Abstract
Skin and osteoarticular infections (SKI and OAI, respectively) account for almost one-third of Streptococcus agalactiae infections in nonpregnant adults. We evaluated the genetic diversity and phylogeny of 58 S. agalactiae strains responsible for adult SKI or OAI and of 61 S. agalactiae strains from cases of adult human colonization (HCol) by serotyping and multilocus sequence typing (MLST). We also assessed the prophage DNA content of the genomes of these strains by a PCR-based method. We found that 63% of SKI and 56% of OAI occurred in people aged 55 years and over. Overall, 71% of SKI strains were of serotype Ia or V, and 91% of OAI strains were of serotype Ia, III, or V. Strains of clonal complexes 1 and 23 (CC1 and CC23) were associated with 79% of SKI cases and 62% of OAI cases. Seven groups of strains, groups A, B, C, D, E, F, and G, were obtained by performing a hierarchical analysis on the basis of prophage DNA-PCR data. We found that 85% of CC1 strains clustered in DNA prophage group D, the group with the highest prophage DNA content (average, 4.4; average of absolute deviations [AVEDEV], 0.9). The CC23 strains displayed the greatest diversity in prophage DNA fragment content, but 47% of CC23 strains clustered in group B, which also had a high average prophage DNA content per strain (average, 2.3; AVEDEV, 0.6). Many (65%) of the OAI strains were in prophage DNA group D, whereas 83% of the SKI strains were in prophage DNA groups B and D. These data suggest that S. agalactiae strains from CC1 and CC23 may be subject to particular transduction mechanisms in gene recombination, rendering them particularly capable of invading the skin, bone, or joints in adults.
Streptococcus agalactiae was initially described in 1887 as an animal pathogen causing bovine mastitis (36). Since the 1960s, when human vaginal carriage of S. agalactiae was first documented, S. agalactiae has frequently been linked to neonatal infections and this bacterium has become the leading neonatal pathogen in developed countries (10, 13, 22, 23, 27, 34). S. agalactiae was rarely isolated from nonpregnant adults until 2 decades ago, when such infections began to be reported, particularly for the elderly and for individuals with underlying conditions such as diabetes mellitus, cancer, and a compromised immune system (4, 15, 37, 40-42). However, such infections have been reported even for adults without a known susceptibility factor (29, 33). Many case reports of clinical skin and osteoarticular infections (SKI and OAI, respectively) due to S. agalactiae in adults have been published in recent years, with these infections accounting for at least one-third of the reported cases of S. agalactiae infection in adults (41, 42).
Many genetic markers have been identified as associated with S. agalactiae clones specifically responsible for meningitis in neonates. Indeed, most of the S. agalactiae strains isolated from the cerebrospinal fluid of neonates belong to clonal complex 17 (CC17) and have particular mobile genetic elements, such as the group II intron GBSi1 (2) and particular prophage DNA fragments (47). No particular markers or virulence factors of S. agalactiae strains have been associated with any other disease.
Phages are important vehicles for horizontal gene exchange within bacterial populations and account for much of the genomic variation observed within bacterial species (8, 11, 28). Temperate phages affect bacterial fitness by modifying anchor points for genomic rearrangements, by disrupting genes, by protecting against lytic infection, by lysing competing strains through prophage induction, and by introducing new fitness factors (8, 19). Prophage acquisition, accounting for much of the molecular diversity of the Streptococcus pyogenes genome, rendered some of the strains of this species virulent through the acquisition of phage-encoded virulence factors and enhanced pathogen survival by improving resistance to host defenses under certain circumstances (1, 9). Little is currently known about S. agalactiae phages. They were first isolated in 1969 (39), and more-recent analyses of sequenced S. agalactiae strains have revealed the presence of abundant regions resembling prophages (16, 44, 45). We recently induced phages from S. agalactiae strains of various phylogenetic lineages, characterized them molecularly, and determined their lytic activities (12). The various molecular phage groups were found to correspond to particular strain lineages, with specific morphological features and lytic activities, suggesting a role for phage-mediated horizontal gene transfer in the evolution of the species and the emergence of lineages with a more specific role in particular diseases.
In this study, we characterized S. agalactiae strains isolated from skin and osteoarticular infections in adults, using serotyping and multilocus sequence typing (MLST) to determine the phylogenetic relationships and molecular features of the strains involved through comparison with the characteristics of strains involved in human colonization (HCol). We used a PCR-based method recognizing S. agalactiae prophages to determine the prophage content of strain genomes (12). The genetic relationships between prophage DNA regions of strains were determined by hierarchical analysis. Correlations between the prophage DNA content, the clinical circumstances of isolation, and the phylogenetic position of S. agalactiae strains were investigated.
MATERIALS AND METHODS
Bacterial isolates.
We studied 119 strains of S. agalactiae. We isolated 24 strains from samples taken from patients with the following skin infections (SKI): whitlows (seven cases), perforating ulcers of the foot (seven cases), cellulitis (two cases), erysipelas (three cases), and cutaneous abscesses (five cases). We studied 34 strains responsible for osteoarticular infections (OAI), isolated from joint fluids in 26 cases and from bone biopsy specimens in 8 cases. All strains were isolated from nonpregnant adult patients (≥18 years of age) admitted to hospitals in various regions of France from 2002 to 2007. We characterized 61 human colonization (HCol) strains previously isolated from nonpregnant adults (48) by the same methods to allow a comparison of the molecular characteristics between these strains and those responsible for SKI and OAI.
Serotyping.
Strains were serotyped by PCR, as previously described (26).
MLST.
The strains were analyzed by MLST, as described by Jones et al. (24). Strains were grouped into clonal complexes (CCs) with the eBURST software program (http://eburst.mlst.net/). A neighbor-joining tree was generated from allelic profile data by Phylodendron (http://pubmlst.org/).
PCR for detection of prophage DNA fragments in the genomes of S. agalactiae strains.
