Abstract
Chromosomal translocation is the best-characterized genetic mechanism for oncogene activation. However, there are documented examples of activation by alternate mechanisms, for example gene dosage increase, though its prevalence is unclear. Here, we answered the fundamental question of the contribution of DNA amplification as a molecular mechanism driving oncogenesis. Comparing 104 cancer lines representing diverse tissue origins identified genes residing in amplification ‘hotspots’ and discovered an unexpected frequency of genes activated by this mechanism. The 3431 amplicons identified represent ~10 per hematological and ~36 per epithelial cancer genome. Many recurrently amplified oncogenes were previously known to be activated only by disease-specific translocations. The 135 hotspots identified contain 538 unique genes and are enriched for proliferation, apoptosis and linage-dependency genes, reflecting functions advantageous to tumor growth. Integrating gene dosage with expression data validated the downstream impact of the novel amplification events in both cell lines and clinical samples. For example, multiple downstream components of the EGFR-family-signaling pathway, including CDK5, AKT1 and SHC1, are overexpressed as a direct result of gene amplification in lung cancer. Our findings suggest that amplification is far more common a mechanism of oncogene activation than previously believed and that specific regions of the genome are hotspots of amplification.
Keywords: gene amplification, array CGH, gene expression, integrative analysis, lung cancer, EGFR signaling
Introduction
Genetic aberration and the consequential activation of oncogenes are key to cancer development. Chromosomal translocation is known as the major event in oncogene activation (Futreal et al., 2004). However, the prevalence of alternate mechanisms, such as DNA amplification, have not been extensively quantified, even though oncogenes have been found in: (i) cytogenetically visible double minutes, which are circular, extrachromosomal elements, a few megabases in size that replicate autonomously, (ii) homogenous staining regions, which are large regions of tandem repeats within a chromosome, thought to be formed by repeated breakage-fusion-bridge cycles and (iii) discrete insertions distributed throughout the genome (Albertson, 2006).
Surprisingly, relatively few oncogenes, when compared to chromosome translocation, have been shown to undergo amplification as mechanism of activation during cancer development. In fact, a recent version (January 22, 2007) of a census of genes causally implicated in cancer (cancer genes) originally described by Futreal et al. (2004) reported only seven oncogenes meeting their criteria as being recurrently amplified in the development of human cancers: AKT2 in ovarian cancer, ERBB2 in breast and ovarian cancer, MYCL1 in small cell lung cancer, MYCN in neuroblastoma, REL in Hodgkin lymphoma, epidermal growth factor receptor (EGFR) in glioma and non-small cell lung cancer (NSCLC), and MYC in numerous cancers. We propose that the low incidence of oncogenes activated by amplification may be attributed to the failure of detection, rather than governed by tumor biology. Unlike copy number gains, which are generated by aneueploidy or unbalanced translocations and affect large chromosomal regions, amplifications are traditionally defined as the increase of chromosome segments 0.5–10 megabases (Mb) in size (Myllykangas et al., 2006). The small size of amplicons may escape detection by conventional cytogenetic methods; consequently, the contribution of DNA amplification to the oncogenic process may be grossly underestimated. With advances in high-resolution whole-genome-profiling technologies (Tonon et al., 2005; Garnis et al., 2006), the complexity of the cancer genome is becoming evident, and the prevalence of DNA amplification as a mechanism in the activation of oncogenes needs to be re-evaluated.
In this study, we determined the precise boundaries of amplified chromosomal segments in 104 cancer cell lines from multiple tissues of origin and deduced novel regions of the genome, which are hotspots for genomic amplification. These hotspots were then analysed for their association with genes involved in tumorigenesis and fragile sites. We assessed the functional impact of a subset of the identified hotspots in a panel of NSCLC cell lines and tumors to determine their effect on gene transcription levels and their contribution to the activation of cellular pathways potentially involved in lung tumorigenesis.
Results
Identification of discrete amplicons in cancer genomes
Twenty-four thousand eight hundred and ninety-two genomic loci were assessed for each of the 104 cell lines, scanning all autosomes at a resolution of ~50 kb (Coe et al., 2007). Altogether, 3431 amplicons were detected across all samples (see Supplementary Methods) with an average size of 0.68Mb and a median of 0.33Mb (Table 1, Supplementary Table 1). The number of amplicons per genome varied from 0 to 199 with an average of 33. Hematological malignancies (leukemia and lymphomas) had ~10 amplicons, whereas epithelial cancers had an average of 36.
Table 1.
Summary and distribution of amplicons by cancer type
| Tissue type | Number of Samples | Total number of amplicons | Amplicons/Tumor | Average size (Mb) | Most frequent amplificationa |
|---|---|---|---|---|---|
| Lung | 53 | 1690 | 31.9 | 0.68 | 8q24.21 (28%) |
| Breast | 17 | 905 | 53.2 | 0.77 | 8q24.21 (59%) |
| Lymphoid | 9 | 101 | 11.2 | 0.54 | 9p13.3, 13q31.3, 18q21.33–22.1 (33%) |
| Cervix | 8 | 45 | 5.6 | 0.65 | 5p15.33 (38%) |
| Skin | 4 | 395 | 98.8 | 0.35 | 7p13.3 & 7q35 (75%) |
| Blood | 3 | 23 | 7.7 | 0.80 | N/A |
| Prostate | 3 | 82 | 27.3 | 0.86 | 14q21.3–22.1 (67%) |
| Bone | 2 | 104 | 52 | 1.02 | N/A |
| Colon | 2 | 22 | 11 | 0.70 | N/A |
| Ovary | 2 | 3 | 1.5 | 0.33 | N/A |
| Liver | 1 | 61 | 61 | 0.83 | N/A |
| Total | 104 | 3431 | 32.8 | 0.68 |
The most frequent regions were determined only if found in >2 samples for the corresponding tissue type.
