Abstract
Respiratory syncytial virus (RSV) is the most commonly identified viral agent in young children with acute respiratory tract infections (ARIs) and often causes repeated infections throughout life. This study investigated the genetic variability of the attachment (G) protein gene among RSV strains prevalent in southwestern China. Reverse transcription-PCR (RT-PCR) for a fragment of the RSV G gene was performed with nasopharyngeal aspirates (NPAs) collected from children with ARIs hospitalized in Chongqing Children's Hospital, Chongqing, China. A total of 1,387 NPA specimens were collected from April 2006 to March 2009, and 439 (31.7%) were positive for RSV. During the study period, subgroup A and B viruses accounted for 79.5% (349/439) and 19.8% (87/439) of the total positive samples, respectively. Both subgroup A and B viruses were identified in three samples. Subgroup A viruses were predominant during two epidemic seasons (2006 to 2008), while subgroup B strains prevailed during the 2008-2009 epidemic season. Phylogenetic analyses showed that all 30 group A strains could be clustered into one genotype, genotype GA2, and 30 group B strains could be clustered into three genotypes, genotypes GB1, GB3, and BA, among which 17 genotype BA strains were detected from 23 group B strains selected during the 2008-2009 epidemic season. The G gene of genotype BA was predicted to encode proteins of five different lengths. These data suggest that group A RSV likely predominated in southwestern China and that a new genotype of subgroup B with a 60-nucleotide insertion, named BA-like virus, became the dominant genotype in the 2008-2009 epidemic season.
Human respiratory syncytial virus (RSV) is the most important viral cause of lower respiratory tract infections in infants and young children and also in immunocompromised and elderly individuals (15). RSV strains vary genetically and antigenically and have been classified into two broad groups, groups A and B (2, 8, 16, 21), with additional variability being detected within the two groups (6, 30). Antigenic variability is thought to contribute to the capacity of the virus to establish reinfections throughout life and may pose a challenge to vaccine development. Future planning for vaccine development requires further understanding of the genetic composition of the RSV strains prevalent in the target population.
RSV consists of 11 viral proteins (14, 21), and the RSV G protein shows the highest degree of divergence both between and within the two groups (16). G-protein variability is concentrated in the ectodomain, which contains two hypervariable regions separated by a conserved 13-amino-acid motif (16). The second variable region, which corresponds to the C-terminal region of the G protein, reflects overall gene variability and has been analyzed in molecular epidemiological studies (24, 25). Recent studies demonstrated a worldwide prevalence of very distinct RSV genotypes with temporal rather than geographical clustering (5, 13). However, less information is available on the distribution pattern of RSV epidemic strains in southwestern China. The aim of the present study was to evaluate the genetic diversity of the gene encoding the G protein of RSV isolates collected from hospitalized infants and young children with acute respiratory tract infections (ARIs) during three consecutive epidemic periods in Chongqing, China. Information on the distribution of RSV genotypes in China will be beneficial to the development and implementation of RSV vaccines.
MATERIALS AND METHODS
Collection of specimens.
From April 2006 to March 2009, nasopharyngeal aspirates (NPAs) were obtained on 3 fixed days each week from all children admitted to Chongqing Children's Hospital with symptoms of ARIs as soon as possible after admission. ARIs were classified according to World Health Organization definitions. The specimens were immediately placed at 4°C in tubes containing 3 ml cold viral transport medium (phosphate-buffered saline, 100 U/μl penicillin, 100 μg/ml streptomycin) and transported to the Department of Virology of Chongqing Children's Hospital within 4 to 6 h. The specimens were processed for direct immunofluorescence assay (DFA) of common viral respiratory pathogens on the same day. The specimens were vortexed and centrifuged at 1,500 × g for 10 min at 4°C. The supernatants of the specimens were frozen and stored at −70°C until further testing. This study was approved by the Ethics and Research Council of Chongqing Children's Hospital, and signed consent was obtained from each child's parent or foster parent.
Chongqing Children's Hospital is a public university-affiliated hospital that cares for children who live in the city of Chongqing and other southwestern provinces of China. This 800-bed facility is a regional medical reference center and serves a large area covering Chongqing as well as the surrounding provinces.
