Abstract
More than 10 million people are thought to be infected with Trypanosoma cruzi, primarily in the Americas. The clinical manifestations of Chagas' disease (CD) are variable, but most subjects remain asymptomatic for decades. Only 15 to 30% eventually develop terminal complications. All current diagnostic tests have limitations. New approaches are needed for blood bank screening as well as for improved diagnosis and prognosis. Sera from subjects with asymptomatic CD (n = 131) were compared to those from uninfected controls (n = 164) and subjects with other parasitic diseases (n = 140), using protein array mass spectrometry. To identify biomarkers associated with CD, sera were fractionated by anion-exchange chromatography and bound to two commercial ProteinChip array chemistries: WCX2 and IMAC3. Multiple candidate biomarkers were found in CD sera (3 to 75.4 kDa). Algorithms employing 3 to 5 of these biomarkers achieved up to 100% sensitivity and 98% specificity for CD. The biomarkers most useful for diagnosis were identified and validated. These included MIP1 alpha, C3a anaphylatoxin, and unusually truncated forms of fibronectin, apolipoprotein A1 (ApoA1), and C3. An antipeptide antiserum against the 28.9-kDa C terminus of the fibronectin fragment achieved good specificity (90%) for CD in a Western blot format. We identified full-length ApoA1 (28.1 kDa), the major structural and functional protein component of high-density lipoprotein (HDL), as an important negative biomarker for CD, and relatively little full-length ApoA1 was detected in CD sera. This work provides proof of principle that both platform-dependent (i.e., mass spectrometry-based) and platform-independent (i.e., Western blot) tests can be generated using high-throughput mass profiling.
Chagas' disease (CD), caused by the protozoan parasite Trypanosoma cruzi, is transmitted by triatomid bugs in resource-poor settings. Over 10 million people are thought to be infected worldwide (51), and vector-mediated transmission can occur from regions in the southernmost United States to temperate South America (4, 7). While some CD patients become acutely ill (e.g., local swelling, fever), most are unaware that they are infected and can remain asymptomatic for decades (24). Only 15 to 30% of patients will eventually develop cardiac or gastrointestinal complications (28). CD can also enter into or be maintained in populations through migration, vertical transmission, organ transplantation (11), and blood transfusion (25).
Transfusion-associated transmission of CD occurs both in countries where CD is endemic and in those where it is not (26, 49). Although serologic screening tests have greatly reduced the risk of transfusion-acquired CD in Latin America (25), serologic methods are imperfect, and >10,000 cases may occur annually in Sao Paulo alone (1). False-positive results can also be a serious concern, resulting in wasted units and permanent losses from the blood donor pool (42). A review of serological procedures in 16 Latin American blood banks from 1997 to 2000 reported false-negative rates of 0.7% to 3.7% and false-positive rates of 0.3% to 3.2%. We recently demonstrated that up to 50% of true CD cases are missed by routine screening in some Venezuelan blood banks (6). Transmission of CD by transfusion/transplantation is also a concern in countries where it is not endemic but where there are large Latin American populations. As many as 100,000 of the 7 to 8 million North American residents of Latin American origin may be silent carriers of T. cruzi (24, 44, 48). Geographic exclusion policies can mitigate risk, but donors do not always provide complete information (32). This situation is further confounded because CD can pass unsuspected from mother to child for several generations (33). In 2007, the CDC reported that 1/4,655 blood donations in the United States was positive for T. cruzi (12), and a small serosurvey of 102 Latin American immigrants in Ontario demonstrated that 1% had CD antibodies (45). The relatively small number of transfusion-transmitted T. cruzi cases reported in North America in the last 2 decades may therefore represent only the tip of the iceberg (22, 25, 35). Several recent reports of CD following organ transplantation further underscore the vulnerability of transfusion and transplantation services in North America (11). A new enzyme-linked immunosorbent assay (ELISA), the Ortho T. cruzi ELISA test system, was approved by the FDA for screening of blood donors and was licensed in December 2006 (21). However, the long history of similar tests in the Americas suggests that new diagnostic strategies should also be pursued.
Surface-enhanced laser desorption ionization-time-of-flight mass spectrometry (SELDI-TOF MS) can provide protein profiles of body fluids. This platform has been used primarily as a discovery tool for biomarkers under neoplastic and inflammatory conditions (53). However, we and others have applied SELDI-TOF MS to studies of human infections, such as African sleeping sickness (37), hepatitis C (43), severe acute respiratory syndrome (SARS) (54), and bacterial endocarditis (19), as well as to studies of fasciolosis in sheep (38) and cysticercosis in pigs (16). There has been some justified criticism of protein profiling efforts, including SELDI-TOF data, when authors have failed to proceed to identification and validation of putative biomarkers (50). In this work, we used SELDI-TOF MS as the first step to compare serum protein profiles of asymptomatic CD-infected subjects, healthy controls, and subjects with other parasitic diseases. Promising biomarkers were independently validated, and second-generation reagents were produced (e.g., antipeptide antibodies). We report on both SELDI-TOF MS platform-based tests and alternative assays derived from this biomarker discovery program (i.e., MS platform independent) that can achieve high sensitivity and specificity for latent CD.
(These data were presented in part at the American Society for Tropical Medicine and Hygiene annual meetings in Miami [7 to 11 November 2004; Proteomic Symposium] and Washington [11 to 15 December 2005; Symposium 17].)
MATERIALS AND METHODS
Samples.
A panel of 435 sera was used, including sera from 120 Venezuelan subjects with asymptomatic CD, identified in blood donor clinics (mean age, 47 ± 12 years; range, 24 to 89 years), and 11 asymptomatic Guatemalan children with CD, identified in a community screening program (mean age, 10 ± 2 years; range, 8 to 13 years) (Table 1). Samples were classified as positive based on at least two different reference assays, including an immunofluorescence assay (IFA), indirect hemagglutination assay (IHA), and ELISA (National Chagas Immunodiagnosis Laboratory, Venezuela; Universidad del Valle, Guatemala; and/or CDC, Atlanta, GA). Six of the 11 Guatemalan children had abnormal or borderline electrocardiograms (EKGs) (hypertrophic cardiomyopathy with incomplete right bundle branch block [RBBB], right-axis deviation, low-voltage and T-wave changes in leads II, III, and F, and ventricular extra systole), and 5 displayed normal tracings. These field readings were confirmed for 10/11 subjects by a pediatric cardiologist (M. Béland, Montreal Children's Hospital, Montreal, Quebec, Canada). The last EKG, read as “normal” in the field, was not available for review. No EKG information was available for the Venezuelan subjects. Healthy control CD-negative sera were obtained from Venezuelan blood banks in areas of CD endemicity (VHC group) (n = 104; mean age, 46 ± 19 years; range, 18 to 82 years) and from nontraveling Canadians (CHC group) (n = 60; mean age, 42 ± 15 years; range, 23 to 68 years). The Venezuelan CD-negative subjects were so classified on the basis of a single negative ELISA. Biomarker specificity was assessed using a panel of 140 sera from patients with other parasitic diseases (OPDs), obtained from the National Reference Laboratory for Parasitology (NRCP; Montreal, Quebec, Canada). These OPD samples included 10 each from the following infections: asymptomatic and acute falciparum malaria, South American cutaneous or mucocutaneous leishmaniasis, babesiosis, latent toxoplasmosis, West African sleeping sickness, trichinellosis, strongyloidiasis, Bancroftian filariasis, toxocariasis, echinococcosis, cysticercosis, schistosomiasis (Schistosoma mansoni), and metorchiasis. A subset of 153 Venezuelan sera (83 CD and 70 healthy control sera) was analyzed on three different ProteinChip PBS IIc systems at different times (NRCP, Montreal, Quebec, Canada; Ciphergen Biomarker Discovery Centre, Malvern, PA; and Concordia University, Montreal, Quebec, Canada).
TABLE 1.
Demographic data for the samples used in this study
| Sample classification by ELISA, microscope examination, and/or EKG | No. of samples |
Age range (yr) | Mean age (yr) | ||
|---|---|---|---|---|---|
| Total | Samples from males | Samples from females | |||
| Venezuelan CD | 120 | 48 | 72 | 29-89 | 47 |
| Guatemalan CD | 11 | 5 | 6 | 8-13 | 10 |
| Venezuelan healthy control | 104 | 57 | 47 | 18-82 | 46 |
| Canadian healthy control | 60 | 28 | 32 | 23-68 | 42 |
| Other parasitic diseases (OPD) | 140 | ||||
| Acute falciparum | 10 | 6 | 4 | 10-36 | 21 |
| Asymptomatic falciparum | 10 | 5 | 5 | 26-45 | 35 |
| Leishmaniasis | 10 | 7 | 3 | 10-50 | 28 |
| Babesiosis | 10 | 28-55 | 41 | ||
| Latent toxoplasmosis | 10 | 2 | 8 | 31-47 | 39 |
| Human African trypanosomiasis | 10 | 4 | 6 | 38-59 | 48 |
| Trichinellosis | 10 | 7 | 3 | 27-42 | 38 |
| Strongyloidiasis | 10 | 7 | 3 | 24-46 | 33 |
| Bancroftian filariasis | 10 | 6 | 4 | 38-62 | 49 |
| Toxocariasis | 10 | 4 | 6 | 20-38 | 27 |
| Echinococcosis | 10 | 7 | 3 | 13-32 | 23 |
| Cysticercosis | 10 | 7 | 3 | 18-58 | 35 |
| Schistosomiasis | 10 | 4 | 6 | 15-47 | 29 |
| Metorchiasis | 10 | 7 | 3 | 26-62 | 44 |
Serum fractionation.