Unexpected frequent amplification of known oncogenes
The most recent version (January 22, 2007) of the Cancer Gene Consensus of the Cancer Genome Project at the Sanger Institute (Futreal et al., 2004) contains 363 cancer genes whose aberration are causal in the development of specific cancers. Of these, 70 are tumor suppressor genes, 292 are oncogenes and one can act as both. Only seven (2%) of these oncogenes were shown to be predominately activated by amplification compared to 268 (92%), which are activated mainly by chromosomal translocation. Our data showed amplification at these loci: MYC (28/104), ERBB2 (10/104), EGFR (7/104), MYCL1 (6/104), AKT2 (3/104) and MYCN (1/104). REL amplification was not detected in our dataset, as Hodgkin Lymphoma, in which this gene is amplified, is not represented in our study.
Unexpectedly, 145 of the 292 oncogenes (~50%) showed amplification, with 78 oncogenes (27%) at ≥2 times (Supplementary Table 2). Of the genes amplified in ≥5 cell lines, only MYC, ERBB2, EGFR and MYCL1 have been reported. The frequent amplification of SS18L1, NTRK1 and PRDM16 are novel findings, as translocation was the known mechanism. Indeed, numerous oncogenes, which are primarily activated by translocation, were commonly amplified in the sample set (Supplementary Table 3). The number of oncogenes amplified per genome also varied with an average of 3.5 genes. Remarkably, the genomes of NSCLC HCC1195 (Supplementary Figure 1) and SCLC line H526, each harbor 22 amplified oncogenes, whereas 25 of the lines had no known oncogenes amplified (Supplementary Table 4).
Novel hotspots of frequent genomic amplification in cancer genomes
The high incidence of oncogene amplification per genome suggested that this is a common mechanism of gene activation. Therefore, the discovery of genomic regions that undergo frequent copy number amplification may lead to the identification of novel oncogenes. The genomic coordinates of all amplicons were determined and aligned for all 104 samples (Figure 1). DNA segments amplified ≥5 times were stringently considered as hotspots; they are found in ~5% of samples (see Supplementary Methods). In total, 135 hotspots covering 3% of genome were identified with an average size of 0.67 Mb. Regions of genomic amplification were distributed on all autosomes except chromosome 4, and in all tumor types analysed (Supplementary Table 5). Amplicons are most frequently localized to 1q21–23, 5p15, 7p13–11, 8q22–24, 11q13, 14q12–21, 14q32, 17q12–21 and 20q13. A total of 538 unique genes were contained within the hotspots (Supplementary Table 6) (see Supplementary Methods). Interestingly, the majority of these hotspots did not contain the 292 known oncogenes.
Figure 1.
Hotspots of amplification in cancer genomes. A histogram summarizing the regions of amplification across all 104 samples with the resulting values scaled to the segment with the highest count (28) and plotted against their corresponding genomic position. Hotspots are denoted by the dark blue shading, whereas the light blue shading represents regions amplified ≤5 times. Triangles mark common fragile sites. Detailed genomic position of hotspots and common fragile sites are provided in Supplementary Tables 5 and 17.
There was no association between amplification hotspots and known fragile sites in the human genome based on χ2 test at the chromosome band level, even though colocalization does exist, for example, the three fragile sites on 8q. Figure 1 summaries the location of the 86 common fragile sites assayed relative to hotspots of amplification.
Novel amplification hotspots contain putative oncogenes
Remarkably, 27 of the top 100 most frequently amplified genes have been previously described to be overexpressed in various cancers (Supplementary Table 7), but aside from MYC and ERRB2, the mechanism leading to overexpression of these genes was largely unknown. To further explore the properties of the amplified genes, functional and biological characteristics were evaluated through the use of Ingenuity Pathways Analysis (Ingenuity Systems, see (see Supplementary Methods). Functional analysis identified a significant association between the amplified genes with genes involved in cancer (P = 6.67E−06–3.03E−02; the two significance values refer to a range of specific sub-functions) and other diseases (Supplementary Tables 8 and 9). Furthermore, canonical pathway analysis was used to determine the main signaling pathways in which the amplified genes were involved (Supplementary Table 10). Neuregulin Signaling (P = 1.12E−02), also known as EGFR-family-signaling, was the most significantly affected with GRB7, SHC1, SRC, EGFR, ERBB2 and AKT1 comprising the amplified genes.
