Abstract
Cytoglobin (CYGB) is a recently discovered vertebrate globin distantly related to myoglobin with unknown function. CYGB is assigned to chromosomal region 17q25, which is frequently lost in multiple malignancies. Previous studies failed to detect evidence for mutations in the CYGB gene. Recent studies provided preliminary evidence for increased methylation of the gene in lung cancer. Our study was aimed at investigating the role of CYGB as a tumor suppressor gene. By nested methylation-specific DNA sequencing analysis of lung and breast cancer cell lines and bronchial and mammary epithelial cell lines, we identified that methylation of a 110-bp CpG-rich segment of the CYGB promoter was correlated with gene silencing. We specifically targeted this sequence and developed a quantitative methylation-specific PCR assay, suitable for high-throughput analysis. We showed that the tumor specificity of CYGB methylation in discriminating patients with and without lung cancer, using biopsies and sputum samples. We further showed the tumor specificity of this assay with multiple other epithelial and hematologic malignancies. To show tumor suppressor activity of CYGB, we performed the following: (a) RNA interference–mediated knockdown of CYGB gene on colony formation in a CYGB expression–positive lung cancer cell line, resulting in increased colony formation; (b) enforced gene expression in CYGB expression–negative lung and breast cancer cell lines, reducing colony formation; and (c) identification of potential proximate targets down-stream of the CYGB genes. Our data constitute the first direct functional evidence for CYGB, the newest member of the globin family, as a tumor suppressor gene.
Introduction
Globins are respiratory proteins that usually bind an oxygen molecule between the iron ion of the porphyrin ring and a histidine of the polypeptide chain (1). Cytoglobin (CYGB; Gene Bank accession no. NM_134268) is a recently discovered vertebrate globin distantly related to myoglobin with unknown function (2). CYGB is found in the nuclei and cytoplasm of most cell types (3). CYGB is assigned to chromosomal region 17q25, which is commonly lost in multiple malignancies (4). Tylosis, a form of palmoplantar keratosis, is associated with esophageal cancer. CYGB is the only gene completely contained within the 42.5-kb minimal region of the tylosis locus (5). However, no CYGB mutations were found in Tylosis patients (6). Transcriptional inactivation of promoters of tumor suppressor genes by DNA hypermethylation has been well-documented in many human cancers (7). Recent pyrosequencing studies provided evidence for higher levels of CYGB promoter methylation in lung and esophageal tumors compared with adjacent nonmalignant tissues (8). Thus, the role of CYGB as a possible tumor suppressor and involvement of promoter methylation in CYGB gene silencing needs to be explored.
For the reasons described above, we studied the role of CYGB methylation and gene silencing, and developed a quantitative methylation-specific PCR (qMSP) assay, suitable for high-throughput analysis. Using this assay, we determined whether CYGB methylation was tumor specific for multiple malignancies. We also provided evidence that CYGB can function as a tumor suppressor gene and also identified some of the potential proximate gene targets.
Materials and Methods
Surgically resected non–small lung and bladder cancers and their adjacent nonmalignant lung tissues were obtained from M. D. Anderson Cancer Center. Surgically resected breast cancers and their adjacent nonmalignant breast tissues and leukemia cases (acute myelogenous leukemias) were obtained from the University of Texas Southwestern Medical Center. Peripheral blood mononuclear cells (PBMC) were obtained from healthy individuals with a family history of cancer. Sputum samples were obtained from 13 patients with non–small cell lung cancer (NSCLC) and 25 cancer-free individuals [that were heavy smokers with chronic obstructive pulmonary disease (COPD)] from the Canisius Wilhelmina Hospital, as described previously (9). Informed consent and Institutional Review Board permission were obtained at each site. Lung cancer cell lines, breast cancer cell lines, and human-immortalized bronchial epithelial cell (HBEC) lines were established by us (10, 11). Human mammary epithelial cell (HMEC) line was obtained from Clonetics.
Gene expression in cell lines
Gene expression studies were conducted as previously described (12) with some modifications. Semiquantitative reverse transcription-PCR (RT-PCR) were carried out by using QuantiTect SYBR Green PCR kit using primers described previously (8). The expression levels were quantitated using the comparative Ct method, and the level of CYGB expression values of HBEC3 cultures was set as a value of 1. 5-Aza-2-deoxycytidine treatment of cell lines was done using a protocol as previously described (13). Conventional RT-PCR analysis of Col1A1 and UCP2 was conducted as previously described with some modification of the protocol (14-16). The number of cycles was adjusted to provide a semiquantitative estimate of relative mRNA abundance in samples with highly significant differences in expression.