RNA extraction and cDNA synthesis.
RNA was extracted directly from 140 μl of frozen clinical samples with an RNeasy minikit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The dried RNA pellet was dissolved in 50 μl DNase-free and RNase-free water. For cDNA synthesis, 100 ng of the total RNA was used as a template for reverse transcription-PCR (RT-PCR) with random primers.
Subgroup identification by PCR analysis.
Subgroup-specific products were amplified with 5′ primer P1, subgroup A-specific 3′ primer P2, and subgroup B-specific 3′ primer P3 (34). Three microliters of cDNA was added to a 22-μl PCR master mixture containing 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 1.5 U of Taq DNA polymerase, and 50 pM each primer. The thermocycling protocol was 94°C for 2 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension for 7 min at 72°C. The amplified products of 250 and 341 bp for subgroups A and B, respectively, were analyzed by electrophoresis on a 1.5% agarose gel. The viruses detected were designated by city/group/year of isolation/isolate number.
PCR amplification of C-terminal half of the gene encoding G protein.
Primer GA480 (5′-ACAAACCACCAAACAAACCC-3′, corresponding to bases 480 to 499 of the gene encoding the G protein of strain A2) or GB496 (5′-GATGATTACCATTTTGAAGTGTTCA-3′, corresponding to bases 496 to 520 of the gene encoding the G protein of strain CH18537) and primer F164 (5′-GTTATGACACTGGTATACCAACC-3′, corresponding to bases 164 to 186 of the F-protein gene of strain 18537, with one mismatch with the gene encoding the G protein of strain A2) were used to amplify the C-terminal half of the gene encoding the G protein (12). DNA amplification was carried out by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min. Five microliters of the amplified DNA was resolved by electrophoresis on a 1.5% agarose gel, and the gel was stained with GoldView nucleic acid stain (SBS Genetech) and visualized under UV light.
DNA sequencing.
The PCR products were purified with a QIAquick gel extraction kit (Qiagen), according to the manufacturer's instructions. The purified PCR products were sequenced in the forward and the reverse directions on an ABI Prism 310 genetic analyzer (Applied Biosystems) by using an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems).
Phylogenetic analysis.
The nucleotide and amino acid sequences of the group A and B viruses were aligned separately with the human RSV (HRSV) sequences available from the GenBank database by using the ClustalX (version 1.81) program (29) and were manually edited with the BioEdit (version 7.0.1) program. Phylogenetic trees were constructed by the neighbor-joining method in MEGA (version 3.1) software (18). The statistical significance of the tree topology was tested by bootstrapping (1,000 replicates). For human RSV of group A, phylogenetic trees were constructed by using 18 unique sequences in the present study along with reference sequences of 28 strains representing all known genotypes by using 270 nucleotides from the C-terminal variable region of the gene encoding the G protein. These reference strains included strain A2 and isolates from Seoul, South Korea (SEL); Kenya (Ken); South Africa (SA); Salvador, Brazil (Sal); the United States (TX, MO, AL, NY); Canada (CN); and elsewhere (16, 19, 21, 23, 28, 30, 32). Phylogenetic trees for the 30 group B strains studied were analyzed on the basis of 252 nucleotides of the C-terminal variable region, and these sequences were compared to the G-protein sequences of 42 reference strains representative of all known group B genotypes available from GenBank. These reference strains included strain CH18537 and isolates from South Africa; the United States (MO; NY; Rochester, NY [CH]; AL); Buenos Aires, Argentina (BA); Quebec, Canada (QUE); Belgium (BE); Niigata, Japan (NG); Montevideo, Uruguay (MON); Beijing, China (Beijing); and elsewhere (16, 17, 19, 23, 24, 28, 30, 33, 35).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences obtained in the present study are GU550444 to GU550503.
RESULTS
Specimen collection.