Impact of amplification on gene expression levels
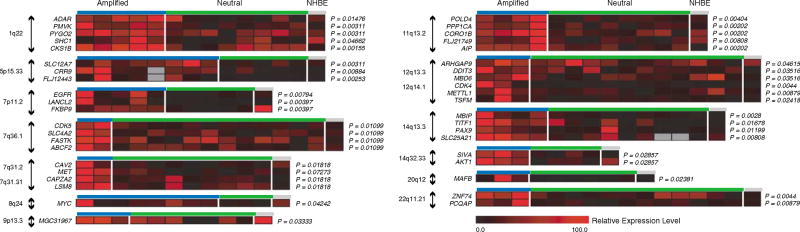
To understand the effects of amplification on gene regulation and transcription, we focused on one type of cancer. Parallel gene expression profiles and array comparative genomic hybridization (CGH) data were integrated for 27 NSCLC cell lines. The expression levels for genes within amplification hotspots (displayed in Supplementary Figure 2) were compared between samples with amplification and those with neutral copy number status using the Mann–Whitney U test (see Supplementary Methods). In total, 221 out of 442 of the amplified genes were expressed at significantly higher levels (P ≤ 0.05) with increased gene dosage (Figure 2 and Supplementary Table 11). For the majority of these genes, amplification is a novel mechanism for activation, although the expression levels of a subset, such as MYC, EGFR, CDK4, MAFB and MET, are known to be affected by increase in gene dosage.
Figure 2.
Impact of amplification on gene transcription levels. The relative expression values for samples with amplification and those with neutral copy number status are plotted as heatmaps for overexpressed genes from representative hotspots. The expression values for each gene have been normalized and scaled across the samples from 0 to 100.
Multiple components of the EGFR-family-signaling pathway are activated by DNA amplification in NSCLC cell lines and clinical tumors
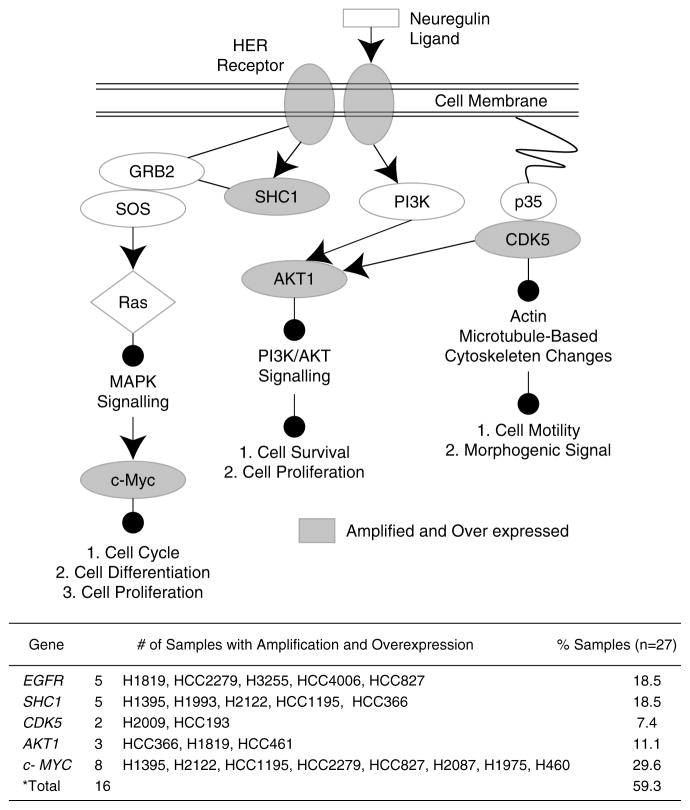
To relate the genes activated by amplification in NSCLC to biological functions, Functional and Canonical Pathway Analysis were performed using IPA software (Supplementary Tables 12 and 13). EGFR-family-signaling was the most affected canonical pathway (P = 6.03E-03) with five genes: AKT1, CDK5, EGFR, MYC and SHC1 amplified and overexpressed in the 27 NSCLCs (Table 2, Figure 2). The amplification and subsequent overexpression of AKT1, CDK5 and SHC1 are novel findings in NSCLC. Figure 3 displays the interaction of these genes during EGFR-family-signaling and the resulting downstream effects of the activation of this pathway, which includes cell proliferation and survival. Interestingly, nearly 60% of the cell lines analysed had one or more components of the EGFR family pathway overexpressed as a result of amplification. The alteration of EGFR and MYC alone could not explain pathway disruption in all cases as ~31% (5/16) of samples with activated downstream components harbored amplification of one or more of CDK5, SHC1 or AKT1 independent of EGFR and MYC. Furthermore, by breaking the NSCLC lines down into their histological subtypes, it was discovered that 14 out of 20 (70%) adenocarcinoma samples—whereas no squamous and only one large cell carcinoma samples—had altered components, suggesting that the disruption of this pathway is prevalent in the adenocarcinoma subtype of lung cancer.
Table 2.
Canonical pathways affected by amplification in non-small cell lung cancer
| Signaling pathway | P-value | Genes |
|---|---|---|
| Neuregulin | 6.0E−03 | MYC, CDK5, SHC1, EGFR and AKT1 |
| Huntington’s disease | 1.1E−02 | SDHA, POLR2H, CDK5, SHC1, POLR2J, EGFR and AKT1 |
| VEGF | 3.1E−02 | VEGF, ARNT, SHC1 and AKT1 |
| Insulin receptor | 3.2E−02 | PARD3, PPP1R3D, PPP1CA, SHC1 and AKT1 |
| Nitric oxide | 3.9E−02 | VEGF, CALM1 and AKT1 |
| Hypoxia | 4.3E−02 | VEGF, ARNT and AKT1 |
Figure 3.
Frequent amplification and overexpression of multiple EGFR-family-signaling components in non-small cell lung cancer (NSCLC). Diagram highlighting the interaction of EGFR, SHC1, CDK5, SHC1 and MYC in the EGFR family-signaling pathway. Altered components are shaded grey. The table summarizes the number and specific samples with amplification and overexpression of each pathway component. The total represents the number of samples with at least one pathway component amplified and overexpressed.