All binding and washing steps were performed using a BioMek 2000 robot (Beckman Coulter, Fullerton, CA) extended by an integrated microplate shaker (MicroMix 5; Diagnostic Products Company, Los Angeles, CA) that holds an array bioprocessor (Bio-Rad Laboratories, Hercules, CA). The samples were fractionated by pH, using a ProteinChip serum fractionation kit (Bio-Rad). The kit consists of a 96-well filtration plate with Q HyperD F anion-exchange beads that require rehydration and equilibration before use. Two hundred microliters of rehydration buffer (50 mM Tris-HCl, pH 9) was added two times to each well and equilibrated three times with U1 buffer [1 M urea, 2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM Tris-HCl, pH 9]. Prior to fractionation, 20 μl of serum sample was mixed with 30 μl of U9 buffer (9 M urea, 2% CHAPS, and 50 mM Tris-HCl, pH 9) in a 96-well V-bottom plate for 20 min. The sample was then diluted with 50 μl of U1 buffer. One hundred microliters of the diluted serum sample was applied to each well, incubated, and mixed on a MicroMix machine for 30 min. The flowthrough was collected by vacuum filtration into V-bottom microplates. The anion-exchange resin was incubated with an additional 100 μl of Tris-HCl buffer (50 mM Tris-HCl, 0.1% octyl β-d-glucopyranoside [OGP], pH 9) for 10 min at room temperature with shaking. The wash was collected by vacuum filtration. This procedure was repeated twice with 100 μl each of appropriate buffers with decreasing pH (pH 7, 5, 4, and 3). The final wash was performed with an organic wash buffer containing 33% (vol/vol) isopropanol and 16.7% (vol/vol) acetonitrile in 0.1% trifluoroacetic acid (TFA) as previously described in detail (38). The six fractions, each collected twice, were pooled. The pooled fractions were divided over two separate standard V-bottom 96-well plates to reduce the number of freeze-thaw cycles. After fractionation, plates were stored at −80°C until analysis.
Preparation of ProteinChip arrays.
For quality control purposes, commercial human reference serum samples from healthy donors (Valley Biomedical, Winchester, VA) and blind duplicates were included with test serum samples. Samples were randomized within and across arrays to reduce analytical bias and were run blind, so that exposure status was unknown. Blank spots were included across the bioprocessor as negative controls.
(i) WCX ProteinChip array preparation (CM10 ProteinChip).
Prior to sample loading, spots were equilibrated two times with 150 μl of CM binding/washing buffer (0.1 M sodium acetate, pH 4). Ten microliters of fractionated serum sample was incubated in 90 μl of binding buffer (0.1 M sodium acetate) for 30 min on a shaker at room temperature. Afterwards, arrays were washed three times with 150 μl binding buffer and two times with deionized water to remove unbound serum proteins. After the arrays were air-dried, 1 μl of energy-absorbing matrix (saturated sinapinic acid [Sigma, St. Louis, MO] in an aqueous solution containing 50% acetonitrile and 0.5% trifluoroacetic acid) was added twice to each spot. The surface was allowed to air dry between each application.
(ii) IMAC ProteinChip array preparation (IMAC30 ProteinChip).
Spots were first charged with 50 μl 0.1 M copper sulfate for 5 min, followed by a first wash with 200 μl deionized water to remove the unbound metal. A second wash was done with 150 μl of neutralization buffer (0.1 M sodium acetate, pH 4), and the last wash was done with deionized water. Each wash was performed by 5 min of incubation at room temperature. Arrays were then incubated two times with 150 μl binding buffer (0.1 M sodium phosphate, 0.5 M NaCl, pH 7) for 5 min. Ten microliters of fractionated serum sample was spotted onto arrays with 90 μl of binding buffer for 30 min on a shaker at room temperature. After the binding step, washing and addition of energy-absorbing matrix were done as described above.
Data acquisition and analysis.
Arrays were analyzed in a ProteinChip biology system reader (model PBS IIc) equipped with an autoloader using ProteinChip software, version 3.2 (both from Bio-Rad Laboratories). Each spot was read at low-energy (LE) and high-energy (HE) laser intensities. Data were collected up to m/z 200,000 as previously described (16). All spectra were subjected to mass calibration based on the settings used to collect the data, using external calibration standards (bovine insulin, 5,733.6 Da; ubiquitin, 8,564.84 Da; cytochrome c, 12,230.9 Da; β-lactoglobulin, 18,363.3 Da; horseradish peroxidase [HRP], 43,240 Da; and IgG, 147,300 Da). The baseline was subtracted using a setting of 15 times the expected peak width. Noise was subtracted at 2,000 Da for the low-energy intensity and at 10,000 Da for the high-energy intensity. All data were normalized by total ion current for either low intensity (2 to 100 kDa) or high intensity (10 to 200 kDa), using an external coefficient of 0.2. Spectra with normalization factors of more than double the mean were deleted. Reproducibility for quality control samples was evaluated by the coefficient of variance (CV = σ/μ × 100) for m/z and peak intensities in intervals of 2 to 100 kDa for the LE intensity and 10 to 200 kDa for the HE intensity, with automatic clustering of m/z values differing by maximums of 0.3 and 2.0%, respectively, using Biomarker Wizard software (Bio-Rad Laboratories). Noise was calculated within the respective mass region, and software settings were a signal-to-noise ratio (S/N) of 5 and a minimum peak threshold of 50%.
Analyses were performed in two steps. In the first step, automated peak detection was applied, using cluster features of Biomarker Wizard software (version 5.0). Peak sets were then generated across multiple spectra in two passes of increasing stringency. A peak cluster was recorded if a given peak was found in ≥10% of the spectra in one group. Automatic peak detection was performed using cutoff settings of three times the S/N for the first pass and two times the S/N for the second pass. The cluster mass window was set at 0.3% of the peak mass for the LE intensity and at 2% of the peak mass for the HE intensity. Initial P values for differences in peak intensities between CD and control groups for each cluster were generated using the Mann-Whitney U test. As a quality control, peak clusters with P values of <0.05 were visually inspected, followed by manual peak relabeling. In the second step, after relabeling, the peak intensity values of the duplicates were averaged, and exact P values for differences in average peak intensity between groups were calculated (Wilcoxon exact test). Peaks with P values of <0.01 (CD versus healthy control and OPD sera) were considered to represent potential biomarkers. We assessed diagnostic performance by estimating sensitivity and specificity and by determining the area under the receiver-operating characteristic (ROC) curve for each potential biomarker (sensitivity versus 1 − specificity). An area under the curve (AUC) of 1 suggests good diagnostic power, while AUC values that approach 0.5 indicate little diagnostic potential. Biomarker pattern software (Bio-Rad Laboratories) analysis was then applied to the cleaned cluster data. This program uses a supervised pattern classification method (classification and regression tree [CART]) to identify peaks with the greatest contribution to discrimination between groups. The CART, a nonparametric procedure, constructs candidate diagnostic algorithms based on a series of binary decision trees that recursively partition a data set into blocks of predicted positive and negative samples (46). The procedure minimizes a cost function that balances prediction errors and the total number of markers used. The relative importance of a peak in each algorithm is measured by the order in which it is selected in the decision tree and the number of correct predictions it is credited for. For the initial CART analysis, data are randomly split into a training data set, which is used to grow the classification tree, and a validation data set, which is used to evaluate the prediction performance of the tree. CART analysis has been used by clinical investigators to identify diabetic patients with poor glycemic control (8), to differentiate high- and low-risk groups following prostate biopsy (20), to classify injury severity in young and middle-aged adults (41) and pediatric motor vehicle trauma patients (34), and to predict prognosis among patients with unknown primary cancers (23).
Purification of candidate biomarkers.