A total of 1,387 specimens were collected from patients with ARIs from April 2006 to March 2009, and 439 (31.7%) were positive for RSV by RT-PCR. Strains of subgroups A and B were identified in 349 (79.5%) and 87 (19.8%) specimens, respectively; and in 3 (0.7%) specimens, both HRSV subgroup A and HRSV subgroup B were identified. Most infected individuals were babies 0 to 6 months old, which accounted for 56.0% of all samples positive for RSV, and babies 6 to 12 months old, which accounted for 18.2% of all positive samples. Children 2 to 5 years old accounted for 8% of positive samples. Some 55.1% of the children infected with RSV subgroup A were less than 6 months old, while the proportion was 59.8% for RSV subgroup B. In terms of gender, 71.4% (249/349) of the RSV subgroup A-infected cases were males, while 69.0% (60/87) of the RSV subgroup B-infected cases were males. The rates of positivity for RSV infection ranged from 24.8% to 36.8% during the three epidemic seasons. The HRSV outbreak activity in Chongqing was high in November, December, and January and reached a peak in December. The detection rate declined in April, and no RSV strain was found in September 2006 or June or July 2007. Subgroup A viruses were predominant during two epidemic seasons (2006 to 2008), and only seven group B RSV strains were detected. Subgroup B viruses prevailed during the 2008-2009 epidemic season, representing 63.5% (80/126) of all RSV isolates detected (Fig. 1).
FIG. 1.
(A) Seasonal distribution of group A and B HRSV strains in infants and children with acute lower respiratory tract infection in Chongqing, China, between April 2006 and March 2009. (B) Monthly distribution of subgroup B genotypes of HRSV in Chongqing, China, between April 2006 and March 2009.
Information on clinical symptoms was available for all the 439 RSV-positive patients. The main clinical presentations included fever (n = 182; 41.5%), cough (n = 435; 99.1%), wheezing (n = 312; 71.1%), and cyanosis (n = 373; 85.0%). The RSV-infected children were diagnosed with bronchiolitis (38.5%), bronchopneumonia (25.1%), interstitial pneumonia (24.8%), bronchial asthma exacerbation (7.5%), acute infectious laryngitis (1.8%), asthmatic bronchitis (1.4%), and bronchitis (0.9%). Fifty patients (11.4%) were diagnosed with severe pneumonia and respiratory failure, including 36 group A-positive and 14 group B-positive patients, and 5 patients were transferred to the intensive care unit (ICU).
Phylogenetic analysis and genotype distribution patterns.
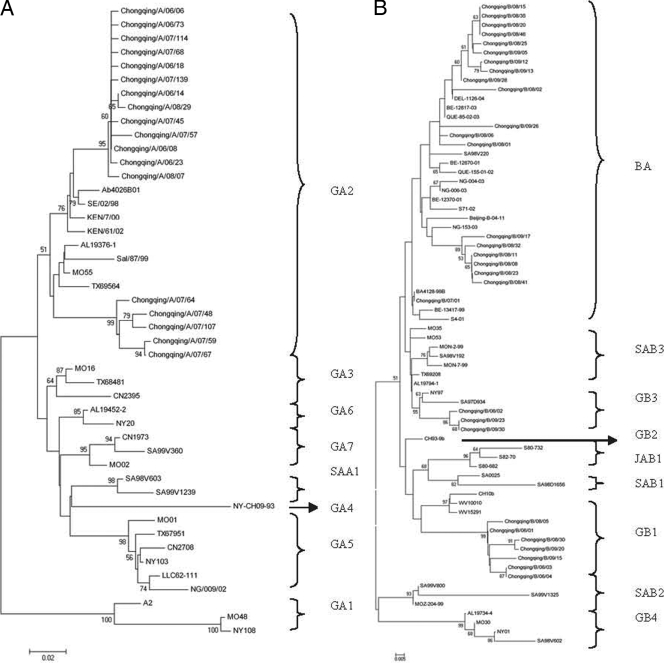
The RSV strains with the unique gene encoding the G protein in each epidemic season were included in the phylogenetic analysis. As a result, the sequences from 30 group A strain-infected patients and 30 group B strain-infected patients were analyzed. Of these 30 group A sequences, 18 unique group A sequences were identified, among which two groups of 4 and 10 strains, respectively, each showed identical sequences of the G-protein gene. Phylogenetic analysis revealed that 18 unique group A strains clustered into one genotype, genotype GA2 (Fig. 2).