To further validate our results, quantitative real-time PCR was performed on select genes, AKT1, CDK5 and SHC1. First, expression levels for these genes were determined using the ΔΔCt method and compared between cell lines and normal lung tissue to confirm their overexpression. Relative to the normal lung reference, AKT1 was 10.49-fold overexpressed in samples with gene amplification compared to 1.94-fold overexpression in NSCLC cells with neutral copy number status for this gene (Supplementary Figure 3). Likewise, CDK5 (Supplementary Figure 4) and SHC1 (Figure 4) also showed higher expression with increase gene dosage, suggesting a strong correlation of gene dosage and transcription levels for these genes. Expression changes held true in clinical specimens as clinical adenocarcinoma samples frequently showed overexpression of AKT1, CDK5 and SHC1 compared to their corresponding matched normal lung tissues (Supplementary Figure 3c, 4c and Figure 4c, respectively). Since these genes were hypothesized to be overexpressed due to DNA amplification, a one-tailed Wilcoxon sign-rank test was used to determine whether overexpression of these genes was significant in the set of matched tumor and normal samples. Indeed, each gene was significantly overexpressed in the tumors compared to their matched normal (P < 0.05), confirming the results from the cell lines (qPCR data is provide in Supplementary Tables 14 and 15).
Figure 4.
SHC1disruption in NSCLC cell lines and clinical tumors. (a) Representative array CGH profiles for samples with and without SHC1 amplification. Vertical lines denote log2 signal ratios from −1 to 1 with copy number increases to the right (red lines) and decreases to the left (green lines) of 0 (purple line), with amplified region shaded orange. Red and green arrows mark clones used in subsequent Fluorescence in situ hybridization (FISH) analysis. (b) SHC1 expression in NSCLC cell lines. The normalized fold change of expression compared to a normal lung reference is plotted for samples with amplification (red) and those with neutral copy number status (black). (c) Overexpression of SHC1 in clinical tumors. The log2 fold change in expression levels of SHC1 relative to their matched normal lung tissue is plotted for each tumor. The P-value from the Wilcoxon sign-rank test is indicated. (d and e) FISH confirmation of SHC1 amplification in H1395. FISH was performed using BAC clones mapping to SHC1 (RP11-624P9) and to an adjacent neutral copy number region (RP11-313J15).
Discussion
Oncogene activation is traditionally associated with translocation events. We hypothesized that DNA amplification is a prevalent, but underestimated, mechanism of oncogene activation in cancer genomes. To our knowledge, no studies to date have assembled a large panel of paired high-resolution copy number and gene expression data to accurately assess this question. In this study, we examined 104 cancer cell lines comprising various tissues of origin (Table 1) at 24 892 autosomal loci per genome, a resolution that detected amplicons as small as 0.05Mb in size (Ishkanian et al., 2004) and discovered that not only is the incidence of oncogene amplification much greater than previously believed, but specific regions of the genome are hotspots for segmental amplification in cancer cells.
Amplification as a major mechanism of oncogene activation
The activation of oncogenes is a hallmark of tumor development. Cancer cells frequently display chromosome rearrangements resulting in the deregulation of gene expression, as well as in the fusion of genes raising oncogenic activity. As such, the majority of known oncogenes, including 92% of those analysed in this study, have been discovered through their involvement in disease-specific chromosomal translocations (Futreal et al., 2004). Thus, the high incidence of amplification we report suggests that oncogenes may have multiple mechanisms of activation, with the increase in gene dosage being a prominent mechanism of activation. This was particularly evident in the fact that genes, which have been shown to be activated primarily by translocation, were frequently amplified (Supplementary Table 3). The majority of these genes has not been shown to be activated by amplification previously, and as such, this data represent a novel finding. For example, t(14;20)(q32;q12) translocation is known to juxtapose IgH enhancers to the MAFB gene locus upregulating its expression in multiple myeloma (Wang et al., 1999; Boersma-Vreugdenhil et al., 2004). We demonstrated that MAFB amplification also occurs in lung (H1395, H1650 and H1666), cervical (SW756) and liver (HepG2) cancer cells. Likewise, NTRK1 is an oncogene frequently activated by translocation (Roccato et al., 2005). Fusion with TPR, TPM3 or TFG results in constitutive tyrosine kinase activity (Pierotti et al., 1996). Remarkably, we also detected NTRK1 copy number increased in lung (HCC366, HCC1833, HCC1195, H82, H526, H2122 and H187) and breast (ZR7530) cancer genomes, suggesting that amplification and subsequent overexpression may be an alternate mechanism of activation.
Existence of an amplifier phenotype
There is evidence suggesting that some cancer cells have a greater propensity to undergo DNA amplification than others and that there is an underlying genetic basis for this ‘amplifier phenotype’ (Albertson, 2006). Our data showed that the number of oncogenes amplified may differ in individual genomes. The variation existed across both general cancer classes and individual tissue types. In addition, amplification of the same genes in the different tumor types suggests that there may be a selective advantage to have certain genes or their related functions elevated in the context of cancer development and that amplifications are not simply byproducts of general genomic instability characteristic of late-stage tumors. It was also common for a sample to simultaneously harbor multiple amplicons on different chromosomes, highlighting the possibility of an underlying genetic basis for amplification development (Supplementary Figure 1). It has been proposed that amplifications are mainly related to solid tumors and are seldom involved in hematological malignancies in which oncogene activation is generally associated with translocations (Mitelman, 2000). Indeed, only 11 known oncogenes were amplified in leukemia or lymphoma genomes in our data set, whereas 144 were amplified in epithelial cases (Supplementary Table 2). Similar results were observed in the number of amplicons in each genome (~10 per hematological, ~36 in epithelial samples). These results suggest that subsets of cancers, such as the epithelial cancers, are driven by an amplifier phenotype, whereas others, typically hematological malignancies, develop mainly through different genetic mechanisms, such as chromosomal translocation.