We previously identified and characterized bacteriophages and prophage remnants from S. agalactiae strain genomes and designed primer pairs recognizing the prophage sequences of S. agalactiae bacteriophages for use in PCR (12, 47). Ten of these primer pairs were used here for the evaluation, by PCR, of the prophage content of S. agalactiae strains (Table 1). PCR was carried out with a Chromo 4 system instrument (Bio-Rad, Hercules, CA) on DNA isolated from the strains. PCR was carried out in a final volume of 25 μl, containing 5 μl of extracted DNA, a 0.5 μM concentration of each primer, and 1× iQ SYBR green Supermix (Qiagen SA, Courtaboeuf, France) including 3 mM MgCl2. Amplification was performed over 40 cycles of 10 s at 94°C, 10 s at the annealing temperature (45°C for F5, 48°C for F7, 49°C for F10 and SAK_2094, 50.5°C for SAJ_2395 and SAK_1326, 52°C for SAK_0748, or 54°C for SAG0566, SAK_2090, and SAK_0738), and 30 s at 72°C. The reaction products were then cooled to 35°C and subjected to a post-PCR melting cycle by increasing the temperature by 0.2°C for each 10-s cycle, up to 95°C.
TABLE 1.
PCR primers and amplicon sizes used for prophage screening
| Prophage DNA fragment | Target gene description | Reference strain(s) | Orientation | PCR primer sequence (5′ → 3′) | Amplicon size (bp) |
|---|---|---|---|---|---|
| F5 | A terminase large subunit | S. pyogenes 10394 | Forward | ATC TTA GCA AGC TCC CAC GA | 341 |
| Reverse | TCA ACG GCT GGT ATG GAT TT | ||||
| F7 | A phage-associated cell wall hydrolase and a phage-associated lysin | S. pyogenes 10394 | Forward | AGG CCG CAA CCT TAA ATC T | 497 |
| SpyM6 | Reverse | CGA GTG AAA ACG TGT CTG G | |||
| F10 | A phage-encoded transcriptional regulator, ArpU family | S. pyogenes 5005 | Forward | TCA GCA GAG GAA GGA AAG GA | 510 |
| Reverse | CAA TCA AAG AGC CCT CCC TA | ||||
| SAG0566 | Single-strand binding protein prophage lambda Sa1 | S. agalactiae 2603 V/R | Forward | GTG CTT TGG TTG GAA TTA C | 132 |
| 18RS21 | Reverse | TCT GTT GTT GGC TAT TGC | |||
| SAK_0738 | DNA methylase | CJB111 | Forward | GGG ATA AGA AAG CCA ATC | 172 |
| Prophage lambda W4 | A909 | Reverse | ACA TAG ATA GAC GCA TCG | ||
| SAK_0748 | Phage major capsid protein HK97 family | CJB111 | Forward | TGA TTT CTC TTA CTA CTG GAT TG | 136 |
| A909 | Reverse | CGC TTC TGG TAG AAC GAG | |||
| SAK_2090 | BRO domain protein, prophage antirepressor | A909 | Forward | TAG AGC ACC AAG GCG AAT G | 102 |
| Prophage Sa05 | H36B | Reverse | AAA CGA CCT CAT CAA CTA AAC G | ||
| CJB111 | |||||
| SAK_2094 | Prophage Sa05 | A909 | Forward | AAA GAG TAA AGC ATT TCG | 526 |
| Site-specific recombinase | H36B | Reverse | CCT AAT CTA TAT TGG AGT TC | ||
| Phage integrase family | CJB111 | ||||
| 18RS21 | |||||
| COH1 | |||||
| SAJ_2395 | Phage terminase-like protein, large subunit (remnant) | 18RS21 | Forward | TGA TAG ATA AGT ATG TGA GAT TC | 251 |
| 515 | Reverse | TTG TCT TTC CGA GTT AGC | |||
| SAK_1326 | Site-specific recombinase, phage integrase family (remnant) | A909 | Forward | TTT GAC CTA CGG GAT TAT G | 261 |
| H36B | Reverse | TGA ACG CCA TCT TAG AAG | |||
| CJB111 |
The genetic relationships between the prophage DNA regions of strain genomes were investigated by a hierarchical analysis based on the Jaccard dichotomy coefficient method, as implemented in the SYSTAT 12 software program. The genetic feature analyzed was the presence of a PCR amplicon corresponding to the prophage sequences studied. An absence of gene amplification was not considered to indicate similarity between the studied strains.
Statistical analysis.
Data were analyzed by chi-square tests and Fisher's exact tests to evaluate associations, with a P value of ≤0.05 considered significant.
RESULTS
Relationships of sex and age with infection or colonization rate.
S. agalactiae skin infections (SKI) were equally distributed between the sexes, like S. agalactiae human colonization (HCol) (Table 2). In contrast, S. agalactiae osteoarticular infections (OAI) were significantly more frequent (P = 0.02) (Table 2) in men (25/34; 74%) than in women (9/34; 26%), whereas no such difference between the sexes was observed for colonization in men (29/61; 48%) and in women (32/61; 52%).
TABLE 2.
S. agalactiae strains from skin and osteoarticular infections and human colonization cases: sexes and ages of individuals and serotypes of strains
| Characteristica | No. of individuals (% prevalence) with S. agalactiae causing: |
||
|---|---|---|---|
| Skin infection | Osteoarticular infection | Human colonization | |
| Sex | |||
| F | 11 (46) | 9 (26) | 32 (52) |
| M | 13 (54) | 25 (74) | 29 (48) |
| Age (yr) | |||
| 18-24 | 2 (8) | 2 (6) | 3 (5) |
| 25-40 | 3 (13) | 3 (9) | 18 (30) |
| 41-54 | 4 (17) | 10 (29) | 23 (38) |
| 55-69 | 6 (25) | 7 (21) | 11 (18) |
| ≥70 | 9 (38) | 12 (35) | 6 (10) |
| Strain serotype | |||
| Ia | 9 (38) | 8 (24) | 11 (18) |
| Ib | 2 (8) | 2 (6) | 11 (18) |
| II | 1 (4) | 3 (5) | |
| III | 3 (13) | 10 (29) | 13 (21) |
| IV | 1 (4) | 1 (3) | 4 (7) |
| V | 8 (33) | 13 (38) | 12 (20) |
| NT | 7 (11) | ||
| Total | 24 | 34 | 61 |
F, female; M, male; NT, nontypeable.