Serum fractions were treated with protein A HyperD resin (Bio-Rad Laboratories) to deplete IgG and either filter concentrated (YM-30 Microcon units; Millipore, Bedford, MA) or purified by reverse-phase chromatography (RPC Poly-Bio beads; Biosepra, Cergy, France). Enriched fractions were purified in NuPAGE precast gels (Invitrogen Life Technologies, Carlsbad, CA), and colloidal blue-stained bands were excised (colloidal blue staining kit; Invitrogen). For proteins of <20 kDa, whole bands were excised, proteins were extracted with 50% formic acid-25% acetonitrile-15% isopropanol-10% water, and molecular masses of extracted proteins were confirmed by SELDI-TOF MS. Extracts were vacuum dried and digested in solution with modified trypsin (Roche Applied Science, Indianapolis, IN). For proteins of ≥20 kDa, concentrated/purified samples were loaded in duplicate lanes. One lane was used for SELDI-TOF confirmation of molecular masses, and the second was used for in-gel digestion. Tryptic digests were analyzed by tandem mass spectrometry (Q-STAR XL; Applied Biosystems/MDS Sciex, Foster City, CA) on a machine equipped with a PCI-1000 ProteinChip interface (Bio-Rad Laboratories). First, spectra were collected from m/z 1,000 to m/z 3,000 Da in single-MS mode. The most significant unique ions not previously identified as ions of bovine trypsin and human keratins were selected for MS/MS analysis. The collision-induced dissociation spectra were submitted to the database mining tool Mascot (version 2.1.2; Matrix Science) and searched against the updated SwissProt or NCBInr databases, using the following search parameters: trypsin, allowing up to 2 missed cleavages (or semitrypsin if the trypsin search was not successful); peptide tolerance of ±50 ppm; MS/MS tolerance of ±0.3 Da; and peptide charge of +1. Spectra were analyzed using databases of human protein sequences as well as databases of protein sequences from all species, including T. cruzi.
Immunoassay confirmation of biomarkers.
The identities of several of the most promising candidate CD biomarkers were confirmed by immunologic assays. Serum MIP1 alpha levels were measured by a commercial ELISA performed according to the manufacturer's instructions (R&D Systems, Minneapolis, MN). For C3a anaphylatoxin and apolipoprotein A1 (ApoA1) fragments, bead-based immunoassays were performed. Briefly, protein A HyperD beads were loaded with appropriate anti-C3a (clone H13; Chemicon International, Temecula, CA) or rabbit anti-ApoA1 (Calbiochem) antibody or with control antibodies. Beads were washed three times with phosphate-buffered saline (PBS) to remove unbound proteins. Two to five microliters of the test serum samples, diluted to 50 μl in PBS, was bound to the beads for 30 min at room temperature. The beads were washed three times with PBS and once with water. Bound proteins were eluted with 12 μl of 0.1 M acetic acid and analyzed by SELDI-TOF MS, using NP20 or H50 ProteinChip arrays (Bio-Rad Laboratories). Resulting spectra were aligned with the corresponding spectra from the profiling study by use of Ciphergen ProteinChip software to determine whether or not (i) the masses of immunoaffinity-captured proteins matched the masses of the selected biomarkers and (ii) the relative abundances of biomarkers in serum samples observed in the profiling study were reproduced by bead-based immunoassay.
Cloning and expression of fibronectin fragment.
The C-terminally truncated fibronectin sequence (CttFbn) was amplified using cDNA produced from cultured HepG2 cells (ATCC HB-8065; ATCC, Bethesda, MD). cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and PCR amplified using Taq DNA polymerase and the following primers: forward, 5′-CCGGAATTCCACCATCATCATCATCATCAGGCTCAGCAAATGGTTCAG- 3′; and reverse, 5′-CGCGGATCCTCAAACATCGGTGAAGGGGCCAGATC-3′. PCR was performed under the following conditions: denaturation for 3 min at 94°C and 35 cycles of 45 s at 94°C, 30 s at 55 to 65°C, and 1 min at 68°C. The purified PCR product was cloned into YEpFLAG-1 (Sigma, St. Louis, MO), expressed in Saccharomyces cerevisiae BJ3505 cells, and purified using anti-FLAG M2 affinity gel (Sigma) as recommended by the manufacturer.
Antibody against fibronectin fragment and Western blotting.
Antipeptide antibodies were generated against the predicted neo-termini of two ApoA1 fragments (TEHLSTLSEKAKPALEDL and AELQEGARQKLHELQEKL [data not shown]) as well as against the CttFbn fragment. Briefly, peptide RHTSVQTTSSGSGPFTDV, corresponding to amino acids 241 to 258 of mature human fibronectin, was synthesized by BioSynthesis Inc. (Lewisville, TX): this peptide represents the 18 amino acids at the predicted carboxyl end of the fibronectin fragment. A polyclonal rabbit antiserum against the synthetic peptide was generated by Cocalico Biologicals (Reamstown, PA).
Western blot analysis.
CD and control sera were separated in 4 to 12% Novex NuPAGE Bis-Tris gradient gels (Invitrogen) under reducing conditions. Separated proteins were transferred to nitrocellulose membranes. Nonspecific sites were blocked with 5% skim milk in 0.05% PBST (0.05% Tween 20 in PBS) for 1 h at room temperature. Membranes were incubated with rabbit anti-CttFbn antibodies at a 1:500 dilution, followed by incubation with HRP-conjugated anti-rabbit IgG at a 1:100,000 dilution (Amersham Biosciences Co., Piscataway, NJ). The membranes were incubated in SuperSignal West Pico detection solution (Pierce, Rockford, IL) and exposed to X-ray film.
RESULTS
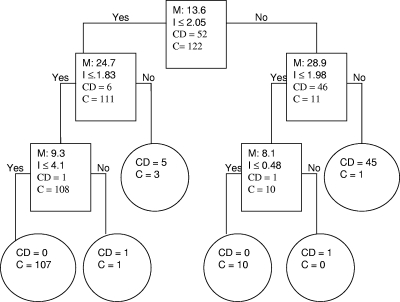
The 435 sera studied yielded a total of 20,880 spectra (6 fractions in duplicate × 2 ProteinChip chemistries × 2 laser intensities). Eighteen classifiers with significant intensity differences between CD and control populations were identified using Biomarker Wizard software (P values of <10−3 to 10−9) (Table 2). Several of these individual biomarkers achieved good sensitivity (84.7 to 98.5%) and specificity (84.9 to 99.0% specificity versus all controls [VHC, CHC, and OPD groups combined], 92.7 to 99.4% specificity versus all healthy control sera [VHC and CHC groups combined], and 72.8 to 99.3% specificity versus sera from other parasitic diseases) (Table 3). To select biomarkers with the greatest discriminatory power, biomarker pattern software was used to generate random training and test data sets and candidate decision trees. This iterative process yielded a total of 45 diagnostic algorithms that were subjected to cross-validation. The six peaks that served as the main “splitters” for each training set (i.e., those with the highest individual predictive rates) were chosen, and a candidate algorithm was built using these peaks alone and evaluated with the validation set. A total of 174 samples (52 CD and 122 control samples) were randomly selected for the training phase. The validity and accuracy of each classification algorithm were then challenged with 261 randomly chosen samples (79 CD and 182 control samples) that constituted the validation set. Diagnostic algorithms that included 2 to 5 of the individual biomarkers generally yielded progressively better sensitivity (86 to 100%) and specificity (76 to 98%). An example of a 5-node decision tree that achieved 98% sensitivity and 100% specificity is presented in Fig. 1, using biomarkers of m/z 8.1, 9.3, 13.6, 24.7, and 28.9 kDa. In effect, each of these algorithms represents a candidate SELDI-based assay for CD (i.e., platform-dependent assay). Six of the most discriminatory biomarkers (8.1, 9.3, 10.1, 13.6, 24.7, and 28.9 kDa) appeared in more than half of the algorithms and differed markedly in intensity between CD-positive subjects and controls (e.g., range of fold differences for CD subjects versus controls [VHC and CHC] or OPD sera of 2.5 to 18.1). The AUCs of the ROC curves for several of these individual biomarkers exceeded 0.80 (Table 2). Potential sample bias was reviewed for each of the differentially expressed biomarkers listed in Tables 2 and 3, but there were no significant differences in the sex or age distributions or in the signal intensities between the groups.
TABLE 2.