Novel amplification hotspots are enriched for putative oncogenes
Figure 1 indicates that regions of frequent copy number amplification are preferentially localized in genome. These results are similar to those found in a bibliomics survey, which looked across 73 different neoplasms using conventional CGH (Myllykangas et al., 2006). However, in this study, we further refined the regions beyond the chromosome band level, determining the exact genes affected by these aberrations (Supplementary Table 6), identifying novel hotspots, such as the discrete amplicons on 14q.
There are two potential factors that may determine the localization of amplification hotspots. First, the selective pressure imposed on the tumor may lead to the selection of amplification of regions containing genes advantageous to tumor growth. Consistent with this theory, we observed a significant enrichment for genes involved in cellular functions and canonical pathways commonly involved in tumorigenesis within the amplification hotspots (Supplementary Tables 8–10). Many genes implicated in key biological processes, such as cell cycle and cell growth and proliferation contained within these regions that may be considered as novel candidate oncogenes. In addition, 27 of the top 100 genes within the hotspots have been previously described to be overexpressed in cancer further supporting their oncogenic role (Supplementary Table 7). Since the mechanism leading to the overexpression of the majority of these genes was previously unknown, our data suggest that amplification may be a key mechanism of their activation.
Second, the intrinsic features of the chromosome regions themselves may be involved in their preferential amplification (Wahl et al., 1984). Mechanistic models, such as breakage-fusion-bridge and episome excision, imply that two double-stranded DNA breaks are required to initiate amplification generation (Myllykangas and Knuutila, 2006). As such, it has been proposed that regions of the genome that are more susceptible to breakage have a greater propensity to undergo amplification, such as, fragile sites (Hellman et al., 2002; Buttel et al., 2004). Conventional CGH studies indicated that many amplification hotspots colocalized with fragile sites, however, this association was not statistically significant on a genome-wide scale, presumably due to inadequate resolution (with chromosome band level comparison) as fragile sites and amplification hotspots covered 30 and 45% of the genome, respectively (Myllykangas et al., 2006). Our analysis addressed this problem as the increased resolution of the platform used in this study allowed the refinement of amplification hotspots, limiting their coverage to ~3% of the genome. However, although we observed a general trend of the colocalization of amplification hotspots and common fragile sites (Figure 1), the global association was still not significant. We speculate that the cloning of fragile sites to determine their specific sequences is needed to complement our array CGH data to accurately assess their association. Nevertheless, there is a strong possibility that the hotspot regions represent damage-prone sites in the genome and further investigation in the future is warranted. Notably, in addition to fragile sites, other genomic features, such as copy number variations and segmental duplications, may also contribute to DNA rearrangement in cancer cells (Squire et al., 2003).
Global impact of amplification on gene expression levels in NSCLC
Although array CGH allows the fine mapping of amplification boundaries at unprecedented resolution, multiple genes may map to an individual amplicon. Therefore, integration of copy number and expression data is needed to distinguish overexpressed genes from bystander genes within amplicons. To accomplish this, we integrated parallel array CGH and expression data for a subset of 27 NSCLC cell lines. Aside from genes known to be activated by amplification in NSCLC, such as EGFR and MYC, our data suggest that the expression of oncogenes, including CDK4, MAFB, BCL11B and MET, is also driven by amplification (Figure 2). Since these genes are typically activated by translocation or missense mutation in malignancies other than NSCLC, these data further supports our hypothesis that amplification is an alternate mechanism of oncogene activation in a subset of cancers.
The integration of the data sets identified expressed genes within novel amplification hotspots in lung cancer that are potentially involved in tumorigenesis. For example, thyroid transcription factor 1 (TITF1) in the novel hotspot at 14q12–q13 is known to be overexpressed specifically in lung adenocarcinoma, the predominate subtype of NSCLC from which the 27 cell lines in our study were derived (Fabbro et al., 1996). This gene encodes a homeodomain transcription factor that is involved in regulating pulmonary development and gene expression (Apergis et al., 1998) and has been proposed to be a lineage marker for tumors arising from the peripheral airway (Stenhouse et al., 2004). Adenocarcinomas that express TITF1 are dependent on its persistent expression for survival (Tanaka et al., 2007). Acquired somatic alteration of this gene during differentiation leads to aberrant lineage-survival pathway signaling. The resulting tumors become addicted to persistent expression of the gene for survival, known as ‘lineage addiction’ (Garraway and Sellers, 2006). Our data suggests that amplification may be a mechanism driving the expression of linage-survival oncogenes in a subset of cancers and further supports a role for TITF1 in lung adenocarcinoma tumorigenesis (Figure 2 and Supplementary Figure 5).
