepatitis E vaccine. N. Engl. J. Med. 356:895-903. [DOI] [PubMed] [Google Scholar]
- 43.Smith, J. L. 2001. A review of hepatitis E virus. J. Food Prot. 64:572-586. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, K., T. Aikawa, and H. Okamoto. 2002. Fulminant hepatitis E in Japan. N. Engl. J. Med. 347:1456. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi, T., S. Nakagawa, T. Hashimoto, K. Takahashi, M. Imai, Y. Miyakawa, and M. Mayumi. 1976. Large-scale isolation of Dane particles from plasma containing hepatitis B antigen and demonstration of a circular double-stranded DNA molecule extruding directly from their cores. J. Immunol. 117:1392-1397. [PubMed] [Google Scholar]
- 46.Takahashi, K., A. Machida, G. Funatsu, M. Nomura, S. Usuda, S. Aoyagi, K. Tachibana, H. Miyamoto, M. Imai, T. Nakamura, Y. Miyakawa, and M. Mayumi. 1983. Immunochemical structure of hepatitis B e antigen in the serum. J. Immunol. 130:2903-2907. [PubMed] [Google Scholar]
- 47.Takahashi, M., T. Nishizawa, H. Miyajima, Y. Gotanda, T. Iita, F. Tsuda, and H. Okamoto. 2003. Swine hepatitis E virus strains in Japan form four phylogenetic clusters comparable with those of Japanese isolates of human hepatitis E virus. J. Gen. Virol. 84:851-862. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi, K., N. Kitajima, N. Abe, and S. Mishiro. 2004. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology 330:501-505. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi, M., S. Kusakai, H. Mizuo, K. Suzuki, K. Fujimura, K. Masuko, Y. Sugai, T. Aikawa, T. Nishizawa, and H. Okamoto. 2005. Simultaneous detection of immunoglobulin A (IgA) and IgM antibodies against hepatitis E virus (HEV) is highly specific for diagnosis of acute HEV infection. J. Clin. Microbiol. 43:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi, M., T. Tanaka, M. Azuma, E. Kusano, T. Aikawa, T. Shibayama, Y. Yazaki, H. Mizuo, J. Inoue, and H. Okamoto. 2007. Prolonged fecal shedding of hepatitis E virus (HEV) during sporadic acute hepatitis E: evaluation of infectivity of HEV in fecal specimens in a cell culture system. J. Clin. Microbiol. 45:3671-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, M., Y. Hoshino, T. Tanaka, H. Takahashi, T. Nishizawa, and H. Okamoto. 2008. Production of monoclonal antibodies against hepatitis E virus capsid protein and evaluation of their neutralizing activity in a cell culture system. Arch. Virol. 153:657-666. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi, M., K. Yamada, Y. Hoshino, H. Takahashi, K. Ichiyama, T. Tanaka, and H. Okamoto. 2008. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 153:1703-1713. [DOI] [PubMed] [Google Scholar]
- 53.Tam, A. W., M. M. Smith, M. E. Guerra, C. Huang, D. W. Bradley, K. E. Fry, and G. R. Reyes. 1991. Hepatitis E virus (HEV): molecular cloning and sequence of the full-length viral genome. Virology 185:120-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura, A., Y. K. Shimizu, T. Tanaka, K. Kuroda, Y. Arakawa, K. Takahashi, S. Mishiro, K. Shimizu, and M. Moriyama. 2007. Persistent infection of hepatitis E virus transmitted by blood transfusion in a patient with T-cell lymphoma. Hepatol. Res. 37:113-120. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka, T., M. Takahashi, E. Kusano, and H. Okamoto. 2007. Development and evaluation of an efficient cell-culture system for Hepatitis E virus. J. Gen. Virol. 88:903-911. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka, T., M. Takahashi, H. Takahashi, K. Ichiyama, Y. Hoshino, S. Nagashima, H. Mizuo, and H. Okamoto. 2009. Development and characterization of a genotype 4 hepatitis E virus cell culture system using a HE-JF5/15F strain recovered from a fulminant hepatitis patient. J. Clin. Microbiol. 47:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tei, S., N. Kitajima, K. Takahashi, and S. Mishiro. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362:371-373. [DOI] [PubMed] [Google Scholar]
- 58.Tsuda, F., S. Naito, E. Takai, Y. Akahane, S. Furuta, Y. Miyakawa, and M. Mayumi. 1984. Low molecular weight (7s) immunoglobulin M antibody against hepatitis B core antigen in the serum for differentiating acute from persistent hepatitis B virus infection. Gastroenterology 87:159-164. [PubMed] [Google Scholar]
- 59.Tyagi, S., H. Korkaya, M. Zafrullah, S. Jameel, and S. K. Lal. 2002. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 277:22759-22767. [DOI] [PubMed] [Google Scholar]
- 60.Tyagi, S., M. Surjit, A. K. Roy, S. Jameel, and S. K. Lal. 2004. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor alpha1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 279:29308-29319. [DOI] [PubMed] [Google Scholar]
- 61.Williams, T. P., C. Kasorndorkbua, P. G. Halbur, G. Haqshenas, D. K. Guenette, T. E. Toth, and X. J. Meng. 2001. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J. Clin. Microbiol. 39:3040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamada, K., M. Takahashi, Y. Hoshino, H. Takahashi, K. Ichiyama, S. Nagashima, T. Tanaka, and H. Okamoto. 2009. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J. Gen. Virol. 90:1880-1891. [DOI] [PubMed] [Google Scholar]
- 64.Yamashita, T., Y. Mori, N. Miyazaki, R. H. Cheng, M. Yoshimura, H. Unno, R. Shima, K. Moriishi, T. Tsukihara, T. C. Li, N. Takeda, T. Miyamura, and Y. Matsuura. 2009. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. USA 106:12986-12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yazaki, Y., H. Mizuo, M. Takahashi, T. Nishizawa, N. Sasaki, Y. Gotanda, and H. Okamoto. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 84:2351-2357. [DOI] [PubMed] [Google Scholar]
- 66.Zhao, C., Z. Ma, T. J. Harrison, R. Feng, C. Zhang, Z. Qiao, J. Fan, H. Ma, M. Li, A. Song, and Y. Wang. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J. Med. Virol. 81:1371-1379. [DOI] [PubMed] [Google Scholar]