S. agalactiae HCol strains were most frequently isolated from individuals under the age of 55 years (44/61; 72%), whereas S. agalactiae SKI and OAI strains were most frequently isolated from individuals aged 55 years and over (15/24 [63%] and 19/34 [56%], respectively; P = 0.003) (Table 2).
Serotyping.
All S. agalactiae strains responsible for SKI or OAI could be serotyped, whereas 7 of the 61 HCol strains (11%) could not be typed (Table 2). More than two-thirds of the strains responsible for SKI (71%) belonged to serotypes Ia (38%) and V (33%), and most of the strains isolated from OAI (91%) were of serotype V (38%), serotype III (29%), or serotype Ia (24%). In contrast, HCol strains were more equally distributed between the various serotypes (5 to 21%) (Table 2).
MLST characterization.
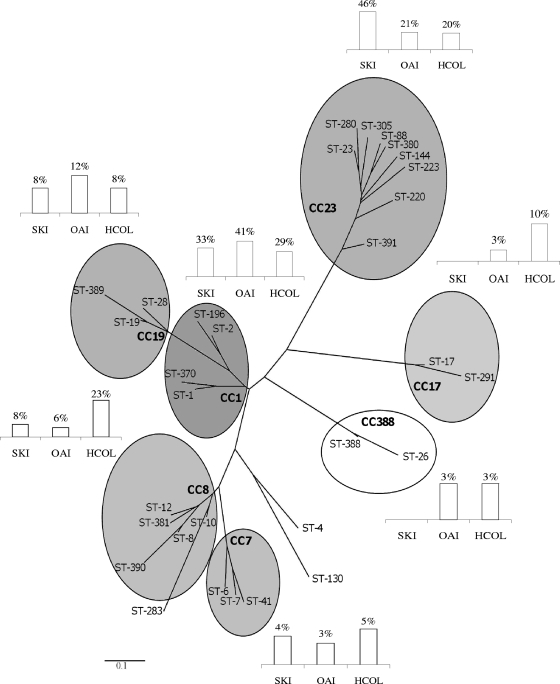
We identified 31 different sequence types (STs) among the 119 strains tested. The eBURST software program assigned 114 strains from 28 of the 31 STs to seven clonal complexes (CCs), CC1, CC7, CC8, CC17, CC19, CC23, and CC388 (Table 3). The genetic relationship between the STs and CC is presented as a dendrogram (Fig. 1).
TABLE 3.
CCs, STs, and serotypes of S. agalactiae strains from skin and osteoarticular infections and cases of human colonizationa
| Clonal complex (no. of isolates) | ST | No. (%) of isolates from: |
No. (%) of isolates of indicated serotype |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SKI | OAI | HCol | Ia | Ib | II | III | IV | V | NT | ||
| CC1 (40) | 8 (33) | 14 (41) | 18 (29) | 2 (7) | 2 (13) | 3 (75) | 2 (8) | 4 (67) | 24 (73) | 3 | |
| 1 (27) | 7 (29) | 11 (32) | 9 (15) | 1 | 2 | 23 | 1 | ||||
| 2 (7) | 2 (6) | 5 (8) | 1 | 1 | 1 | 2 | 2 | ||||
| 196 (5) | 1 (4) | 1 (3) | 3 (5) | 1 | 4 | ||||||
| 370 (1) | 1 (2) | 1 | |||||||||
| CC7 (5) | 1 (4) | 1 (3) | 3 (5) | 2 (13) | 1 (4) | 1 (3) | 1 | ||||
| 41 (3) | 1 (4) | 2 (3) | 1 | 1 | 1 | ||||||
| 6 (1) | 1 (3) | 1 | |||||||||
| 7 (1) | 1 (2) | 1 | |||||||||
| CC8 (18) | 2 (8) | 2 (6) | 14 (23) | 11 (73) | 1 (4) | 1 (17) | 2 (6) | 3 | |||
| 8 (9) | 2 (8) | 7 (11) | 8 | 1 | |||||||
| 10 (5) | 1 (3) | 4 (7) | 1 | 2 | 2 | ||||||
| 12 (2) | 2 (3) | 1 | 1 | ||||||||
| 381 (1) | 1 (3) | 1 | |||||||||
| 390 (1) | 1 (2) | 1 | |||||||||
| CC17 (7) | 1 (3) | 6 (10) | 6 (23) | 1 (17) | |||||||
| 17 (6) | 1 (3) | 5 (8) | 6 | ||||||||
| 291 (1) | 1 (2) | 1 | |||||||||
| CC19 (11) | 2 (8) | 4 (12) | 5 (8) | 1 (4) | 1 (25) | 8 (31) | 1 (3) | ||||
| 19 (8) | 1 (4) | 4 (12) | 3 (5) | 1 | 7 | ||||||
| 28 (2) | 1 (4) | 1 (2) | 1 | 1 | |||||||
| 389 (1) | 1 (2) | 1 | |||||||||
| CC23 (30) | 11 (46) | 7 (21) | 12 (20) | 24 (86) | 6 (23) | ||||||
| 23 (18) | 7 (29) | 6 (18) | 5 (8) | 14 | 4 | ||||||
| 88 (2) | 2 (8) | 2 | |||||||||
| 144 (2) | 1 (4) | 1 (4) | 2 | ||||||||
| 220 (2) | 2 (3) | 2 | |||||||||
| 223 (2) | 2 (3) | 2 | |||||||||
| 280 (1) | 1 (3) | 1 | |||||||||
| 380 (1) | 1 (4) | 1 | |||||||||
| 305 (1) | 1 (2) | 1 | |||||||||
| 391 (1) | 1 (2) | 1 | |||||||||
| CC388 (3) | 1 (3) | 2 (3) | 3 (9) | ||||||||
| 26 (1) | 1 (3) | 1 | |||||||||
| 388 (2) | 2 (3) | 2 | |||||||||
| Singletons (5) | 4 (12) | 1 (2) | 1 (4) | 2 (8) | 2 (6) | ||||||
| 4 (1) | 1 (3) | 1 | |||||||||
| 130 (2) | 1 (3) | 1 (2) | 2 | ||||||||
| 283 (2) | 2 (6) | 2 | |||||||||
| Total | 24 | 34 | 61 | 28 | 15 | 4 | 26 | 6 | 33 | 7 | |
SKI, skin infection; OAI, osteoarticular infection; HCol, human colonization; NT, nontypeable.