On a global scale, we found amplification had a strong impact on transcription levels as 50% of the genes from amplification hotspots within these samples showed enhanced expression as a consequence of alteration. This falls within observations of previous studies that reported 19.3–62% of amplified genes being overexpressed (Hyman et al., 2002; Pollack et al., 2002; Wolf et al., 2004; Heidenblad et al., 2005). Nevertheless, since this is the first study to integrate high-resolution copy number and gene expression profiles in lung cancer on a whole-genome scale, future analysis will be needed to confirm the functional impact of the genes in each amplicon.
Novel disruptions of the EGFR-family-signaling pathway in NSCLC by gene amplification
Our data indicate that multiple components of the EGFR-family-signaling pathway are frequently amplified and overexpressed in NSCLC (Figure 3). Deregulation of this network commonly occurs in cancer and specifically NSCLC. In NSCLC, the mechanism of deregulation is usually attributed to receptor overexpression or point mutations in the catalytic domain, resulting in ligand-independent constitutive receptor activation and signaling (Bublil and Yarden, 2007). However, in cases with normal levels of wild-type receptor, aberrant constitutive signaling may also occur. Strikingly, our data suggest that amplification of downstream signaling components may be an alternate mechanism of pathway activation in a subset of tumors. Although alteration of this pathway was detected in 59% of the cell lines analysed, only 31% of these cases could be explained by EGFR activation. Indeed, the majority of cell lines with pathway perturbation (69%) contained amplification of key signaling components downstream of the receptor level (Figure 3). The overexpression of these components was confirmed in NSCLC tumors, highlighting their clinical significance (Figure 4 and Supplementary Figures 3 and 4). Although MYC amplification has been previously reported, this is the first study to describe the frequent amplification and overexpression of SHC1, AKT1 and CDK5 in NSCLC. Since these genes are involved in the activation of mitogenic-signaling pathways involved in EGFR-induced transformation and their overexpression has been previously implicated in cancer, these results suggest that their direct genetic activation may play a causal role in NSCLC tumorigenesis and highlights the impact of these novel amplifications on cancer biology (Hennessy et al., 2005).
The direct amplification of downstream components may also have substantial effect on the response to clinical treatment strategies. The high frequency of EGFR family overexpression in cancer has led to the development of targeted therapeutics aimed at inhibiting receptor function. For example, anti-ErbB2 antibodies are currently used for breast cancer treatment and EGFR-specific tyrosine kinase inhibitors, such as Gefitinib and Erlotinib are used in NSCLC therapy (Bublil and Yarden, 2007). The receptor-independent activation of downstream signaling components would impact the effectiveness of these treatment strategies as constitutive activation of signaling pathways would occur regardless of receptor inhibition. Previous studies have shown that downregulation of the AKT/PI3K signaling pathway is required for EGFR tyrosine kinase inhibitors to induce apoptosis in cancer cells (Hemstrom et al., 2006). In addition, activated AKT/PI3K signaling due to MET amplification has been shown to lead to Gefitinib resistance in NSCLC cells (Engelman et al., 2007) Thus, the direct activation of AKT1 and CDK5 would lead to resistance in a NSCLC due to maintained AKT/PI3K signaling in the presence of inhibitor. Likewise, in a drug-resistant NSCLC cell line, alterations of adaptor-protein-mediated signal transduction from EGFR, such as those initiated by SHC1, has been proposed as a possible mechanism of resistance to Gefitinib (Koizumi et al., 2005). Our findings highlight the need to assess the activation status of downstream signaling components and suggest that amplification AKT1, SHC1, CDK5 and MYC may be used for this process.
Conclusion
Since alterations at the DNA level potentially represent causal events in the development of cancer, the genes deregulated as a result of amplification in NSCLC can be viewed as the primary oncogenic targets in a tumor that lead to downstream pathway abrogation. The gain-of-function effect of gene amplifications makes them ideal targets for therapeutic intervention due to the direct nature of their activation and the fact that a tumor can become addicted to their enhanced expression (Weinstein, 2002). Although cancer cell lines were used in this study, the genomic and transcriptional characteristics of such models have been shown to mirror primary tumors and are appropriate systems to identify molecular features that predict or indicate response to targeted therapies (Neve et al., 2006; Greshock et al., 2007). Furthermore, validation of a subset of amplified genes in primary tumors confirmed their clinical importance. Future studies of additional primary tumors will be required to further validate the role of the hotspots in clinical specimens and confirm that they are not artifacts of in vitro culture. Our discovery of high incidence of amplification suggests that it is a major mechanism of oncogene activation in cancer and will provide essential starting points for the discovery of novel oncogenes.
Materials and methods
Whole-genome profiling
DNA copy number profiles for 104 cancer cell lines of lung, breast, prostate, cervical, skin, ovarian, liver and hematological origins were used in this study (Supplementary Table 16). DNA was isolated by proteinase K digestion followed by phenol–chloroform extraction. Array hybridization was performed as previously described (Lockwood et al., 2007), using SMRT array v.2 (Ishkanian et al., 2004; Watson et al., 2007). Array images were analysed using SoftWoRx Tracker Spot Analysis software (Applied Precision, Issaquah, WA, USA). Systematic biases were removed using the stepwise normalization procedure CGH Norm (Khojasteh et al., 2005). SeeGH software allowed visualization of log2 ratio plots in karyograms (Chi et al., 2004). All raw array data files have been made publicly available through the System for Integrative Genomic Microarray Analysis (SIGMA), which can be accessed at http://sigma.bccrc.ca (Chari et al., 2006).