FIG. 2.
Phylogenetic trees of Chinese RSV group A (A) and group B (B) nucleotide sequences from the second variable region of the gene encoding the G protein. The numbers at the branch nodes represent the number of bootstrap probabilities. Reference sequences for each genotype (genotypes GA1 to GA7, SAA1, GB1 to GB4, SAB1, and SAB3) were obtained from GenBank. Additional sequences from around the world were included in the comparison by selecting representatives of distinct clusters found in previous studies and selecting isolate sequences from GenBank that gave the best hits in BLAST searches with each of the Chinese clusters. Prototype strains (strain A2 for group A and strain CH18537 for group B) were used as the outgroup sequences in the tree. The genotype assignment is indicated by the braces on the right.
The rates of divergence between prototype strain A2 and the Chinese group A strains were 13% to 14.8% at the nucleotide level and 20.5% to 26.1% at the amino acid level. Differences of up to 9.9% at the nucleotide level and 20.5% at the amino acid level were observed among the Chinese group A strains.
The group B strains were found to comprise three genotypes. Three group B strains belonged to genotype GB3, 7 strains belonged to genotype GB1, and the other 20 strains belonged to the newly identified genotype, genotype BA, which had a 60-nucleotide duplication in the second variable region of the gene encoding the G protein (1, 12, 19, 22, 27, 36) (Fig. 2). The first genotype BA strain was detected in December 2007, and only three BA genotype strains were detected among all the seven group B strains in the epidemic seasons of 2006 to 2008. Seventeen genotype BA strains were detected from among 23 group B strains selected in the subsequent 2008-2009 epidemic season, during which group B was dominant. In addition, 6 nucleotides after residue 489 were deleted in 15 genotype BA-like sequences, resulting in a 2-amino-acid deletion. In one BA-like sequence, 6 nucleotides were deleted after residue 489 and 6 nucleotides were inserted after residue 694. Genotype BA strains with 6 nucleotides deleted after residue 489 were first identified on mainland China. Additionally, 9 nucleotides were inserted after residue 721 in one group B sequence (Chongqing/B/09/15). Among the 30 group B sequences, there were 26 unique group B sequences. There was 12.9 to 16.5% divergence at the nucleotide level and 21.5 to 28.8% divergence at the amino acid level between the group B strains and prototypic genotype B strain CH18537. Among the group B strains, up to 12.4% nucleotide differences and up to 15.4% amino acid differences were observed. Among the 200 genotype BA strains, 18 unique sequences were identified. The BA strains and prototypic genotype BA strain BA/4128/99B showed 2.6 to 4.7% divergence at the nucleotide level and 5 to 12.4% divergence at the amino acid level. The genotype BA strains and the Beijing/B/04/11 strain, which was the BA genotype strain first detected on mainland China (33), showed 5 to 6.8% divergence at the nucleotide level and 9.6 to 14.4% divergence at the amino acid level. In addition, comparisons of the duplicated region within each virus revealed that in every instance there were two to three nucleotide sequence differences resulting in one or two amino acid-coding changes.
For the HRSV genotype B strains, a significant difference in the average ages of the patients infected with genotypes BA and GB1 was observed (0.5 ± 0.4 years and 1.1 ± 0.6 years, respectively; P < 0.05), but the average ages of the patients infected with genotype GA2, HRSV genotype B or A, and HRSV genotype B strains as a whole were not statistically different.
Amino acid analysis.