Mass (m/z), mean signal intensities, and AUCs for selected individual differentially expressed peptides/proteins between Chagasic patients (CD), healthy controls (HC; Venezuelan healthy controls and Canadian healthy controls), and control patients infected with other parasites (OPD)
| m/z (/1,000) | Fractions and chemistries | P value (CD vs HC), AUC for ROC curve (fold CD/HC) | Mean signal intensity ± SD |
P value (CD vs OPD), AUC for ROC curve (fold CD/OPD) | ||
|---|---|---|---|---|---|---|
| CD (n = 131) | HC (n = 164) | OPD (n = 140) | ||||
| 3.7 | F1 IMAC; F3,6 WCX | 0.003, 0.80 (+5.9) | 1.87 ± 0.36 | 0.31 ± 0.16 | 0.62 ± 0.28 | 0.00001, 0.78 (+3) |
| 4.4 | F1,5 IMAC; F1,2 WCX | 0.001, 0.81 (+2.7) | 2.87 ± 0.92 | 1.06 ± 0.20 | 1.49 ± 0.46 | 0.00005, 0.75 (+1.9) |
| 5.4a | F1 IMAC; F2,3 WCX | 0.005, 0.20 (−2.4) | 1.29 ± 1.38 | 3.11 ± 2.0 | 2.46 ± 1.70 | 0.001, 0.32 (−1.9) |
| 6.3b | F1,3,6 IMAC | 0.0007, 0.80 (+2) | 1.99 ± 1.21 | 0.99 ± 0.42 | 0.73 ± 0.08 | 0.0002, 0.82 (+2.7) |
| 6.6 | F1,5 WCX | 0.006, 0.79 (+2.4) | 2.56 ± 0.98 | 1.06 ± 0.97 | 0.48 ± 0.05 | 0.0001, 0.83 (+5.3) |
| 7.5b | F1,3 WCX | 0.0001, 0.86 (+6) | 3.16 ± 2.73 | 0.52 ± 0.25 | 1.12 ± 0.6 | 0.0003, 0.75 (+2.8) |
| 7.8 | F1,3 IMAC; F3,6 WCX | 0.0004, 0.79 (3.4) | 3.78 ± 1.57 | 1.11 ± 0.85 | 1.3 ± 0.92 | 0.006, 0.76 (+2.9) |
| 8.1 | F1,2,3 IMAC; F3,4 WCX | 0.0006, 0.83 (+5) | 1.68 ± 0.64 | 0.37 ± 0.53 | 0.39 ± 0.53 | 0.0001, 0.80 (+2.5) |
| 9.3 | F1 IMAC; F1 WCX | 0.0006, 0.78 (+6) | 8.37 ± 3.69 | 1.38 ± 2.38 | 0.46 ± 0.58 | 0.00002, 0.80 (+18.1) |
| 10.1 | F4 IMAC; F1,4 WCX | 0.0003, 0.80 (+5.1) | 2.01 ± 0.41 | 0.39 ± 0.61 | 0.33 ± 0.59 | 0.000000007, 0.88 (+6) |
| 12.7 | F1 IMAC | 0.002, 0.79 (+3.3) | 2.15 ± 0.52 | 0.65 ± 1.62 | 0.71 ± 0.98 | 0.005, 0.79 (+3) |
| 13.6 | F1 IMAC; F4,5 WCX | 0.0001, 0.82 (+5.8) | 2.41 ± 0.39 | 0.41 ± 0.63 | 0.65 ± 0.4 | 0.0001, 0.80 (+3.7) |
| 15.2b | F5 WCX | 0.001, 0.84 (+9.6) | 1.26 ± 0.15 | 0.13 ± 0.11 | 0.16 ± 0.09 | 0.0009, 0.71 (+7.5) |
| 16.3 | F1,3 IMAC | 0.002, 0.80 (+3.2) | 1.08 ± 021 | 0.33 ± 0.41 | 0.38 ± 0.67 | 0.004, 0.78 (+2.8) |
| 24.7 | F3,4 WCX | 0.001, 0.79 (+5.9) | 2.37 ± 0.42 | 0.40 ± 0.59 | 0.24 ± 0.33 | 0.000006, 0.80 (+9.8) |
| 28.1a | F1,3,5 WCX | 0.00005, 0.18 (−5.7) | 0.69 ± 0.24 | 3.97 ± 0.48 | 3.12 ± 0.31 | 0.004, 0.23 (−4.2) |
| 28.9 | F1 IMAC; F1 WCX | 0.000001, 0.86 (+12.8) | 2.45 ± 0.39 | 0.19 ± 0.41 | 0.22 ± 0.30 | 0.00001, 0.82 (+11.1) |
| 75.4 | F6 IMAC | 0.0003, 0.90 (+1.9) | 0.09 ± 0.03 | 0.04 ± 0.02 | 0.05 ± 0.02 | 0.00002, 0.80 (+1.7) |
Downregulated protein in CD-positive subjects.
Upregulated protein in Guatemalan CD-positive children with abnormal EKG readings.
TABLE 3.
Characteristics of selected differentially expressed serum biomarkersc
| m/z (kDa) | Mean intensity (cutoff) | No. of positive samples |
% Sensitivity | % Specificity vs VHC, CHC, and OPD groups | % Specificity vs VHC and CHC groups | % Specificity vs OPD group | |||
|---|---|---|---|---|---|---|---|---|---|
| CD (n = 131) | VHC (n = 104) | CHC (n = 60) | OPD (n = 140) | ||||||
| 3.7 | 1.65 | 126 | 2 | 2 | 4 | 96.9 | 97.4 | 97.6 | 97.1 |
| 4.4 | 2.87 | 125 | 3 | 0 | 4 | 95.4 | 97.7 | 97.6 | 97.1 |
| 5.4a | 1.29 | 9 | 101 | 60 | 135 | 93.2 | 97.4 | 98.2 | 96.4 |
| 6.3b | 1.75 | 13 | 1 | 0 | 2 | 9.9 | 99.0 | 99.4 | 98.6 |
| 6.6 | 2.23 | 126 | 1 | 1 | 5 | 96.1 | 97.7 | 98.8 | 96.4 |
| 7.5b | 2.96 | 11 | 1 | 1 | 1 | 8.4 | 99.0 | 98.8 | 99.3 |
| 7.8 | 3.67 | 128 | 6 | 2 | 38 | 97.7 | 84.9 | 95.1 | 72.8 |
| 8.1 | 1.21 | 129 | 3 | 2 | 10 | 98.4 | 95.0 | 96.9 | 92.8 |
| 9.3 | 4.1 | 129 | 4 | 2 | 6 | 98.5 | 96.0 | 96.3 | 95.7 |
| 10.1 | 1.74 | 129 | 4 | 2 | 6 | 98.5 | 96.0 | 96.3 | 95.7 |
| 12.7 | 1.97 | 128 | 2 | 0 | 2 | 97.7 | 98.7 | 98.8 | 98.6 |
| 13.6 | 2.05 | 111 | 10 | 2 | 16 | 84.7 | 90.8 | 92.7 | 88.6 |
| 15.2b | 1.26 | 14 | 1 | 1 | 1 | 10.7 | 99.0 | 98.8 | 99.3 |
| 16.3 | 1.06 | 126 | 2 | 2 | 6 | 96.2 | 96.7 | 97.6 | 95.7 |
| 24.7 | 1.83 | 124 | 10 | 1 | 3 | 94.6 | 95.4 | 93.3 | 97.8 |
| 28.1a | 2.89 | 7 | 104 | 60 | 136 | 94.6 | 98.7 | 98.8 | 97.1 |
| 28.9 | 1.98 | 125 | 2 | 0 | 2 | 95.4 | 98.7 | 98.8 | 98.6 |
| 75.4 | 0.89 | 125 | 4 | 2 | 6 | 95.4 | 96.0 | 96.3 | 95.7 |
Downregulated protein in CD-positive subjects.
Upregulated protein in Guatemalan CD-positive children with abnormal EKG readings.
Sera were obtained from CD-positive subjects (CD), including all Guatemalan children (with normal and abnormal EKGs) and Venezuelan adults identified in blood donor clinics, from Venezuelan healthy control blood donors (VHC), from Canadian healthy controls (CHC), and from control patients infected with other parasites (OPD).
FIG. 1.
Biomarker pattern software based on CART analysis was used to generate candidate diagnostic algorithms. The CART procedure seeks to minimize a cost function that balances prediction errors in either sense (e.g., false-positive or false-negative results) and the total number of biomarkers used. Equal weight is given to false-positive and false-negative results. The relative importance of each peak in any given algorithm is measured by the order in which it is selected in the decision tree and the number of correct predictions credited to it. An example of decision tree classification in the training data set, using CD and control (C) (VHC, CHC, and OPD) samples, is shown. The numbers in the root node (top box), the descendant nodes (lower boxes), and the terminal nodes (ovals) represent the biomarker peak mass (M [kDa]), the peak intensity criterion (I), and the class (CD and C), respectively. In this algorithm, the intensities of the 8.1-, 9.3-, 13.6-, 24.7-, and 28.9-kDa biomarkers establish the splitting rules. Cases that follow the rule are placed in the left daughter node (yes), and samples that do not follow the rule go to the right (no) daughter. For example, the first splitting-rule question asks the following: “Does the peak at 13.6 kDa have an intensity of ≤2.05?”