FIG. 1.
Phylogenetic tree showing the relationship between sequence type (ST) and clonal complex (CC) obtained by analyzing MLST data from 119 S. agalactiae strains isolated from skin infections (SKI), osteoarticular infections (OAI), and cases of human colonization (HCol). Columns indicate the percentages of SKI, OAI, and HCol strains in each CC.
HCol strains were more genetically diverse than the strains responsible for infections; the 61 HCol strains belonged to 23 different STs, with a maximum of 9 strains for a single ST (15%, for ST-1) (Table 3).
The distributions of SKI and HCol strains between the various STs (Table 3) differed significantly (P < 0.00001). SKI strains were less diverse, with the 24 strains belonging to only 10 STs. SKI strains were more frequently classified as ST-1 (7/24; 29%) or ST-23 (7/24; 29%) than were HCol strains (9/61 [15%] ST-1 and 5/61 [8%] ST-23). Similarly, the distributions of SKI and HCol strains between the various CCs differed significantly (P < 0.00001) (Table 3). SKI strains were more likely to belong to CC1 or CC23 (19/24; 79%) than HCol strains (30/61; 49%).
The distributions of OAI and HCol strains between the various STs (Table 3) differed significantly (P < 0.00001). The 34 OAI isolates belonged to 14 STs. OAI strains were more frequently of ST-1 (11/34; 32%) and ST-23 (6/34; 18%) than were HCol strains (9/61 [15%] ST-1 and 5/61 [8%] ST-23). Nevertheless, the distributions of OAI strains and of HCol strains between CCs did not differ significantly (P = 0.2), although OAI strains were more likely to belong to CC1 or CC23 (21/34; 62%) than HCol strains (30/61; 49%).
S. agalactiae strains of each serotype were distributed between several STs (Table 3), but the strains of serotypes Ia, Ib, IV, and V were mostly of ST-23 (50%), ST-8 (53%), ST-196 (67%), and ST-1 (70%), respectively. Serotype Ia, Ib, and V isolates belonged principally to CC23 (86%), CC8 (73%), and CC1 (73%), respectively.
Prophage DNA fragments in the S. agalactiae genome.
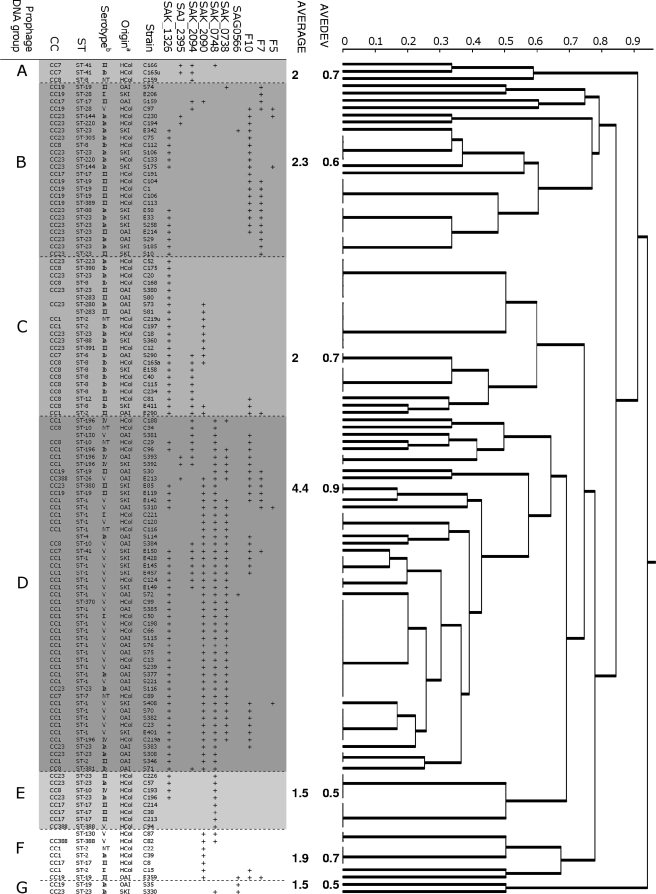
The prophage DNA fragments studied here were not detected by PCR in four S. agalactiae strains, all isolated from HCol cases. For each of the remaining 115 strains, PCR amplified 1 to 7 of the 10 prophage DNA fragments studied. The genetic relationships between the prophage DNA regions of strain genomes were represented as a dendrogram (Fig. 2). This analysis assigned the strains to seven major prophage DNA groups, A to G. The natures and frequencies of the prophage DNA fragments amplified from the strains differed significantly between prophage DNA groups (Table 4) (P < 0.00001). Three different patterns of prophage DNA fragment amplification were observed in these groups (Table 4). For the 49 strains of prophage group D, all the prophage targets studied were found in at least one strain and PCR amplified a large number of prophage DNA fragments in each strain (average, 4.4) (Fig. 2). For the 24 strains of prophage group B, a large number of prophage DNA fragments were amplified (only one prophage sequence was never amplified [SAK_0748]) and the mean number of prophage DNA fragments amplified per strain was 2.3 (Fig. 2). Only one or two prophage DNA fragments were amplified for 35 strains from the 42 strains of the other five prophage DNA groups (A, C, E, F, and G). In each of these groups, the mean number of prophage DNA fragments amplified per strain was ≤2 (Fig. 2).