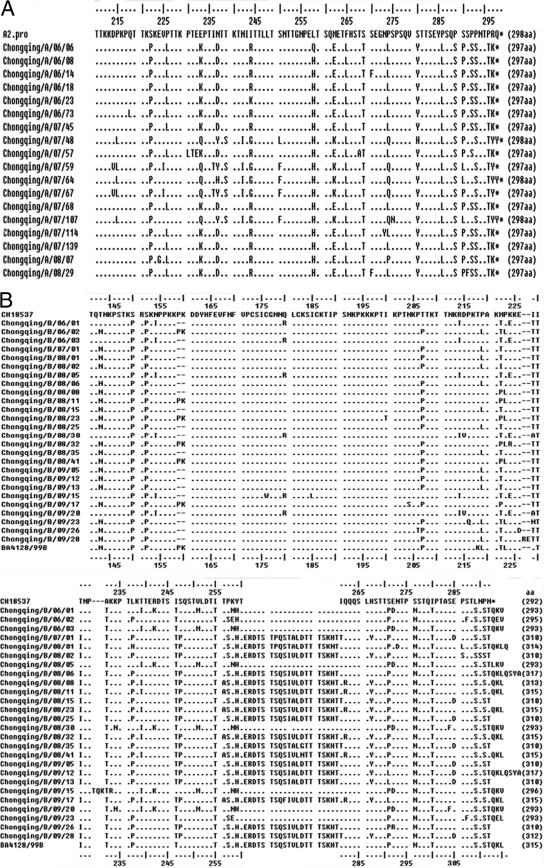
The predicted amino acid sequences of the group A and group B strains were compared to those of the prototype strains A2 and CH18537, respectively (Fig. 3). Twenty-seven of the 30 Chinese group A viruses exhibited changes in the stop codon position compared with that for prototypic strain A2, which had 298 amino acids in the deduced G-protein sequence. Three Chinese group A strains had a predicted G protein of 298 amino acids, while 27 Chinese group A strains had a predicted G protein of 297 amino acids.
FIG. 3.
Amino acid alignment of the G protein of Chinese group A (A) and group B (B) strains. Alignments relative to the sequences of prototype strains A2 and CH18537 are shown. The relative lengths (in number of amino acids [aa]) for these regions are indicated in parentheses on the right.
There were significant differences in the lengths of the deduced G-protein sequences among the group B strains. The predicted complete G proteins of the Chinese group B strains were of nine different amino acid lengths (293, 295, 296, 310, 312, 313, 314, 315, and 317 amino acids) (Fig. 3). Differences in the G-protein lengths were caused by the use of alternative termination codons and by the presence of in-frame duplications, deletions, and insertions. Among the 30 group B strains, 6 viruses were deduced to have a G protein of 293 amino acids, 1 virus was deduced to have a G protein of 295 amino acids, and 1 virus (Chongqing/B/09/15) was deduced to have a G protein of 296 amino acids and 9 nucleotides inserted after residue 721. The G-protein genes of the 20 genotype BA strains were predicted to encode proteins of six different lengths: 310, 312, 313, 314, 315, and 317 amino acids. In addition, in 15 genotype BA strains, 6 nucleotides were deleted after residue 489, resulting in a 2-amino-acid deletion. Among these strains was one strain in which 6 nucleotides were also inserted after residue 694, resulting in a 2-amino-acid insertion. A Ser 247-to-Pro amino acid change was observed in the 20-amino-acid duplicated region in all the Chinese genotype BA strains compared to the sequence of the prototype genotype BA strain.
DISCUSSION
Knowledge of the molecular epidemiology of HRSV has mainly been based on studies done in developed countries (7, 11, 13, 19, 20, 27, 35). In this regard, few studies have been conducted in China (9, 10, 33). The findings of our epidemiological study indicate that large epidemics of RSV occurred in Chongqing during three consecutive epidemic seasons (April 2006 to March 2009). A total of 1,387 specimens were collected, and 439 (31.7%) were positive for RSV by RT-PCR. Our results, which showed that cases of infection with RSV group A predominated over cases of infection with RSV group B, are in agreement with the findings of the majority of such typing studies (23, 33). RSV was detected through all three seasons, and in Chongqing, the RSV outbreak activity was during the winter months (November to January), which was similar to the findings of Agrawal et al. (1).
In previous studies of RSV strain circulation patterns, both antigenic group A and antigenic group B were noted to cocirculate in the same city during the epidemic periods and had various patterns of predominance. In the present study, subgroup A viruses were predominant during two epidemic seasons (2006 to 2008), while subgroup B viruses prevailed during the subsequent 2008-2009 epidemic season. In a community, a shift in the dominance of an RSV subgroup can occur at different time intervals, and different patterns of subgroup A and B prevalence have been described. Similar subgroup occurrence patterns have been seen in Kenya, Uruguay, and the United States, where HRSV subgroup B became predominant after two consecutive seasons with a high prevalence of subgroup A (3, 7, 26), while in Niigata, Japan, epidemiological analysis of RSV subgroups showed that group B was predominant in the 2002-2003 season and but that this shifted to a predominance of group A from 2003 to 2007 (27). The shift in the predominant group was correlated in part with variability in the G-protein gene.