Gene expression profiling
RNA samples from 27 NSCLC cell lines and normal human bronchial epithelial cells were analysed using the Affymetrix Gene Chips HG-U133A and HG-U133B (Henderson et al., 2005; Zhou et al., 2006) (Supplementary Table 16). These arrays together represent 23 583 unique genes based on Unigene build 173. The identity of these cell lines has been verified by DNA fingerprint, using the Powerplex 1.2 system (Promega). Data normalization and microarray analysis was performed using Affymetrix Microarray Suite 5.0 as described previously(Zhou et al., 2006). The microarray data have been uploaded to GEO (Gene Expression Omnibus, accession number GSE-4824).
Statistical analysis of array data
Detailed methods describing the statistical analysis used for the identification of amplicons, amplification hotspots, amplification hotspots and fragile site colocalization, functional assessment of amplified genes and integration of genomic, and gene expression data are provided in Supplementary Methods.
Gene-specific quantitative real-time reverse transcriptase PCR analysis
TaqMan gene expression assays (AKT1 (Hs00178289_m1), SHC1 (Hs00427539_m1), CDK5 (Hs00358991_g1) and 18 s rRNA (Hs99999901_s1) were performed using 100 ng of cDNA samples in a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The ΔΔCt method was used for expression quantification using the average cycle threshold of 18S rRNA for normalization (Coe et al., 2006) and human lung total RNA (AM7968, Ambion, Austin, TX, USA) as a reference. For clinical samples, total RNA was isolated from 10 microdissected frozen lung adenocarcinoma and matched normal tissue obtained from Vancouver General Hospital using RNeasy Mini Kits (QIAGEN Inc., Mississauga, ON, USA) and 1 μg was converted to cDNA for gene-specific quantitative PCR for AKT1, SHC1, CDK5 and 18S rRNA. Cycle thresholds comparison yielded expression changes in the tumors. Because these genes were hypothesized to be overexpressed, owing to DNA amplification, a one-tailed Wilcoxon sign-rank test was used to determine whether overexpression was significant in the set of matched tumor and normal samples.
Fluorescence in situ hybridization
Fluorescence in situ hybridization was performed as described previously (Watson et al., 2004). Briefly, 100 ng of linker-mediated PCR-amplified BAC DNA was labeled through a random priming reaction with Spectrum Green or Red dUTP (Vysis, Markham, ON, USA). Hybridization was performed in a 50% formamide buffer at 37 °C for 18 h and imaged with Q Capture imaging software (Q Imaging, Burnaby, BC, USA).
Supplementary Material
Acknowledgments
This work was supported by funds from CIHR, Genome Canada/BC, Lung Cancer SPORE P50CA70907, DOD VITAL, the Gillson Longenbaugh and Anderson Charitable Foundations as well as scholarships from NSERC, CIHR and MSFHR to WWL, RC and BPC.
Footnotes
Data deposition: Gene Expression Omnibus, accession number GSE-4824.
Supplementary Information accompanies the paper on the Oncogene (http://www.nature.com/onc).
References
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Apergis GA, Crawford N, Ghosh D, Steppan CM, Vorachek WR, Wen P, et al. A novel nk-2-related transcription factor associated with human fetal liver and hepatocellular carcinoma. J Biol Chem. 1998;273:2917–2925. doi: 10.1074/jbc.273.5.2917. [DOI] [PubMed] [Google Scholar]
- Boersma-Vreugdenhil GR, Kuipers J, Van Stralen E, Peeters T, Michaux L, Hagemeijer A, et al. The recurrent translocation t(14;20)(q32;q12) in multiple myeloma results in aberrant expression of MAFB: a molecular and genetic analysis of the chromosomal breakpoint. Br J Haematol. 2004;126:355–363. doi: 10.1111/j.1365-2141.2004.05050.x. [DOI] [PubMed] [Google Scholar]
- Bublil EM, Yarden Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr Opin Cell Biol. 2007;19:124–134. doi: 10.1016/j.ceb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Buttel I, Fechter A, Schwab M. Common fragile sites and cancer: targeted cloning by insertional mutagenesis. Ann NY Acad Sci. 2004;1028:14–27. doi: 10.1196/annals.1322.002. [DOI] [PubMed] [Google Scholar]
- Chari R, Lockwood WW, Coe BP, Chu A, Macey D, Thomson A, et al. SIGMA: a system for integrative genomic microarray analysis of cancer genomes. BMC Genomics. 2006;7:324. doi: 10.1186/1471-2164-7-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B, DeLeeuw RJ, Coe BP, MacAulay C, Lam WL. SeeGH—a software tool for visualization of whole genome array comparative genomic hybridization data. BMC Bioinformatics. 2004;5:13. doi: 10.1186/1471-2105-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Lockwood WW, Girard L, Chari R, Macaulay C, Lam S, et al. Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br J Cancer. 2006;94:1927–1935. doi: 10.1038/sj.bjc.6603167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL. Resolving the resolution of array CGH. Genomics. 2007;89:647–653. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Fabbro D, Di Loreto C, Stamerra O, Beltrami CA, Lonigro R, Damante G. TTF-1 gene expression in human lung tumours. Eur J Cancer. 1996;32A:512–517. doi: 10.1016/0959-8049(95)00560-9. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnis C, Lockwood WW, Vucic E, Ge Y, Girard L, Minna JD, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- Greshock J, Nathanson K, Martin AM, Zhang L, Coukos G, Weber BL, et al. Cancer cell lines as genetic models of their parent histology: analyses based on array comparative genomic hybridization. Cancer Res. 2007;67:3594–3600. doi: 10.1158/0008-5472.CAN-06-3674. [DOI] [PubMed] [Google Scholar]