FIG. 2.
Distribution of 119 S. agalactiae strains isolated from skin infections (SKI), osteoarticular infections (OAI), and human colonization (HCol) cases into prophage DNA groups on the basis of PCR evaluations of the prophage content of strains. Jaccard analysis generated a dendrogram of similarity values for the results of the 10 prophage sequences studied. The average number of prophage DNA fragments amplified by PCR from strains and the average of absolute deviations (AVEDEV) were calculated for each prophage DNA group of strains. a, anatomic origin of strains; b, serotype of strains; ST, sequence-type; CC, clonal complex; NT, nontypeable.
TABLE 4.
Distribution of the S. agalactiae strains of various origins, serotypes, and clonal complexes into prophage DNA groups displayed by SYSTAT 12 softwarea
| Characteristic (no. of strains) | No. of strains (%) in indicated prophage DNA group |
|||||||
|---|---|---|---|---|---|---|---|---|
| NP (4) | A (3) | B (24) | C (22) | D (49) | E (8) | F (7) | G (2) | |
| Prophage DNA fragment | ||||||||
| F5 | 114 (96) | 3 (3) | 2 (2) | |||||
| F7 | 95 (83) | 15 (13) | 1 (1) | 7 (6) | 1 (1) | |||
| F10 | 73 (61) | 18 (15) | 3 (3) | 23 (19) | 2 (2) | |||
| SAG0566 | 114 (96) | 1 (1) | 1 (1) | 1 (1) | 2 (2) | |||
| SAK_0738 | 78 (66) | 1 (1) | 40 (34) | |||||
| SAK_0748 | 58 (49) | 1 (1) | 49 (41) | 8 (7) | 2 (2) | 1 (1) | ||
| SAK_2090 | 59 (50) | 1 (1) | 11 (9) | 41 (34) | 7 (6) | |||
| SAK_2094 | 90 (76) | 3 (3) | 2 (2) | 9 (8) | 15 (13) | |||
| SAJ_2395 | 112 (94) | 2 (2) | 2 (2) | 3 (3) | ||||
| SAK_1326 | 43 (36) | 13 (11) | 22 (18) | 37 (31) | 4 (3) | |||
| Anatomic origin | ||||||||
| SKI (24) | 9 (38) | 3 (13) | 11 (46) | 1 (4) | ||||
| OAI (34) | 4 (12) | 6 (18) | 22 (65) | 1 (3) | 1 (3) | |||
| HCol (61) | 4 (7) | 3 (5) | 11 (18) | 13 (21) | 16 (26) | 8 (13) | 6 (10) | |
| Serotype | ||||||||
| Ia (28) | 1 (4) | 12 (43) | 5 (18) | 5 (18) | 2 (7) | 1 (4) | 2 (7) | |
| Ib (15) | 1 (7) | 1 (7) | 1 (7) | 10 (67) | 2 (13) | |||
| II (4) | 1 (25) | 2 (50) | 1 (25) | |||||
| III (26) | 1 (4) | 9 (35) | 6 (23) | 4 (15) | 4 (15) | 2 (8) | ||
| IV (6) | 1 (17) | 4 (67) | 1 (17) | |||||
| V (33) | 1 (3) | 1 (3) | 28 (85) | 1 (3) | 2 (6) | |||
| NT (7) | 1 (14) | 1 (14) | 4 (57) | 1 (14) | ||||
| Sequence type | ||||||||
| 1 (27) | 27 (100) | |||||||
| 2 (7) | 3 (43) | 1 (14) | 3 (43) | |||||
| 8 (9) | 1 (11) | 1 (11) | 7 (78) | |||||
| 10 (5) | 1 (20) | 3 (60) | 1 (20) | |||||
| 17 (6) | 2 (33) | 3 (50) | 1 (17) | |||||
| 19 (8) | 4 (50) | 2 (25) | 1 (13) | 1 (13) | ||||
| 23 (18) | 8 (44) | 3 (17) | 3 (17) | 3 (17) | 1 (6) | |||
| 196 (5) | 5 (100) | |||||||
| Others (34) | 3 (9) | 2 (6) | 9 (26) | 9 (26) | 8 (24) | 1 (3) | 2 (6) | |
| Clonal complex | ||||||||
| 1 (40) | 3 (8) | 34 (85) | 3 (8) | |||||
| 7 (5) | 2 (40) | 1 (20) | 2 (40) | |||||
| 8 (18) | 2 (11) | 1 (6) | 1 (6) | 9 (50) | 4 (22) | 1 (6) | ||
| 17 (7) | 1 (14) | 2 (29) | 3 (43) | 1 (14) | ||||
| 19 (11) | 7 (64) | 2 (18) | 1 (9) | 1 (9) | ||||
| 23 (30) | 1 (3) | 14 (47) | 7 (23) | 4 (13) | 3 (10) | 1 (3) | ||
| 388 (3) | 1 (33) | 1 (33) | 1 (33) | |||||
| Others (5) | 2 (40) | 2 (40) | 1 (20) | |||||
NP, no prophage amplification; SKI, skin infection; OAI, osteoarticular infection; HCol, human colonization; NT, nontypeable.
Strains of the three different clinical origins (SKI, OAI, and HCol) were differentially distributed between prophage DNA groups A to G (Table 4) (P = 0.003). HCol strains were evenly distributed between prophage DNA groups A to F, whereas 65% of OAI strains (22/34 strains) (Table 4) belonged to prophage DNA group D, the members of which had the largest numbers of amplified prophage DNA fragments. Similarly, 83% of SKI strains (20/24 strains) (Table 4) belonged to prophage DNA groups B and D, which displayed the highest numbers of amplified prophage DNA fragments. In contrast, 17 of the 18 strains of prophage groups A, E, and F, in which the number of amplified prophage DNA fragments was low, were HCol strains.