Among the RSVs collected, the genetic variability of the group A strains (up to 14.8%) was comparatively higher than that of the group B strains (up to 12.4%). The higher degree of genetic variability among the group A viruses may also be responsible for the predominance of this virus group worldwide (7, 24). Previous investigations have reported that a replacement of the predominating HRSV genotype occurs each year (4, 24). However, GA2 remained the dominant HRSV genotype in three consecutive epidemic seasons during the study period. The finding that GA2 was the most common genotype of RSV group A is consistent with the results of long-term studies from other geographic areas, including the Beijing area of China (23, 33).
Among the 30 group B viruses in circulation, there were 26 unique group B sequences, of which 23 were found once and 7 were grouped into three sets of identical sequences of 2 to 3 sequences each. A new group B genotype, genotype BA, with a 60-nucleotide G-protein gene duplication first appeared in Argentina in 1999 (30). The first genotype BA strain detected in mainland China was identified in Beijing in 2004, and only three BA strains have so far been found in China (10, 33). In recent years, Shobugawa et al. (27) and Agrawal et al. (1) also separately indentified 143 and 22 BA strains in Japan and eastern India, respectively. No studies, however, were designed to track this new group B genotype in mainland China after 2004. In the present study, the genotype BA strain detected in Chongqing first appeared in December 2007 and became the dominant genotype in the 2008-2009 epidemic season. That was the first report that a genotype BA strain had become dominant during the RSV epidemic season in mainland China. It is remarkable that viruses with the largest duplications described so far have been prevalent for at least 10 years, i.e., since 1999. Some recent reports also showed that genotype BA had replaced other prevailing genotypes in certain places, including Argentina and India (23, 31). The selective advantage of genotype BA viruses over viruses of other genotypes is unknown. An antigenic change in the G protein and the avoidance of host immune responses may be the selective advantage (31).
The predicted G proteins of the subgroup B strains had nine different amino acid lengths, among which five different amino acid lengths (296, 310, 313, 314, and 317 amino acids) were first detected in mainland China. The mechanisms responsible for this variability include amino acid substitutions, insertions, deletions, duplications, and changes in stop codon usage. Among the 20 genotype BA strains, 6 nucleotides were deleted after residue 489 in 15 strains, which resulted in a 2-amino-acid deletion. Agrawal et al. also reported that for all 22 genotype BA strains detected in the 2005-2006 epidemic season in eastern India, 6 nucleotides were deleted after residue 489 (1). This finding reinforces the notion of the transfer of viruses between close communities.
In summary, the occurrence of various human RSV subgroups was observed among hospitalized patients in Chongqing, China, during three recent consecutive epidemic seasons. A new genotype of subgroup B with a 60-nucleotide insertion, named the genotype BA-like virus, became the dominant genotype in the 2008-2009 epidemic season. This study thus contributes to better knowledge of the molecular epidemiology of RSV in mainland China. Comprehensive information on the prevalence profile of RSV is important for the selection of appropriate vaccine strains and may thus contribute to vaccine development.
Acknowledgments
We thank all family members for their enrollment in this study. We appreciate Qiang-lin Duan from Tongji Hospital for critical reading of the manuscript.
This work was supported in part by the Program for New Century Excellent Talents in University (grant NCET-05-0774) and the Chongqing Excellent Young Scientists Fund (CSCT; grant 2008BA5040).