DNA extraction, bisulfite modification, and DNA sequencing
Genomic DNA was extracted from cell lines, primary tumors, nonmalignant cells, and sputum samples as previously described (9). Sodium bisulfite treatment was performed as described previously (17). The modified DNA was used as a template for qPCR analysis. DNA sequencing was carried out as previously described, using Applied Biosystems prism dye terminator cycle sequencing method (Perkin-Elmer Co.; ref. 18). Primers were designed to exclude CpG sites, thereby rendering DNA amplification independent of the methylation status. The primer sequences were as follows: Forward, 5′-GGGAATTGATTTAAAGTTTAAT-3′; Reverse, 5′-TAACCCCCCAAACCTAA-3′. The amplicon sequences were confirmed by sequencing in both directions.
qMSPanalysis
qPCR analysis was performed using the Chromo4MJ Research Real-time PCR system. Sodium bisulfite–treated genomic DNA was amplified by fluorescence-based real-time MSP using TaqMan technology as described previously (9, 19). In brief, CYGB promoter region primers, 5′-CGAGGTCGATCGTTAGTTCGTTC-3′ ( forward), 5′-CCAACGACTAACTCGAAAACGCG-3′ (reverse), and 5′-FAM CGGCGGTCGTCGTGGATTTAGT BHQ-1-3′ (probe), were designed to specifically amplify bisulfite-converted DNA within the region of the test genes that was differentially methylated between CYGB expression–positive and CYGB expression–negative cell lines (Fig. 1). The nonmethylated form of MYOD1 was used as an internal reference standard (9). Sputum DNA samples were coded and shipped from the Netherlands and analyzed in Dallas in a blinded fashion.
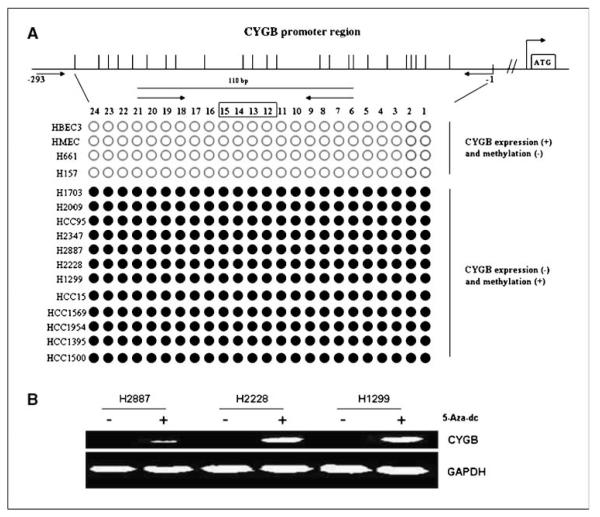
Figure 1.
A, direct sequencing of the bisulfite PCR product from the promoter region of the CYGB gene for 10 lung cancer cell lines and a bronchial epithelial cell line (HBEC3). •,methylated CpG sites; ○,unmethylated CpG sites. A, the region in the promoter (nucleotide positions labeled as −1to −293) represent the amplicon sequenced and encompasses 24 CpG sites. The bent arrow on the far right denotes the start and direction of transcription. Hashed lines, separation of transcription start site from the CpG island region of the promoter used for the sequence analysis. The expression levels of CYGB were quantitated using comparative Ct method. For expression analysis,the mean of expression values for the HBEC was considered to have a value of 1. Expression positive,relative expression of ≥1; expression negative, relative expression of <0.50. Sequencing and expression analyses were conducted as described in Materials and Methods. Arrows, the locations of primers; open box, the location of the probe. B, effect of 5-aza-2′-deoxycytidine treatment of lung cell lines on reactivation of CYGB gene expression. The expression analysis was conducted on cDNA using the primers previously described (8) and analyzing the PCR amplicon on 2% agarose gel. GAPHD, glyceraldehyde-3-phosphate dehydrogenase.