The distribution of strains from the various serotypes, STs, and CCs between prophage DNA groups was not random (Table 4) (P < 0.00001). Strains of the two major lineages, ST-1 and ST-23, and of the corresponding clonal complexes, CC1 and CC23, which were frequently implicated in SKI and OAI, had particular characteristics in terms of their prophage DNA content. All 27 ST-1 strains (100%) and 34 of the 40 CC1 strains (85%) (Table 4), most of which were of serotype V (Fig. 2), belonged to prophage DNA group D. And the six CC1 strains that did not belong to prophage DNA group D were rarely isolated from patients with infectious disease (only one strain, from a case of OAI) (Fig. 2). The CC8 and CC19 strains mostly belonged to prophage groups C (9/18; 50%) and B (7/11; 64%), respectively (Table 4 and Fig. 2). The strains of phylogenetic lineage ST-23 and its corresponding clonal complex, CC23, were mostly of serotype Ia and were distributed between five of the seven prophage DNA groups (groups B, C, D, E, and G) (Fig. 2 and Table 4). Therefore, they displayed considerable diversity in terms of the prophage DNA fragments amplified. Nevertheless, 14 of the 30 strains (47%) of CC23 belonged to prophage DNA group B, which included 64% of the CC19 strains.
DISCUSSION
S. agalactiae infections in nonpregnant adults have greatly increased in frequency over the last 2 decades in the United States and Europe (4, 15, 37, 40). Many clinical cases of S. agalactiae SKI and OAI have been reported. Our data confirm that S. agalactiae SKI and OAI are significantly more frequent in older people (≥55 years of age) (14, 15, 40), that most of the strains responsible for SKI (71%) belong to serotypes Ia and V, and that serotypes V, III, and Ia predominate among OAI strains (91%) (5, 14, 18, 46). Nevertheless, no other study to date has specifically focused on molecular characterization, including evaluation of the prophage content of S. agalactiae strains responsible for SKI and OAI.
Only strains from a particular phylogenetic lineage, initially recognized by multilocus enzyme electrophoresis (MLEE) (35, 38) and more recently defined as ST-17 by MLST, have been shown to be associated with a particular disease, due to a higher likelihood of their invading the central nervous system (CNS) of neonates (3, 24, 25, 30, 31). Our data indicate that strains of two other major lineages, CC1 and CC23, found in 79% of SKI and 62% of OAI, are frequently associated with skin, bone, and joint infections. The similar phylogenetic characteristics of strains isolated from skin and bone or joint infections provide support for the hypothesis that the skin and soft tissues may be a potential portal of entry for bone and joint infections, as previously suggested (14).
The high percentages of CC1 and CC23 strains observed in SKI and OAI suggested that strains of these lineages have enhanced invasiveness for skin, bone, and joints. As for the propensity of ST-17 strains that invade the CNS of neonates, the pathogenic features of the strains of the two phylogenetically distant lineages CC1 and CC23, which may account for the particular ability of these strains to invade skin, bone, and joints, are unknown. In terms of evolution, the strains of these two lineages probably acquired virulence through the acquisition of different genetic elements. Indeed, the strains of these two lineages displayed a high degree of phylogenetic divergence (6, 20, 21, 24, 31, 32, 43, 48). In addition, the CC23 lineage contains strains of bovine origin isolated during the 1960s and strains subsequently isolated from humans (7, 21). In contrast, the CC1 lineage contains strains that have emerged since the 1990s, responsible for infections in both adults and neonates (6, 17, 32). Thus, the bacteria of these two clones have been exposed to different environmental and nutritional backgrounds during evolution. These constraints may have subjected the bacteria to different stressful conditions, resulting in the induction of different mutations, as observed for housekeeping genes, and probably also resulting in differences in horizontal gene transfer events, leading to marked differences in the virulence proprieties of the strains of these two clones.
Our data are consistent with the hypothesis that horizontal gene transfers related to transduction mechanisms may have played a major role in the emergence of clones able to infect skin, bone, and joints. Indeed, on the basis of the prophage content of the S. agalactiae strain genomes, we identified two major groups of strains responsible for SKI and OAI with (i) very distinctive prophage DNA fragment contents, resulting in clustering into two particular prophage DNA groups, and (ii) the largest number of prophage DNA fragments per strain in their genome (groups B and D) (Fig. 2).
Prophage DNA group D had the largest number of amplified prophage DNA fragments per strain and contained all the strains of ST-1 and 85% of the CC1 strains, this phylogenetic lineage being the most frequently implicated in SKI and OAI. In addition, CC1 strains from prophage DNA groups other than group D were rarely isolated from SKI or OAI cases, with only six such isolations observed. These strains belonged to prophage DNA groups from which only one or two prophage DNA fragments were amplified, consistent with a low prophage content in the genome. Therefore, lysogeny, which has been shown to play an important role in bacterial virulence and genome diversification in the genus Streptococcus (1, 8), may be a key genetic event affecting the virulence of CC1 S. agalactiae strains, leading to the emergence of strains particularly able to infect skin, bone, and joints.