- Heidenblad M, Lindgren D, Veltman JA, Jonson T, Mahlamaki EH, Gorunova L, et al. Microarray analyses reveal strong influence of DNA copy number alterations on the transcriptional patterns in pancreatic cancer: implications for the interpretation of genomic amplifications. Oncogene. 2005;24:1794–1801. doi: 10.1038/sj.onc.1208383. [DOI] [PubMed] [Google Scholar]
- Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith DI, et al. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Hemstrom TH, Sandstrom M, Zhivotovsky B. Inhibitors of the PI3-kinase/Akt pathway induce mitotic catastrophe in non-small cell lung cancer cells. Int J Cancer. 2006;119:1028–1038. doi: 10.1002/ijc.21927. [DOI] [PubMed] [Google Scholar]
- Henderson LJ, Coe BP, Lee EH, Girard L, Gazdar AF, Minna JD, et al. Genomic and gene expression profiling of minute alterations of chromosome arm 1p in small-cell lung carcinoma cells. Br J Cancer. 2005;92:1553–1560. doi: 10.1038/sj.bjc.6602452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- Khojasteh M, Lam WL, Ward RK, MacAulay C. A stepwise framework for the normalization of array CGH data. BMC Bioinformatics. 2005;6:274. doi: 10.1186/1471-2105-6-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi F, Shimoyama T, Taguchi F, Saijo N, Nishio K. Establishment of a human non-small cell lung cancer cell line resistant to gefitinib. Int J Cancer. 2005;116:36–44. doi: 10.1002/ijc.20985. [DOI] [PubMed] [Google Scholar]
- Lockwood WW, Coe BP, Williams AC, MacAulay C, Lam WL. Whole genome tiling path array CGH analysis of segmental copy number alterations in cervical cancer cell lines. Int J Cancer. 2007;120:436–443. doi: 10.1002/ijc.22335. [DOI] [PubMed] [Google Scholar]
- Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462:247–253. doi: 10.1016/s1383-5742(00)00006-5. [DOI] [PubMed] [Google Scholar]
- Myllykangas S, Himberg J, Bohling T, Nagy B, Hollmen J, Knuutila S. DNA copy number amplification profiling of human neoplasms. Oncogene. 2006;25:7324–7332. doi: 10.1038/sj.onc.1209717. [DOI] [PubMed] [Google Scholar]
- Myllykangas S, Knuutila S. Manifestation, mechanisms and mysteries of gene amplifications. Cancer Lett. 2006;232:79–89. doi: 10.1016/j.canlet.2005.07.045. [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierotti MA, Bongarzone I, Borello MG, Greco A, Pilotti S, Sozzi G. Cytogenetics and molecular genetics of carcinomas arising from thyroid epithelial follicular cells. Genes Chromosomes Cancer. 1996;16:1–14. doi: 10.1002/(SICI)1098-2264(199605)16:1<1::AID-GCC1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccato E, Bressan P, Sabatella G, Rumio C, Vizzotto L, Pierotti MA, et al. Proximity of TPR and NTRK1 rearranging loci in human thyrocytes. Cancer Res. 2005;65:2572–2576. doi: 10.1158/0008-5472.CAN-04-4294. [DOI] [PubMed] [Google Scholar]
- Squire JA, Pei J, Marrano P, Beheshti B, Bayani J, Lim G, et al. High-resolution mapping of amplifications and deletions in pediatric osteosarcoma by use of CGH analysis of cDNA microarrays. Genes Chromosomes Cancer. 2003;38:215–225. doi: 10.1002/gcc.10273. [DOI] [PubMed] [Google Scholar]
- Stenhouse G, Fyfe N, King G, Chapman A, Kerr KM. Thyroid transcription factor 1 in pulmonary adenocarcinoma. J Clin Pathol. 2004;57:383–387. doi: 10.1136/jcp.2003.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl GM, Robert de Saint Vincent B, DeRose ML. Effect of chromosomal position on amplification of transfected genes in animal cells. Nature. 1984;307:516–520. doi: 10.1038/307516a0. [DOI] [PubMed] [Google Scholar]
- Wang PW, Eisenbart JD, Cordes SP, Barsh GS, Stoffel M, Le Beau MM. Human KRML (MAFB): cDNA cloning, genomic structure, and evaluation as a candidate tumor suppressor gene in myeloid leukemias. Genomics. 1999;59:275–281. doi: 10.1006/geno.1999.5884. [DOI] [PubMed] [Google Scholar]
- Watson SK, deLeeuw RJ, Horsman DE, Squire JA, Lam WL. Cytogenetically balanced translocations are associated with focal copy number alterations. Hum Genet. 2007;120:795–805. doi: 10.1007/s00439-006-0251-9. [DOI] [PubMed] [Google Scholar]
- Watson SK, deLeeuw RJ, Ishkanian AS, Malloff CA, Lam WL. Methods for high throughput validation of amplified fragment pools of BAC DNA for constructing high resolution CGH arrays. BMC Genomics. 2004;5:6. doi: 10.1186/1471-2164-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Wolf M, Mousses S, Hautaniemi S, Karhu R, Huusko P, Allinen M, et al. High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia. 2004;6:240–247. doi: 10.1593/neo.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.