Gene transfection
A plasmid with a sequence verified CYGB cDNA cloned within pCMVNeo vector (Origene) was transfected into NCI-H2228 cells, NCI-2887 cells (lung cancer), and HCC 1569 cells (breast cancer) at 60% confluency, using Fugene HD transfection reagent (Roche). A plasmid with vector alone was transfected as a negative control. The treatment was stopped at 48 h for transient transfection, whereas for stable transfection (NCI-H2228, NCI-2887), cells were replated in quadruplet in 6-well plates (1,000 cells per well), and the cells were selected in medium containing G418 (Sigma Co.) for 14 d. The plasmid with CYGB shRNA (CTCAACACTGTCGTGGAGAACCTGCATGA) cloned within pRSPuro vector (Origene) was transfected into NCI-H661 cells at 60% confluency, using Fugene HD transfection reagent (Roche). Plasmid with a shRNA cassette targeted against green fluorescent protein (GFP) was transfected as a negative control (TGACCACCCTGACCTACGGCGTGCAGTGC). After 72 h, the cells were plated in triplicate in 6-well plates (1,000 cells per well) and selected in medium containing puromycin for 14d. Pilot dose-response studies were performed before initiation of transfection experiments to determine the lowest dose of G418 or puromycin that can be used to 100% kill untrasfected cells. At concentration levels for the two drugs used in our study, cells without resistance markers did not survive beyond 1 wk.
RT2 Profiler PCR array system
RT2 Profiler PCR Array (SuperArray Bioscience Corporation) technology for gene expression analysis features the profiling capabilities of microarray and the quantitative nature of real-time PCR. The results are highly reproducible within the same run or between different runs. The expression profile of 43 genes was analyzed following the manufacture’s instructions. The custom-made array represented 43 key genes involved in various pathways frequently deregulated in cancers and 5 internal control genes. The data were analyzed using Ct values for the gene of interest and housekeeping gene, and using the software program provided by the manufacturer.
Western blot
Western blots were performed as previously described by Suzuki and colleagues (20) using mouse polyclonal antibody to cytoglobin (Abcam).
Liquid colony formation assay
Cells were harvested after transfection and were plated (1,000 cells per well) in triplicate or quadruplet using 6-well plates. After 14d in selection medium, the surviving colonies were stained with 0.25% crystal violet in 50% ethanol and were counted.
Statistical analysis
Receptor operating characteristics (ROC) curves, a plot of the sensitivity versus 1-specificity across all possible cutoff values, were used to identify the accuracy of a marker in discriminating cancer from nonmalignant tissue. The quantitative methylation data for the gene were correlated with tumor stage using the Mann-Whitney U test, which does not require parametric assumption on the distribution of quantitative methylation. P values of <0.05 were considered significant. Statistical differences between groups were examined using Fisher’s exact test or Student’s t test. P values of <0.05 were considered significant.
Results
We sequenced the promoter region of the CYGB gene from 10 non–small lung cancer cell lines, 4 breast cancer cell lines (HCC1569, HCC1954, HCC1395, and HCC1500), a human bronchial epithelial cell line (HBEC3; ref. 10), and an HMEC line (10, 11), and also analyzed these cell lines for CYGB expression (Fig. 1A). The results showed that all the 24CpG sites within the CYGB promoter region analyzed were methylated in 8 of 10 lung cancer cell lines and 4 of 4breast cancer cell lines, whereas all the sites were unmethylated in the other two cancer cell lines (NCI-H661 and NCIH157) and in the nonmalignant HBEC3 and HMEC cells (Fig. 1A). All methylation-positive cell lines had significantly reduced expression of CYGB (relative expression, <0.50). The four methylation negative cell lines were positive for CYGB expression (relative expression, >1). 5-Aza-2′-deoxycytidine treatment of three CYGB-methylated lung cell lines (NCI-H2887, NCI-H2228, and NCI-H1299) resulted in induction of CYGB expression in all the three cell lines (Fig. 1B). In an independent study, NCI-H661 (CYGB expression positive) was treated with 5-Aza-2′-deoxycytidine, and effect on expression was determined using qPCR. No further increase in the endogenous CYGB expression was observed after the drug treatment (data not shown). The results suggest that methylation of the specific sites is related to gene silencing. Based on this expression methylation relationship, the specific CpG region was considered a potential target to develop an assay to discriminate between cancer cells and normal cells.