CC23 was the second most frequently implicated lineage in SKI and OAI. Analysis of the prophage content of the genome of CC23 strains resulted in the grouping together of half these strains (14/30 strains) in prophage DNA group B, one of the prophage DNA groups with the largest number of similar amplified prophage DNA fragments, suggesting a role for lysogeny in the specialization of this group of strains. Within this prophage DNA group, the CC23 strains clustered with 64% of the CC19 strains (7/11 strains). These two groups of strains may therefore have been subjected to similar ecological conditions or similar constraints leading to lysogenization by similar phages. However, the CC23 strains of prophage DNA group B were frequently associated with SKI and OAI (10/14 strains), whereas the CC19 strains of this prophage DNA group were rarely isolated from patients with these diseases (2/7 strains). Thus, the prophage characteristics of prophage DNA group B recognized by PCR in this study either play no role in the propensity of CC23 strains to infect skin, bone, and joints or may modulate other virulence factors specifically carried by the genomes of the strains of the CC23 lineage. Our data tend to support the second hypothesis. Indeed, CC23 strains from prophage DNA groups B and D, which had the greatest amplified prophage DNA content, were frequently isolated from SKI and OAI (14/18 strains), whereas CC23 strains from other prophage DNA groups, in which prophage DNA fragment amplification was less frequent, were mostly isolated from cases of HCol (8/12 strains; P = 0.014).
In conclusion, our data suggest a role for S. agalactiae strains of two phylogenetic lineages, CC1 and CC23, in SKI and OAI in adults, particularly for strains exposed to particular transduction mechanisms in gene recombination. The impacts of lysogeny on the virulence of the strains of these two lineages may be markedly different. Further studies are therefore required to assess in detail the role of the observed prophage-like elements in the emergence of S. agalactiae clones displaying a particular tropism for skin, bone, or joints. Are these elements involved in the importation of new phage-encoded virulence factors or the modification of transcriptional control mechanisms for chromosomal virulence genes? Particular host signals, linked to risk factors for development of SKI or OAI, may also induce changes in gene expression due to phage transduction.
Acknowledgments
N.V.D.M.-M. and R.Q. conceived and designed the experiments. M.S. (serotyping, MLST, prophage PCR, and PFGE) and A.-S.D. and L.A. (prophage PCR) performed the experiments. M.S., N.V.D.M.-M, and R.Q. analyzed the data. M.S. and R.Q. wrote the paper.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Banks, D. J., S. B. Beres, and J. M. Musser. 2002. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10:515-521. [DOI] [PubMed] [Google Scholar]
- 2.Bidet, P., N. Brahimi, C. Chalas, Y. Aujard, and E. Bingen. 2003. Molecular characterization of serotype III group B-Streptococcus isolates causing neonatal meningitis. J. Infect. Dis. 188:1132-1137. [DOI] [PubMed] [Google Scholar]
- 3.Bisharat, N., N. Jones, D. Marchaim, C. Block, R. M. Harding, P. Yagupsky, T. Peto, and D. W. Crook. 2005. Population structure of group B Streptococcus from a low-incidence region for invasive neonatal disease. Microbiology 151:1875-1881. [DOI] [PubMed] [Google Scholar]
- 4.Blancas, D., M. Santin, M. Olmo, F. Alcaide, J. Carratala, and F. Gudiol. 2004. Group B streptococcal disease in nonpregnant adults: incidence, clinical characteristics, and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 23:168-173. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173:365-373. [DOI] [PubMed] [Google Scholar]
- 6.Bohnsack, J. F., A. Whiting, M. Gottschalk, D. M. Dunn, R. Weiss, P. H. Azimi, J. B. Philips III, L. E. Weisman, G. G. Rhoads, and F.-Y. C. Lin. 2008. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. academic centers from 1995 to 1999. J. Clin. Microbiol. 46:1285-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brochet, M., E. Couvé, M. Zouine, T. Vallaeys, C. Rusniok, M. C. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 8:1227-1243. [DOI] [PubMed] [Google Scholar]
- 8.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Currie, B. J. 2006. Group A streptococcal infections of the skin: molecular advances but limited therapeutic progress. Curr. Opin. Infect. Dis. 19:132-138. [DOI] [PubMed] [Google Scholar]
- 10.Davies, H. D., S. Raj, C. Adair, J. Robinson, and A. McGeer. 2001. Population-based active surveillance for neonatal group B streptococcal infections in Alberta, Canada: implications for vaccine formulation. Pediatr. Infect. Dis. J. 20:879-884. [DOI] [PubMed] [Google Scholar]
- 11.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brussow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 12.Domelier, A. S., N. van der Mee-Marquet, P. Y. Sizaret, G. H. Arnaud, M. F. Lartigue, L. Méreghetti, and R. Quentin. 2009. Molecular characterization and lytic activities of Streptococcus agalactiae bacteriophages and determination of lysogenic strain features. J. Bacteriol. 191:4776-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eickhoff, T., J. O. Klein, A. K. Daly, D. Ingall, and M. Finland. 1964. Neonatal sepsis and other infections due to group B beta-hemolytic streptococci. N. Engl. J. Med. 271:1221-1228. [DOI] [PubMed] [Google Scholar]
- 14.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 15.Farley, M. M., R. C. Harvey, T. Stull, J. D. Smith, A. Schuchat, J. D. Wenger, and D. S. Stephens. 1993. A population-based assessment of invasive disease due to group B Streptococcus in nonpregnant adults. N. Engl. J. Med. 328:1807-1811. [DOI] [PubMed] [Google Scholar]
- 16.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, L. H., D. M. Dwyer, and J. A. Johnson. 1995. Emergence of serotype V group B streptococcal infection among infants and adults. J. Infect. Dis. 171:513. [DOI] [PubMed] [Google Scholar]