All six of these highly discriminatory biomarkers were of host origin (Table 4) and included (i) a C-terminal truncation of C3a desArg (8.1 kDa; also evident as a doubly charged ion at m/z 4.4 kDa and a dimer at m/z 16.3 kDa), (ii) three N-terminally truncated forms of apolipoprotein A1 (9.3 kDa, 10.1 kDa, and 13.6 kDa), (iii) a C-terminally truncated form of apolipoprotein A1 (24.7 kDa), and (iv) a C-terminal truncation of fibronectin (28.9 kDa). The predicted identities and masses of the tryptic fragments of these 6 biomarkers are presented in Table 3. The identities of several of these biomarkers were verified by either commercial ELISA (e.g., MIP1 alpha) or immunobead capture (9.3-, 10.1-, and 13.6-kDa ApoA1 fragments and 8.1-kDa fragment of C3a desArg). Representative data for the 9.3-kDa ApoA1 fragment are presented in Fig. 2 (other data not shown). No biomarkers of parasite origin were identified. One of the biomarkers downregulated in CD subjects (versus CHC, VHC, and OPD groups) was identified as full-length ApoA1 (28.1 kDa) (P < 00005) (Table 2). Very few of the CD-positive subjects had a strong 28.1-kDa ApoA1 peak (7/131 samples [5%]), whereas 300/304 control samples (98.7%) had a peak intensity of >2.89 (Table 3).
TABLE 4.
Sample identification of selected candidate biomarkers
| m/z value (/1,000) | Protein identity | Positions | No. of ions sequenced (% sequence coveragea) | Predicted mass (Da) | Predicted pI |
|---|---|---|---|---|---|
| 8.1 | C-terminal truncation of C3a anaphylatoxin desArg (Ctt-C3a) | 1-68 | 4 (50) | 8,126.52 | 9.38 |
| 9.3 | N-terminal truncation of apolipoprotein A1 (Ntt-ApoA1a) | 161-243 | 8 (84) | 9,306.59 | 6.77 |
| 10.1 | N-terminal truncation of apolipoprotein A1 (Ntt-ApoA1b) | 154-243 | 8 (78) | 10,069.46 | 7.15 |
| 13.6 | N-terminal truncation of apolipoprotein A1 (Ntt-ApoA1c) | 124-243 | 6 (54) | 13,570.40 | 6.6 |
| 24.7 | C-terminal truncation of apolipoprotein A1 (Ctt-ApoA1) | 1-214 | 7 (36) | 24,756.76 | 5.15 |
| 28.9 | C-terminal truncation of fibronectin (Ctt-Fbn) | 1-258 | 7 (43) | 28,730.95 | 7.49 |
Percentage of predicted tryptic peptides sequenced by tandem mass spectrometry.
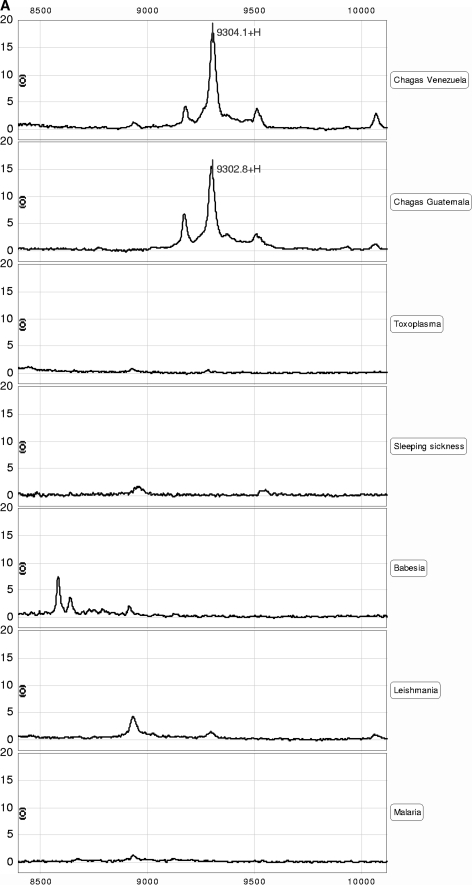
FIG. 2.
SELDI-TOF MS identification of the 9.3-kDa candidate biomarker. (A) Representative spectral view of IMAC spectra of the Chagas' disease sera and the parasite controls (Toxoplasma, African sleeping sickness, Babesia, Leishmania, and malaria). The 9.3-kDa protein, detected in Chagas' disease but not other diseases, is shown. (B) The 9.3-kDa protein was purified from serum samples by anion-exchange chromatography followed by reverse-phase chromatography and, finally, 16% Tricine SDS-PAGE. Coomassie blue-stained bands were extracted and reprofiled on NP20 arrays. A band corresponding to the 9.3-kDa protein was digested in solution with trypsin, and the digest was analyzed by tandem mass spectrometry. Eight peptides were identified as components of a C-terminal fragment of ApoA1. (C) Amino acid sequence of human ApoA1. The amino acid sequence encompassed by the peptides identified by tandem mass spectrometry is highlighted in bold. The calculated molecular mass for the highlighted sequence is 9,306.59 Da. (D) Immunocapture of C-terminal fragments from serum samples, using rabbit polyclonal antibody against ApoA1.
Although SELDI platform-dependent assays could eventually have clinical utility, we sought to exploit the unanticipated discovery of abnormally truncated host proteins to develop a diagnostic assay that does not rely on this platform. By Western blotting, rabbit antipeptide sera against the C-terminal sequence of the 28.9-kDa Fbn (Fig. 3A and B) fragment detected multiple Fbn fragments in both CD and healthy control sera (25, 40, and 45 kDa), but the 28.9-kDa SELDI biomarker was predominantly observed in CD sera (arrows in Fig. 4A and B). The 28.9-kDa Fbn fragment was not detected in any of the 140 OPD sera tested (Fig. 4C). Although the sensitivity (78%) and specificity (90%) of this initial candidate “second-generation” assay were modest, these data nonetheless provide proof of principle that MS platform-independent assays can also be generated using a biomarker approach. The antipeptide rabbit polyclonal antibodies directed against either the 24.7-kDa C-terminal fragment or the 13.6-kDa N-terminal fragment of ApoA1 confirmed the presence of these protein fragments in sera of CD subjects but were not as specific as the anti-Fbn antiserum in Western blots (data not shown).
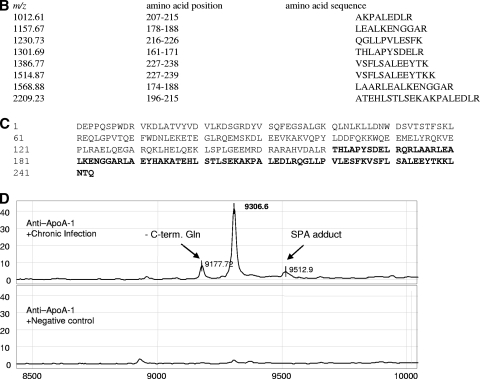
FIG. 3.
SELDI-TOF MS identification of 28.9-kDa candidate biomarker. (A) The 28.9-kDa protein was enriched from serum samples by anion-exchange chromatography (flowthrough) and protein A chromatography (flowthrough), concentrated/desalted using YM30 filtration, and finally purified by 12% Bis-Tris SDS-PAGE. Coomassie blue-stained bands were extracted and reprofiled on NP20 arrays. A band corresponding to the 28.9-kDa protein was digested in solution with trypsin, and the digest was analyzed by tandem mass spectrometry. Seven peptides were identified as components of the N-terminal fragment of fibronectin. (B) Sequence of the first 300 amino acids of human fibronectin. The full-length sequence of fibronectin, consisting of 2,355 amino acids, is not shown. The fragments identified by MS/MS are highlighted in bold. Importantly, one fragment with m/z 1,707.78 Da had a nontryptic cut at the C terminus, indicating that this was the C-terminal end of the 28.9-kDa protein. The position of the peptide used to generate the rabbit antiserum is underlined. The calculated molecular mass of the sequence from the N terminus to Val258, with the N-terminal Gln modified to pyrrolidone carboxylic acid and with nine disulfide bonds, is 28,931 Da. The putative C-terminal Val258 is bold and underlined.
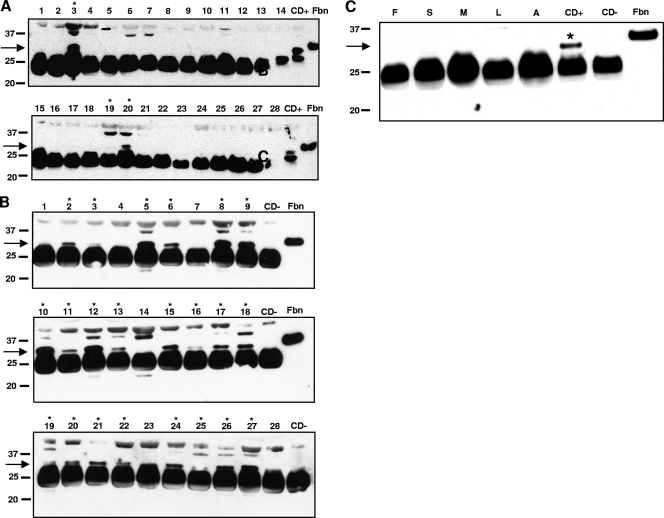
FIG. 4.