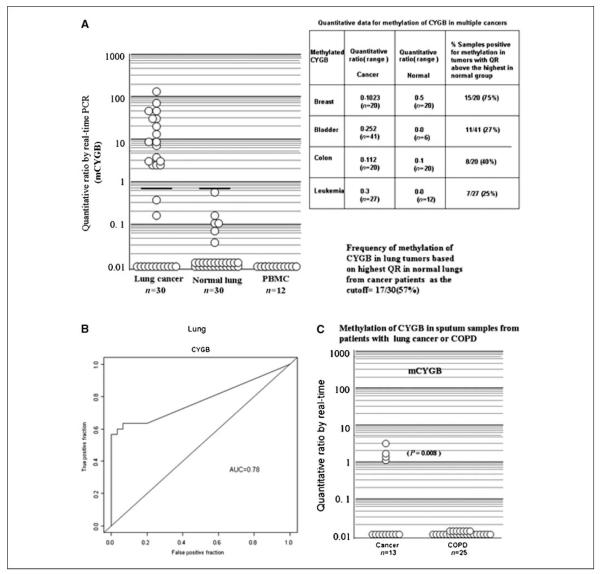
We tested 30 NSCLC tumors (18 adenocarcinomas and 12 squamous carcinomas) and their adjacent nonmalignant lung tissues for methylation of CYGB by qPCR assay. Additionally, PBMCs from 12 healthy subjects recruited for genetic epidemiology studies were also quantitatively analyzed. Figure 2A shows the quantitative methylation data for NSCLC and their adjacent nonmalignant lung tissue. We found detectable levels of methylation of CYGB in 19 of 30 (63%) primary lung tumors, whereas detectable but very low levels (in 6 of 30, 18%) at adjacent lung tissue. Based on the highest quantitative ratio (QR) in nonmalignant tissue as the cutoff, 17 of 30 (57%) of primary NSCLC were found to be methylated for CYGB. Additionally, PBMCs from 12 healthy subjects recruited for genetic epidemiology studies were analyzed for CYGB methylation and found to be below the levels of detection. Figure 2B shows the ROC curve, which provides evidence for the excellent discriminatory capacity of CYGB methylation in separating cancer from adjacent normal lung tissue. We correlated quantitative methylation data for CYGB with tumor stage using the Mann-Whitney U test. No relation was observed between methylation levels and tumor stages 1 to 3 (P > 0.05%; data not shown). There was trend toward higher frequency of CYGB methylation in lung adenomcarinoma compared with that in squamous carcinoma [5 of 18 (28%) versus 1 of 12 (8%)]. However, the differences did not reach statistical significance. We analyzed 38 sputum DNA samples (13 from cancer and 25 noncancer patients) for methylation of CYGB, and we found 4of 13 (30%) cancer sputa (QR, ranged from 0–3.10) and 0 of 25 (0%) noncancer sputa ( from patients with COPD) showed methylation of CYGB (P < 0.008; Fig. 2C).
Figure 2.
A, methylation levels of mCYGB in NSCLC tumors,adjacent nonmalignant lung,and PBMCs from cancer-free individuals. Methylation levels were quantitated by semiquantitative real-time PCR. Real-time analysis was performed as described in Materials and Methods. Quantitative ratio is defined as the ratio of the fluorescence emission intensity values for the PCR products of the biomarker gene to those of PCR products of MYOD1 multiplied by 100. The ratio is a measure of the relative level of methylation in an individual sample. Because values are expressed on a log scale,completely negative values are expressed as values of 0.01. Solid horizontal bar, the threshold above the samples are considered positive for methylation. Table (insert) summarizes quantitative data for methylation of mCYGB in breast cancer, bladder cancer, colon cancer, acute myelogenous leukemia, and corresponding nonmalignant tissues. B, ROC curves for mCYGB at separating cancer from the adjacent nonmalignant lung. ROC curves are plots of the true-positive rate (vertical axis) against the false-positive rate (horizontal axis) for the different possible cutoff points of a diagnostic test. The closer the curve follows the left-hand border and then the top border of the ROC space, the more accurate the test. AUC, 0.78. C, methylation levels of mCYGB in sputum from NSCLC patients and in sputa from patients without malignancy (COPD). Methylation levels were quantitated as described in Materials and Methods.
We found high levels of methylation of CYGB in 26 of 30 (87%) breast cancers and in much lower or undetectable levels in (13 of 30; 40%) adjacent nonmalignant breast tissue (Fig. 2, insert). Based on the highest QR in nonmalignant tissue as the cutoff, 15 of 20 (75%) of primary breast tumors were found to have CYGB methylation values over normal tissue value. Thus, CYGB methylation has excellent discriminatory capacity at separating cancer from adjacent nonmalignant breast tissue [ROC curve; area under the curve (AUC), 0.91].