- 18.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, and A. Schuchat. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland Emerging Infection Program. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 19.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Héry-Arnaud, G., G. Bruant, P. Lanotte, S. Brun, A. Rosenau, N. van der Mee-Marquet, R. Quentin, and L. Mereghetti. 2005. Acquisition of insertion sequences and the GBSi1 intron by Streptococcus agalactiae isolates correlates with the evolution of the species. J. Bacteriol. 187:6248-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Héry-Arnaud, G., G. Bruant, P. Lanotte, S. Brun, B. Picard, A. Rosenau, N. van der Mee-Marquet, P. Rainard, R. Quentin, and L. Mereghetti. 2007. Mobile genetic elements provide evidence for a bovine origin of clonal complex 17 of Streptococcus agalactiae. Appl. Environ. Microbiol. 73:4668-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt, D. E., S. Halket, J. de Louvois, and D. Harvey. 2001. Neonatal meningitis in England and Wales: 10 years on. Arch. Dis. Child. Fetal Neonatal Ed. 84:F85-F89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood, M., A. Janney, and G. Dameron. 1961. Beta hemolytic streptococcus group B associated with problems of perinatal period. Am. J. Obstet. Gynecol. 82:809-818. [DOI] [PubMed] [Google Scholar]
- 24.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, N., K. A. Oliver, J. Barry, R. M. Harding, N. Bisharat, B. G. Spratt, T. Peto, and D. W. Crook. 2006. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B Streptococcus is independent of capsular serotype. Clin. Infect. Dis. 42:915-924. [DOI] [PubMed] [Google Scholar]
- 26.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lancefield, R. C., and R. Hare. 1935. The serological differentiation of pathogenic and non-pathogenic strains of haemolytic streptococci from parturient women. J. Exp. Med. 61:335-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence, J. G., R. W. Hendrix, and S. Casjens. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9:535-540. [DOI] [PubMed] [Google Scholar]
- 29.Lee, N. Y., J. J. Yan, J. J. Wu, H. C. Lee, K. H. Liu, and W. C. Ko. 2005. Group B streptococcal soft tissue infections in non-pregnant adults. Clin. Microbiol. Infect. 11:577-579. [DOI] [PubMed] [Google Scholar]
- 30.Lin, F. Y., A. Whiting, E. Adderson, S. Takahashi, D. M. Dunn, R. Weiss, P. H. Azimi, J. B. Philips III, L. E. Weisman, J. Regan, P. Clark, G. G. Rhoads, C. E. Frasch, J. Troendle, P. Moyer, and J. F. Bohnsack. 2006. Phylogenetic lineages of invasive and colonizing strains of serotype III group B streptococci from neonates: a multicenter prospective study. J. Clin. Microbiol. 44:1257-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luan, S. L., M. Granlund, M. Sellin, T. Lagergård, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning, S. D., A. C. Springman, E. Lehotzky, M. A. Lewis, T. S. Whittam, and H. D. Davies. 2009. Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 47:1143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinaud, C., T. Gaillard, B. Graffin, Y. Muzellec, and P. Brison. 2008. Vertebral osteomyelitis due to Streptococcus agalactiae ST-17. Ann. Biol. Clin. (Paris) 66:87-89. (In French.) [DOI] [PubMed] [Google Scholar]
- 34.Mayon-White, R. T. 1985. The incidence of GBS disease in neonates in different countries. Antibiot. Chemother. 35:17-27. [DOI] [PubMed] [Google Scholar]
- 35.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. U. S. A. 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nocard, M., and R. Mollereau. 1887. Sur une mammite contagieuse des vaches laitieres. Ann. Inst. Pasteur 1:109. [Google Scholar]
- 37.Phares, C. R., R. Lynfield, M. M. Farley, J. Mohle-Boetani, L. H. Harrison, S. Petit, A. S. Craig, W. Schaffner, S. M. Zansky, K. Gershman, K. R. Stefonek, B. A. Albanese, E. R. Zell, A. Schuchat, and S. J. Schrag for the Active Bacterial Core Surveillance/Emerging Infections Program Network. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA 299:2056-2065. [DOI] [PubMed] [Google Scholar]
- 38.Quentin, R., H. Huet, F. S. Wang, P. Geslin, A. Goudeau, and R. K. Selander. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 33:2576-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell, H., N. L. Norcross, and D. E. Kahn. 1969. Isolation and characterization of Streptococcus agalactiae bacteriophage. J. Gen. Virol. 5:315-317. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz, B., A. Schuchat, M. J. Oxtoby, S. L. Cochi, A. Hightower, and C. V. Broome. 1991. Invasive group B streptococcal disease in adults—a population-based study in metropolitan Atlanta. JAMA 266:1112-1114. [PubMed] [Google Scholar]
- 41.Sendi, P., L. Johansson, and A. Norrby-Teglund. 2008. Invasive group B streptococcal disease in non-pregnant adults: a review with emphasis on skin and soft-tissue infections. Infection 36:100-111. [DOI] [PubMed] [Google Scholar]
- 42.Skoff, T. H., M. M. Farley, S. Petit, A. S. Craig, W. Schaffner, K. Gershman, L. H. Harrison, R. Lynfield, J. Mohle-Boetani, S. Zansky, B. A. Albanese, K. Stefonek, E. R. Zell, D. Jackson, T. Thompson, and S. J. Schrag. 2009. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin. Infect. Dis. 49:85-91. [DOI] [PubMed] [Google Scholar]
- 43.Sun, Y., F. Kong, Z. Zhao, and G. L. Gilbert. 2005. Comparison of a 3-set genotyping system with multilocus sequence typing for Streptococcus agalactiae (group B Streptococcus). J. Clin. Microbiol. 43:4704-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyrrell, G. J., L. D. Senzilet, J. S. Spika, D. A. Kertesz, M. Alagaratnam, M. Lovgren, and J. A. Talbot. 2000. Invasive disease due to group B streptococcal infection in adults: results from a Canadian, population-based, active laboratory surveillance study—1996. Sentinel Health Unit Surveillance System Site Coordinators. J. Infect. Dis. 182:168-173. [DOI] [PubMed] [Google Scholar]
- 47.van der Mee-Marquet, N., A. S. Domelier, L. Mereghetti, P. Lanotte, A. Rosenau, W. van Leeuwen, and R. Quentin. 2006. Prophagic DNA fragments in Streptococcus agalactiae strains in association with neonatal meningitis. J. Clin. Microbiol. 44:1049-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Mee-Marquet, N., L. Fourny, L. Arnault, A. S. Domelier, M. Salloum, M. F. Lartigue, and R. Quentin. 2008. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J. Clin. Microbiol. 46:2906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]