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Agrawal, A. S., M. Sarkar, S. Ghosh, M. Chawla-Sarkar, N. Chakraborty, M. Basak, and T. N. Naik. 2009. Prevalence of respiratory syncytial virus group B genotype BA-IV strains among children with acute respiratory tract infection in Kolkata, Eastern India. J. Clin. Virol. 45:358-361. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, L. J., J. C. Hierholzer, C. Tsou, R. M. Hendry, B. F. Fernie, Y. Stone, and K. McIntosh. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626-633. [DOI] [PubMed] [Google Scholar]
- 3.Arbiza, J., A. Delfraro, and S. Frabasile. 2005. Molecular epidemiology of human respiratory syncytial virus in Uruguay: 1985-2001—a review. Mem. Inst. Oswaldo Cruz 100:221-230. [DOI] [PubMed] [Google Scholar]
- 4.Cane, P. A., D. A. Matthews, and C. R. Pringle. 1994. Analysis of respiratory syncytial virus strain variation in successive epidemics in one city. J. Clin. Microbiol. 32:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cane, P. A., and C. R. Pringle. 1995. Molecular epidemiology of respiratory syncytial virus: a review of the use of reverse transcription-polymerase chain reaction in the analysis of genetic variability. Electrophoresis 16:329-333. [DOI] [PubMed] [Google Scholar]
- 6.Cane, P. A. 2001. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 11:103-116. [DOI] [PubMed] [Google Scholar]
- 7.Coggins, W. B., E. J. Lefkowitz, and W. M. Sullender. 1998. Genetic variability among group A and group B respiratory syncytial viruses in a children's hospital. J. Clin. Microbiol. 36:3552-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cristina, J., J. A. Lopez, C. Albo, B. Garcia-Barreno, J. Garcia, J. A. Melero, and A. Portela. 1990. Analysis of genetic variability in human respiratory syncytial virus by the RNase A mismatch cleavage method: subtype divergence and heterogeneity. Virology 174:126-134. [DOI] [PubMed] [Google Scholar]
- 9.Deng, J., Q. Yuan, Z. Ru-nan, W. Fang, and Z. Lin-qing. 2006. Surveillance for respiratory syncytial virus subtypes A and B in children with acute respiratory infections in Beijing during 2000 to 2006 seasons. Chin. J. Pediatr. 44:924-927. [PubMed] [Google Scholar]
- 10.Deng, J., Z. Ru-nan, Q. Yuan, Z. Lin-qing, and W. Fang. 2006. Sequence analysis of G glycoprotein of human respiratory syncytial virus subtype B strains isolated from children with acute respiratory infections in Beijing, China in years 2000-2004. Chin. J. Microbiol. Immunol. 26:1-5. [Google Scholar]
- 11.Frabasile, S., A. Delfraro, L. Facal, C. Videla, M. Galiano, M. J. de Sierra, D. Ruchansk, N. Vitureira, M. Berois, G. Carballal, J. Russi, and J. Arbiza. 2003. Antigenic and genetic variability of human respiratory syncytial viruses (group A) isolated in Uruguay and Argentina: 1993-2001. J. Med. Virol. 71:305-312. [DOI] [PubMed] [Google Scholar]
- 12.Galiano, M. C., C. Palomo, C. M. Videla, J. Arbiza, J. A. Melero, and G. Carballal. 2005. Genetic and antigenic variability of human respiratory syncytial virus (groups a and b) isolated over seven consecutive seasons in Argentina (1995 to 2001). J. Clin. Microbiol. 43:2266-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García, O., M. Martín, J. Dopazo, J. Arbiza, S. Frabasile, J. Russi, M. Hortal, P. Perez-Breña, I. Martínez, and B. García-Barreno. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J. Virol. 68:5448-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, C. B. 2001. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 344:1917-1928. [DOI] [PubMed] [Google Scholar]
- 15.Holberg, C. J., A. L. Wright, F. D. Martinez, C. G. Ray, L. M. Taussig, and M. D. Lebowitz. 1991. Risk factors for respiratory syncytial virus-associated lower respiratory illnesses in the first year of life. Am. J. Epidemiol. 133:1135-1151. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. U. S. A. 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong, X., J. Gao, H. Shou, and Z. Jiang. 2001. The study of respiratory syncytial virus infection of children. Chin. J. Microbiol. Immunol. 21:184-185. [Google Scholar]