Additionally, we tested for methylation of CYGB in bladder cancers, colon cancers, and leukemias with corresponding nonmalignant tissues (Fig. 1, table insert). In all cases, CYGB methylation seems to have excellent discriminatory capacity at separating cancer from nonmalignant tissue. For bladder cancer, CYGB methylation had excellent tumor specificity, but no relation was observed between methylation levels and invasiveness (Mann-Whitney U test).
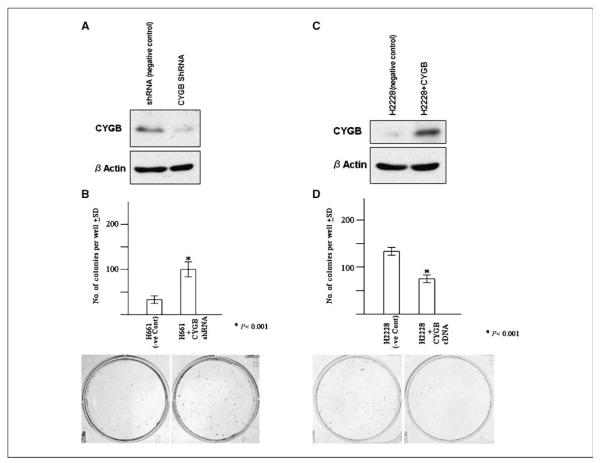
To study the potential of CYGB to regulate lung cancer growth, we tested the effect of RNA interference (RNAi)-mediated CYGB knockdown on NSCLC cell line NCI-H661 (CYGB expression positive). Western Blot data showed loss of CYGB expression in NCI-H661 treated with shRNA but not in the vector control (Fig. 3A). We found significantly increased number of colonies as a result of CYGB knockdown in the transfected cells [mean ± SD: NCI-H661 (negative control), 97.0 ± 7; H661+CGB shRNA, 35 ± 3; P < 0.001; Fig. 3B].
Figure 3.
A, Western blot data for NCI-H661 cells, transfected with plasmid with CYGB shRNA cloned within pRSPuro vector or plasmid with noneffective shRNA cassette against GFP was transfected as a negative control. The treatment was stopped at 48 h for mCYGB protein analysis. B, liquid colony formation assay of NCI-H661 cells, transfected with plasmid containing CYGB shRNA cloned within pRSPuro vector (Origene) or plasmid with noneffective shRNA cassette against GFP (negative control). After 72 h, the cells were plated in triplicate in 6-well plates (1,000 cells per well) and selected in medium containing puromycin for 14 d. The cells were stained with crystal violet after 14 d in selection medium, and the surviving colonies were counted. Columns, mean of triplicates; bars, SD. Representative plates of the colony formation assay comparing vector control with CYGB shRNA–transfected group are shown. C, Western blot data for NCI-H2228 cells, transient transfected with plasmid containing CYGB c-DNA cloned within pCMV vector or with vector alone. D, liquid colony formation assay of NCI-H2228 cells transfected with plasmid containing CYGB cDNA cloned within pCMVNeo vector or Neo vector alone for 48 h; the cells were replated in quadruplet in 6-well plates (1,000 cells per well), and selected in medium containing G418 for 14 d. The cells were stained with crystal violet after 14 d in selection medium, and the surviving colonies were counted. Columns, mean of quadruplets; bars, SD. Representative plates of the colony formation assay comparing vector alone with CYGB cDNA–transfected group are shown.
In another experiment, after CYGB shRNA treatment, allowing the CYGB–knocked down cells to grow in the same wells and subjecting them to selection medium (rather than replating in new wells for colony assay) led to cells forming multilayer foci, in which cells grew on the top of each other (Fig. 1; Supplementary Data).