Western blot analysis of sera from patients with CD or other parasitic diseases, probed with anti-CttFbn peptide sera. (A) Analysis of 28 CD-negative control sera shows that the C-terminally truncated fragment of Fbn (28.9 kDa) was detected in only 3/28 sera (11%). (B) Analysis of 28 CD-positive sera shows that the 28.9-kDa band was detected in 22/28 sera (79%). Fbn, control recombinant Fbn fragment; CD+, pool of 8 positive Chagasic sera; CD−, pooled Canadian negative sera. Lanes in which the 28.9-kDa band was present are marked by asterisks. (C) Analysis of pools of sera from patients with fasciolosis (F), schistosomiasis (S), malaria (M), leishmaniasis (L), African trypanosomiasis (A), or Chagas' disease (CD+) or from Canadian negative controls (CD−). The antipeptide serum detected a band near 28.9 kDa only in the CD+ serum pool.
Three of the highly specific CD biomarkers (6.3, 7.5, and 15.2 kDa) were discovered only through analysis of the Guatemalan sera and were found only in subjects with EKG changes. These three biomarkers had poor sensitivities overall among all CD sera (9.9, 8.4, and 10.7%, respectively) but were very specific for CD complicated by silent cardiac involvement (ROC values in the range of 0.80 to 0.86 versus HC or OPD groups assumed to be free of cardiac disease; range of P values, <2 × 10−5 to 10−6). These biomarkers were present in 4 to 6.6% of the CD-positive samples from the asymptomatic Venezuelan blood donors (data not shown) but in very few of the 304 healthy and OPD control samples (∼1%). These biomarkers have not yet been identified but may be associated with the early cardiac complications of CD.
DISCUSSION
There is no gold standard assay for CD, but tests in current use include ELISAs, IFA, PCR, and even xenodiagnosis. Better tests are needed to protect transfusion and transplant recipients worldwide and to address the needs of frontline physicians in regions where CD is endemic. Our study provides the first evidence that discrete biomarker peaks (m/z values) can be combined in MS platform-based diagnostic algorithms to achieve high sensitivity and specificity for CD. We further demonstrate that knowledge of such biomarkers can be used to generate novel diagnostic reagents for use in standard assay formats. Antibodies directed against one of the candidate biomarkers identified, a 28.9-kDa fibronectin fragment, proved to be highly specific for CD in Western blots.
WHO and Pan American Health Organization (PAHO) guidelines promote the use of serologic screening in countries where CD is endemic to limit the risk of transfusion-associated disease (6). However, several groups have reported PCR-positive and even microscopy-positive results for seronegative patients (40, 52). Indeed, 100% concordance between multiple serologic assays is rarely achieved (6). Discordant results may arise due to technical issues or as a result of the complex host-parasite interaction in CD. It is therefore promising that specific biomarker peaks (m/z values) could be combined in algorithms to achieve a sensitivity of >98% and a specificity of >94%. These findings were reproducible using three different PBS IIc machines in three geographic locations and at different times (data not shown). Although sera from subjects with nonparasitic infections or inflammatory/neoplastic conditions were not included in the current study, at least two of our candidate biomarkers that might logically occur in such illnesses (MIP1 alpha and C3) were not found in the OPD group, which included cases of symptomatic falciparum malaria and babesiosis. Furthermore, these biomarkers have not been reported (by m/z score, at least) in published biomarker studies of African trypanosomiasis (37) or bacterial endocarditis (19). However, a conceptually similar approach based on proinflammatory cytokine profiles (including MIP1 alpha) has been suggested for the early diagnosis of various cancers (27, 30).
The sera used in our study were fractionated without prior depletion of the most abundant serum proteins. Since several of the most useful biomarkers proved to be fragments of these abundant proteins, this strategy was clearly justified. However, high-abundance proteins can mask the presence of lower-abundance proteins of either host or parasite origin in MS studies. The identification of one or more parasite-derived serum biomarkers would greatly enhance the specificity of any candidate MS, pattern-based assay, even if the absolute contribution to sensitivity was limited. Studies are ongoing to deplete abundant proteins (14) in pursuit of parasite-origin biomarkers. However, the expectation of a relatively low parasite burden in chronic CD and the rapid antibody-mediated clearance of parasite proteins (47) will likely complicate this effort.
The discovery of unusually truncated host proteins as valuable biomarkers of CD was unanticipated. Given the complexities of the host-parasite interaction in CD, there are several plausible mechanisms for the generation of these host protein fragments. These include specific cleavage or bystander metabolism by parasite-encoded proteases (e.g., cruzipain) (5) or unusual cleavage by host proteases (e.g., matrix metalloproteinase) at the host-parasite interface (i.e., the result of host defenses or pathology) (18). Several lines of evidence suggest that T. cruzi may specifically target some of the host proteins implicated as biomarkers in our study. For example, Fbn is a multifunctional glycoprotein found in the extracellular matrix (ECM), in connective tissue, on cell surfaces, and in plasma and other body fluids. As a prerequisite for host cell invasion, T. cruzi infective forms must cross the basement membrane and ECM barriers (29). Calvet et al. have shown that the RGD sequence of Fbn is an important adhesive molecule for T. cruzi surface receptors in cardiomyocytes (9). It has been proposed that either soluble or fixed Fbn could act as a bridge between the parasite and the cell (36).
We also identified full-length C3a and several C3 derivatives (e.g., dimeric C3 and C3a desArg) as useful serum biomarkers for CD. The human complement system provides broad-spectrum protection against invading microorganisms, including parasites. Of the 25 known soluble complement proteins, C3 is the most abundant in serum (1.2 g/liter), supporting all 3 pathways of complement activation (39). Once activated by molecules on the surfaces of microorganisms or nonself signals, native C3 (185 kDa) is cleaved into C3b (176 kDa) and C3a (9 kDa). C3a is typically short lived in serum and is cleaved immediately to form the more stable C3a desArg (8.9 kDa) by carboxypeptidases. As a result, it was not particularly surprising that C3 and C3a desArg were included among our candidate biomarkers for CD. However, the relative specificity of these observations for CD (e.g., versus OPD) and the presence of the unusually truncated form of C3a desArg (8.1 kDa) are both intriguing observations (Tables 2 and 3).
ApoA1 was among the most powerful candidate biomarkers for CD that we identified. In addition to the discovery of several distinct C- and N-terminally truncated fragments as useful positive biomarkers, we also identified full-length ApoA1 (28.1 kDa) as an important negative biomarker for CD. Relatively little full-length ApoA1 was detected in CD sera compared to that in VHC, CHC, and OPD controls (Tables 2 and 3). ApoA1 is the major structural and functional protein component of high-density lipoprotein (HDL), where it serves as a cofactor for esterification (15) of plasma cholesterol. Interaction between T. cruzi and ApoA1 could occur at several levels. The possibility that HDL serves some nutritional need of the parasite is supported by the observations that epimastigote growth can be slowed significantly in vitro by lipid depletion (M. Ndao and B. J. Ward, unpublished observations) and that the transition of T. cruzi trypomastigotes to amastigotes is accompanied by a shift from carbohydrate- to lipid-dependent energy metabolism (2). Furthermore, we recently demonstrated that exposure of human HDL to cruzipain during in vitro and in vivo infections of mice with T. cruzi can generate several of the same truncated ApoA1 fragments that we have identified as candidate biomarkers (data not shown). Human adipocytes may serve as tissue reservoirs for this parasite (13), and it is interesting that a human apolipoprotein subset (ApoL1) has a specific lytic effect on some African trypanosomes (3). Relatively little is known about lipid homeostasis in subjects infected with CD. However, the report by Cano et al. (10) that serum ApoA1 levels are low in asymptomatic CD patients is certainly consistent with our data. The known cardioprotective effects of HDL/ApoA1 (31) and the fact that the majority of subjects who succumb to CD die from cardiac causes (17, 28) lend some urgency to further studies of the interaction between T. cruzi and HDL. Whether or not the 3 biomarkers we identified in young CD subjects with EKG changes prove to be prognostically useful will be resolved by ongoing studies of samples from both CD and non-CD subjects with defined cardiac conditions.
Many biomarker discovery programs have foundered on the shoals of biological and clinical relevance. Programs exclusively based on the SELDI-TOF platform, in particular, have struggled with the issues of sensitivity and reproducibility (50). Although our discovery program for parasitic diseases begins with SELDI-TOF, we have been conservative in our interpretation of the protein profiling data and have taken a cautious, stepwise approach to the identification and validation of candidate biomarkers. After demonstrating reproducibility on several machines at different times and in different geographic locations, we used a range of immunologic techniques to validate the most important putative biomarkers. We also generated antipeptide polyclonal sera against the predicted neo-termini of several of the truncated host proteins and demonstrated the presence of the targeted protein fragments by Western blotting. Efforts are now under way to generate monoclonal reagents that will discriminate between the full-length and truncated host proteins for use in traditional immunoassay formats (e.g., solid- and liquid-phase enzyme immunoassays [EIAs], Western blotting).