- 18.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 19.Kuroiwa, Y., K. Nagai, L. Okita, I. Yui, T. Kase, T. Nakayama, and H. Tsutsumi. 2005. A phylogenetic study of human respiratory syncytial viruses group A and B strains isolated in two cities in Japan from 1980 to 2002. J. Med. Virol. 76:241-247. [DOI] [PubMed] [Google Scholar]
- 20.Muelenaer, P. M., F. W. Henderson, V. G. Hemming, E. E. Walsh, L. J. Anderson, G. A. Prince, and B. R. Murphy. 1991. Group-specific serum antibody responses in children with primary and recurrent respiratory syncytial virus infections. J. Infect. Dis. 164:15-21. [DOI] [PubMed] [Google Scholar]
- 21.Mufson, M. A., C. Orvell, B. Rafnar, and E. Norrby. 1985. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 66:2111-2124. [DOI] [PubMed] [Google Scholar]
- 22.Nagai, K., H. Kamasaki, Y. Kuroiwa, L. Okita, and H. Tsutsumi. 2004. Nosocomial outbreak of respiratory syncytial virus subgroup B variants with the 60 nucleotides-duplicated G protein gene. J. Med. Virol. 74:161-165. [DOI] [PubMed] [Google Scholar]
- 23.Parveen, S., W. M. Sullender, K. Fowler, E. J. Lefkowitz, S. K. Kapoor, and S. Broor. 2006. Genetic variability in the G protein gene of group A and B respiratory syncytial viruses from India. J. Clin. Microbiol. 44:3055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peret, T. C., C. B. Hall, K. C. Schnabel, J. A. Golub, and L. J. Anderson. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79:2221-2229. [DOI] [PubMed] [Google Scholar]
- 25.Peret, T. C., C. B. Hall, G. W. Hammond, P. A. Piedra, G. A. Storch, W. M. Sullender, C. Tsou, and L. J. Anderson. 2000. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J. Infect. Dis. 181:1891-1896. [DOI] [PubMed] [Google Scholar]
- 26.Scott, P. D., R. Ochola, M. Ngama, E. A. Okiro, D. J. Nokes, G. F. Medley, and P. A. Cane. 2004. Molecular epidemiology of respiratory syncytial virus in Kilifi District, Kenya. J. Med. Virol. 74:344-354. [DOI] [PubMed] [Google Scholar]
- 27.Shobugawa, Y., R. Saito, Y. Sano, H. Zaraket, Y. Suzuki, A. Kumaki, I. Dapat, T. Oguma, M. Yamaguchi, and H. Suzuki. 2009. Emerging genotypes of human respiratory syncytial virus subgroup A among patients in Japan. J. Clin. Microbiol. 47:2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullender, W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trento, A., M. Galiano, C. Videla, G. Carballal, B. Garcia-Barreno, J. A. Melero, and C. Palomo. 2003. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J. Gen. Virol. 84:3115-3120. [DOI] [PubMed] [Google Scholar]
- 31.Trento, A., M. Viegas, M. Galiano, C. Videla, G. Carballal, A. S. Mistchenko, and J. A. Melero. 2006. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J. Virol. 80:975-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venter, M., S. A. Madhi, C. T. Tiemessen, and B. D. Schoub. 2001. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J. Gen. Virol. 82:2117-2124. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y., W. Xu, K. Shen, Z. Xie, L. Sun, Q. Lu, C. Liu, G. Liang, J. A. Beeler, and L. J. Anderson. 2007. Genetic variability of group A and B human respiratory syncytial viruses isolated from 3 provinces in China. Arch. Virol. 152:1425-1434. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, R. N., X. H. Geng, and Z. L. Wang. 1998. Identification of subgroups of respiratory syncytial virus by reverse transcription polymerase chain reaction. Chin. J. Pediatr. 36:538-540. [Google Scholar]
- 35.Zlateva, K. T., P. Lemey, E. Moes, A. M. Vandamme, and M. Van Ranst. 2005. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J. Virol. 79:9157-9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zlateva, K. T., L. Vijgen, N. Dekeersmaeker, C. Naranjo, and M. Van Ranst. 2007. Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons. J. Clin. Microbiol. 45:3022-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]