Our data provide proof of principle that both MS platform-dependent and platform-independent biomarker-based assays can be useful for subjects with latent CD. Rather than replacing antibody- and nucleic acid-based testing, it seems more likely that MS-based or MS-derived assays will provide complementary information (e.g., confirmatory testing, tests for cure or prognosis). This approach may also give unique insights into the complex and prolonged host-parasite interaction that characterizes Chagas' disease.
Acknowledgments
We acknowledge the assistance of Nidia Rizzo and Byron Arana at the Universidad del Valle Guatemala and the Medical Entomology Research and Training Unit, Guatemala City, Guatemala, for collecting samples and Marie Béland for assistance in the interpretation of electrocardiograms (McGill Pediatric Cardiology, Montreal Children's Hospital, Montreal, Quebec, Canada).
This work was supported by grants provided by the Canadian Institutes of Health Research (200402UOP-UI-130124-B and 200401NTA-126473-DAI-CFAC-41490) and by McGill University. T. W. Spithill held a Canada Research Chair in Immunoparasitology.
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Antunes, C. M. F. 1999. Chagas disease (American trypanosomiasis): the epidemiology of Chagas disease, p. 351-369. In H. M. Gilles (ed.), Protozoal diseases. Arnold, London, United Kingdom.
- 2.Atwood, J. A., III, D. B. Weatherly, T. A. Minning, B. Bundy, C. Cavola, F. R. Opperdoes, R. Orlando, and R. L. Tarleton. 2005. The Trypanosoma cruzi proteome. Science 309:473-476. [DOI] [PubMed] [Google Scholar]
- 3.Baral, T. N., S. Magez, B. Stijlemans, K. Conrath, B. Vanhollebeke, E. Pays, S. Muyldermans, and P. De Baetselier. 2006. Experimental therapy of African trypanosomiasis with a nanobody-conjugated human trypanolytic factor. Nat. Med. 12:580-584. [DOI] [PubMed] [Google Scholar]
- 4.Beard, C. B., G. Pye, F. J. Steurer, R. Rodriguez, R. Campman, A. T. Peterson, J. Ramsey, R. A. Wirtz, and L. E. Robinson. 2003. Chagas disease in a domestic transmission cycle, Southern Texas, U.S.A. Emerg. Infect. Dis. 9:103-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berasain, P., C. Carmona, B. Frangione, J. J. Cazzulo, and F. Goni. 2003. Specific cleavage sites on human IgG subclasses by cruzipain, the major cysteine proteinase from Trypanosoma cruzi. Mol. Biochem. Parasitol. 130:23-29. [DOI] [PubMed] [Google Scholar]
- 6.Berrizbeitia, M., M. Ndao, J. Bubis, M. Gottschalk, A. Ache, S. Lacouture, M. Medina, and B. J. Ward. 2007. Field evaluation of four novel enzyme immunoassays for Chagas' disease in Venezuela blood banks: comparison of assays using fixed-epimastigotes, fixed-trypomastigotes or trypomastigote excreted-secreted antigens from two Trypanosoma cruzi strains. Transfus. Med. 16:419-431. [DOI] [PubMed] [Google Scholar]
- 7.Bradley, K. K., D. K. Bergman, J. P. Woods, J. M. Crutcher, and L. V. Kirchhoff. 2000. Prevalence of American trypanosomiasis (Chagas disease) among dogs in Oklahoma. J. Am. Vet. Med. Assoc. 217:1853-1857. [DOI] [PubMed] [Google Scholar]
- 8.Breault, J. L., C. R. Goodall, and P. J. Fos. 2002. Data mining a diabetic data warehouse. Artif. Intell. Med. 26:37-54. [DOI] [PubMed] [Google Scholar]
- 9.Calvet, C. M., M. Meuser, D. Almeida, M. N. Meirelles, and M. C. Pereira. 2004. Trypanosoma cruzi-cardiomyocyte interaction: role of fibronectin in the recognition process and extracellular matrix expression in vitro and in vivo. Exp. Parasitol. 107:20-30. [DOI] [PubMed] [Google Scholar]
- 10.Cano, R. C., E. R. Rubiolo, and N. O. Santamarina. 1985. Levels of apolipoproteins and cholesterol of low and high density lipoproteins in asymptomatic Chagas disease. Medicina 45:269-272. [PubMed] [Google Scholar]
- 11.CDC. 2006. Chagas disease after organ transplantation—Los Angeles, California, 2006. Morb. Mortal. Wkly. Rep. 55:798-800. [PubMed] [Google Scholar]
- 12.CDC. 2007. Blood donor screening for Chagas disease—United States, 2006-2007. Morb. Mortal. Wkly. Rep. 56:141-143. [PubMed] [Google Scholar]
- 13.Combs, T. P., Nagajyothi, S. Mukherjee, C. J. G. de Almeida, L. A. Jelicks, W. Schubert, Y. Lin, D. S. Jayabalan, D. Zhao, V. L. Braunstein, S. Landskroner-Eiger, A. Cordero, S. M. Factor, L. M. Weiss, M. P. Lisanti, H. B. Tanowitz, and P. E. Scherer. 2005. The adipocyte as an important target cell for Trypanosoma cruzi infection. J. Biol. Chem. 280:24085-24094. [DOI] [PubMed] [Google Scholar]
- 14.Darde, V. M., M. G. Barderas, and F. Vivanco. 2007. Depletion of high-abundance proteins in plasma by immunoaffinity subtraction for two-dimensional difference gel electrophoresis analysis. Methods Mol. Biol. 357:351-364. [DOI] [PubMed] [Google Scholar]
- 15.Davidson, W. S., and R. A. Silva. 2005. Apolipoprotein structural organization in high density lipoproteins: belts, bundles, hinges and hairpins. Curr. Opin. Lipidol. 16:295-300. [DOI] [PubMed] [Google Scholar]
- 16.Deckers, N., P. Dorny, K. Kanobana, J. Vercruysse, A. E. Gonzalez, B. Ward, and M. Ndao. 2008. Use of ProteinChip technology for identifying biomarkers of parasitic diseases: the example of porcine cysticercosis (Taenia solium). Exp. Parasitol. 120:320-329. [DOI] [PubMed] [Google Scholar]
- 17.de Oliveira, R. A., S. de Moraes, R. R. de Andrede, and M. R. Montenegra. 2000. Clinical and pathological assessment of 82 patients with cardiovascular diseases undergoing autopsy at the Hospital das Clinicas of the Faculdade de Medicina de Botucatu from 1988 to 1993. Arq. Bras. Cardiol. 75:303-312. [DOI] [PubMed] [Google Scholar]
- 18.Eberini, I., L. Calabresi, R. Wait, G. Tedeschi, A. Pirillo, L. Puglisi, C. R. Sirtori, and E. Gianazza. 2002. Macrophage metalloproteinases degrade high-density-lipoprotein-associated apolipoprotein A-I at both the N- and C-termini. Biochem. J. 362:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenollar, F., A. Goncalves, B. Esterni, S. Azza, G. Habib, J. P. Borg, and D. Raoult. 2006. A serum protein signature with high diagnostic value in bacterial endocarditis: results from a study based on surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. J. Infect. Dis. 194:1356-1366. [DOI] [PubMed] [Google Scholar]
- 20.Garzotto, M., T. M. Beer, R. G. Hudson, L. Peters, Y. C. Hsieh, E. Barrera, T. Klein, and M. Mori. 2005. Improved detection of prostate cancer using classification and regression tree analysis. J. Clin. Oncol. 23:4322-4329. [DOI] [PubMed] [Google Scholar]
- 21.Gorlin, J., S. S. F. Rossmann, N. Hirschler, K. A. Nguyen, R. Gilcher, H. Fernandes, S. Alvey, P. Ajongwen, P. Contestable, and H. Warren. 2008. Evaluation of a new Trypanosoma cruzi antibody assay for blood donor screening. Transfusion 48:531-540. [DOI] [PubMed] [Google Scholar]
- 22.Grant, I. H., J. Gold, M. Wittner, H. B. Tanowitz, C. Nathan, K. Mayer, L. Reich, N. Wollner, L. Steinherz, and F. Ghavimi. 1989. Transfusion-associated acute Chagas disease acquired in the United States. Ann. Intern. Med. 111:849-851. [DOI] [PubMed] [Google Scholar]
- 23.Hess, K. R., M. C. Abbruzzese, R. Lenzi, M. N. Raber, and J. L. Abbruzzese. 1999. Classification and regression tree analysis of 1000 consecutive patients with unknown primary carcinoma. Clin. Cancer Res. 5:3403-3410. [PubMed] [Google Scholar]
- 24.Kirchhoff, L. V. 1993. American trypanosomiasis (Chagas' disease)—a tropical disease now in the United States. N. Engl. J. Med. 329:639-644. [DOI] [PubMed] [Google Scholar]
- 25.Leiby, D. A., M. H. Fucci, and R. J. Stumpf. 1999. Trypanosoma cruzi in a low- to moderate-risk blood donor population: seroprevalence and possible congenital transmission. Transfusion 39:310-315. [DOI] [PubMed] [Google Scholar]
- 26.Leiby, D. A., B. A. Lenes, M. A. Tibbals, and M. T. Tames-Olmedo. 1999. Prospective evaluation of a patient with Trypanosoma cruzi infection transmitted by transfusion. N. Engl. J. Med. 341:1237-1239. [DOI] [PubMed] [Google Scholar]
- 27.Linkov, F., A. Lisovich, Z. Yurkovetsky, A. Marrangoni, L. Velikokhatnaya, B. Nolen, M. Winans, W. Bigbee, J. Siegfried, A. Lokshin, and R. L. Ferris. 2007. Early detection of head and neck cancer: development of a novel screening tool using multiplexed immunobead-based biomarker profiling. Cancer Epidemiol. Biomarkers Prev. 16:102-107. [DOI] [PubMed] [Google Scholar]
- 28.Maguire, J. H. 2006. Chagas' disease—can we stop the deaths? N. Engl. J. Med. 355:760-761. [DOI] [PubMed] [Google Scholar]
- 29.Marino, A. P., A. A. Silva, R. T. Pinho, and J. Lannes-Vieira. 2003. Trypanosoma cruzi infection: a continuous invader-host cell cross talk with participation of extracellular matrix and adhesion and chemoattractant molecules. Braz. J. Med. Biol. Res. 36:1121-1133. [DOI] [PubMed] [Google Scholar]
- 30.Maurer, M., and E. von Stebut. 2004. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 36:1882-1886. [DOI] [PubMed] [Google Scholar]
- 31.Meyers, C. D., and M. L. Kashyap. 2005. Pharmacologic augmentation of high-density lipoproteins: mechanisms of currently available and emerging therapies. Curr. Opin. Cardiol. 20:307-312. [DOI] [PubMed] [Google Scholar]
- 32.Mungai, M., G. Tegtmeier, M. Chamberland, and M. Parise. 2001. Transfusion-transmitted malaria in the United States from 1963 through 1999. N. Engl. J. Med. 344:1973-1978. [DOI] [PubMed] [Google Scholar]
- 33.Negrette, O. S., M. C. Mora, and M. A. Basombrio. 2005. High prevalence of congenital Trypanosoma cruzi infection and family clustering in Salta, Argentina. Pediatrics 115:e668-e672. [DOI] [PubMed] [Google Scholar]
- 34.Newgard, C. D., R. J. Lewis, and B. T. Jolly. 2002. Use of out-of-hospital variables to predict severity of injury in pediatric patients involved in motor vehicle crashes. Ann. Emerg. Med. 39:481-491. [DOI] [PubMed] [Google Scholar]
- 35.Nickerson, P., P. Orr, M. L. Schroeder, L. Sekla, and J. B. Johnston. 1989. Transfusion-associated Trypanosoma cruzi infection in a non-endemic area. Ann. Intern. Med. 111:851-853. [DOI] [PubMed] [Google Scholar]
- 36.Ouaissi, M. A., J. Cornette, and A. Capron. 1986. Identification and isolation of Trypanosoma cruzi trypomastigote cell surface protein with properties expected of a fibronectin receptor. Mol. Biochem. Parasitol. 19:201-211. [DOI] [PubMed] [Google Scholar]
- 37.Papadopoulos, M. C., P. M. Abel, D. Agranoff, A. Stich, E. Tarelli, B. A. Bell, T. Planche, A. Loosemore, S. Saadoun, P. Wilkins, and S. Krishna. 2004. A novel and accurate diagnostic test for human African trypanosomiasis. Lancet 363:1358-1363. [DOI] [PubMed] [Google Scholar]
- 38.Rioux, M. C., C. Carmona, D. Acosta, B. Ward, M. Ndao, B. F. Gibbs, H. P. Bennett, and T. W. Spithill. 2008. Discovery and validation of serum biomarkers expressed over the first twelve weeks of Fasciola hepatica infection in sheep. Int. J. Parasitol. 38:123-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahu, A., and J. D. Lambris. 2001. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol. Rev. 180:35-48. [DOI] [PubMed] [Google Scholar]
- 40.Salomone, O. A., A. L. Basquiera, A. Sembaj, A. M. Aguerri, M. E. Reyes, M. Omelianuk, R. A. Fernandez, J. Enders, A. Palma, J. M. Barral, and R. J. Madoery. 2003. Trypanosoma cruzi in persons without serologic evidence of disease, Argentina. Emerg. Infect. Dis. 9:1558-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheetz, L. J., J. Zhang, and J. Kolassa. 2009. Classification tree modeling to identify severe and moderate vehicular injuries in young and middle-aged adults. Artif. Intell. Med. 45:1-10. [DOI] [PubMed] [Google Scholar]
- 42.Schmunis, G. A., and J. R. Cruz. 2005. Safety of the blood supply in Latin America. Clin. Microbiol. Rev. 18:12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwegler, E. E., L. Cazares, L. S. Steele, B. L. Adam, D. A. Johnson, O. J. Semmes, T. M. Block, J. A. Marrero, and R. R. Drake. 2005. SELDI-TOF MS profiling of serum for detection of the progression of chronic hepatitis C to hepatocellular carcinoma. Hepatology 41:634-642. [DOI] [PubMed] [Google Scholar]
- 44.Statistics Canada. 2005. Census of Canada. Statistics Canada, Ottawa, Ontario, Canada. http://www12.statcan.ca/english/census01/home/index.cfm.
- 45.Steele, L. S., D. W. MacPherson, J. Kim, J. S. Keystone, and B. D. Gushulak. 2007. The sero-prevalence of antibodies to Trypanosoma cruzi in Latin American refugees and immigrants to Canada. J. Immigr. Minor. Health 9:43-47. [DOI] [PubMed] [Google Scholar]
- 46.Steinberg, D., and P. Colla. 1995. CART: tree-structured non-parametric data analysis. Salford Systems, San Diego, CA.
- 47.Umekita, L. F., S. M. Carneiro, A. Sesso, and I. Mota. 1999. One fate of bloodstream trypomastigote forms of Trypanosoma cruzi after immune clearance: an ultrastructural study. J. Parasitol. 85:867-872. [PubMed] [Google Scholar]
- 48.U.S. Census Bureau. 2005. QuickFacts. U.S. Census Bureau, Washington, DC. http://quickfacts.census.gov/qfd/states/00000.html.
- 49.Wendel, S., and A. L. Gonzaga. 1993. Chagas' disease and blood transfusion: a New World problem? Vox Sang. 64:1-12. [DOI] [PubMed] [Google Scholar]
- 50.Whelan, L. C., K. A. Power, D. T. McDowell, J. Kennedy, and W. M. Gallagher. 2008. Applications of SELDI-MS technology in oncology. J. Cell. Mol. Med. 12:1535-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.WHO. 2000. Certification of interruption of transmission. Wkly. Epidemiol. Rec. 75:10-12.10697461 [Google Scholar]
- 52.Wincker, P., M. F. Bosseno, C. Britto, N. Yaksic, M. A. Cardoso, C. M. Morel, and S. F. Breniere. 1994. High correlation between Chagas' disease serology and PCR-based detection of Trypanosoma cruzi kinetoplast DNA in Bolivian children living in an endemic area. FEMS Microbiol. Lett. 124:419-423. [DOI] [PubMed] [Google Scholar]
- 53.Xiao, Z., D. Prieto, T. P. Conrads, T. D. Veenstra, and H. J. Issaq. 2005. Proteomic patterns: their potential for disease diagnosis. Mol. Cell. Endocrinol. 230:95-106. [DOI] [PubMed] [Google Scholar]
- 54.Yip, T. T. C., J. W. M. Chan, W. C. S. Cho, T. T. Yip, Z. Wang, T. L. Kwan, S. C. K. Law, D. N. C. Tsang, J. K. C. Chan, K. C. Lee, W. W. Cheng, V. W. S. Ma, C. Yip, C. K. P. Lim, R. K. C. Ngan, J. S. K. Au, A. Chan, W. W. L. Lim, and Queen Elizabeth Hospital/Hong Kong Government Virus Unit/Ciphergen SARS Proteomics Study Group. 2005. Protein chip array profiling analysis in patients with severe acute respiratory syndrome identified serum amyloid A protein as a biomarker potentially useful in monitoring the extent of pneumonia. Clin. Chem. 51:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]