We then exogenously expressed CYGB in non–small lung cancer cell lines (NCI-H2228 and NCI-H2887) and a breast cancer cell line (HCC1569) lacking CYGB expression with CYGB cDNA. At 48 h after transfection, Western blot analysis of the CYGB-transfected compared with vector-transfected cell lines showed no detectable CYGB, whereas CYGB-transfected cells showed clearly detectable protein. This has been illustrated in Fig. 3C, for NCI-H2228. RNA was tested from vector control (CYGB expression negative) and CYGB transfected (CYGB expression positive) for all the 3 cell lines at 48 h for expression of 43 genes by the highly sensitive and quantitative RT2Profiler PCR array. Of the 3 CYGB-transfected (CYGB expression positive) cell lines, only 7 of 43 genes were up-regulated or down-regulated >2-fold compared with vector controls. The analysis was repeated on independently transfected cells ( from different wells in the 6-well plate) in the same experiment, and the trends in gene expression were highly reproducible for all the three cell lines. The change in expression was considered significant, only if it was up-regulated or down-regulated >2-fold in each of the two independently transfected samples. When analyzed for gene expression, CYGB was up-regulated, whereas COL1A1, PRPF40A, UCP2, DNMT1, and DAPK were down-regulated in NCI-H2228 transfected with CYGB compared with vector control cells. Similarly, CYGB was up-regulated whereas COL1A1, PRPF40A, and UCP2 were down-regulated in NCI-H2887 transfected with CYGB compared with vector control cells. CYGB was up-regulated, whereas COL1A1, PRPF40A, UCP2, and PYCARD were down-regulated in HCC1569 transfected with CYGB compared with vector control cells. Table 1 shows individual ratios of CYGB-transfected cells: vector-transfected cells with numbers indicative of fold differences for all the 43 genes.
Table 1.
Transient expression of CYGB in NCI-H2228, NCI-H2887, and HCC 1569 cells and its effect on gene expression profile
| Gene Symbol | Description | Accession no. | H2228 |
H2887 |
HCC1569 |
|
|---|---|---|---|---|---|---|
| XH2228+CYGB | XH2887+CYGB | XHCC1569+CYGB | ||||
| 1 | RASSF1A | Ras association domain family 1 | NM_007182 | 1.15 | 0.81 | 0.91 |
| 2 | 3-OST-2 | Heparan sulfate(glucosamine) 3-O-sulfotrasferase2 |
NM_006043 | 1 | 1 | 1 |
| 3 | 3-OST-3 | Heparan sulfate(glucosamine) 3-O-sulfotrasferase3B1 |
NM_006041 | 0.8 | 0.7 | 0.82 |
| 4 | CDKN2A | Cyclin-dependent kinase inhibitor 2A | NM_000077 | 1 | 1 | 0.9 |
| 5 | SOCS1 | Suppressor of cytokine signaling 1 | NM_003745 | 1.33 | 0.9 | 1.39 |
| 6 | SOCS3 | Suppressor of cytokine signaling 3 | NM_003955 | 0.6 | 1.25 | 1.1 |
| 7 | CDH1 | Cadherin1 | NM_004360 | 0.73 | 0.59 | 0.72 |
| 8 | CDH13 | Cadherin13 | NM_001257 | 1 | 1 | 1 |
| 9 | APC | Adenomatosis polyposis coli | NM_000038 | 1.41 | 0.62 | 1.01 |
| 10 | DAPK | Death-associated protein kinase 1 | NM_004938 | 0.4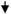
|
1 | 1 |
| 11 | TCF21 | Transcription factor 21 | NM_198392 | 1 | 1 | 1 |
| 12 | PYCARD | PYD and CARD domain containing | NM_013258 | 0.72 | 1 | 0.47 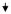
|
| 13 | CDKN2B | Cyclin-dependent kinase inhibitor 2B | NM_004936 | 1 | 1.06 | 1.31 |
| 14 | PTPN6 | Protein tyrosine phosphatase, nonreceptor type 6 | NM_002831 | 0.8 | 1.28 | 0.79 |
| 15 | RUNX3 | Runt-related transcription factor 3 | NM_004350 | 0.75 | 0.84 | 0.7 |
| 16 | TMEFF2 | Transmembrane protein with EGF-like and two follistatin-like domain 2 |
NM_016192 | 1 | 1 | 1 |
| 17 | TIMP3 | TIMP metallopeptidase inhibitor 3 | NM_000362 | 1 | 1 | 1 |
| 18 | RRAD | Ras-related associatiated with diabetes | NM_004165 | 0.6 | 1.34 | 1 |
| 19 | TNFRSF10C | Tumor necrosis factor receptor superfamily, member 10c |
NM_003841 | 1.64 | 0.91 | 1.19 |
| 20 | TNFRSF10D | Tumor necrosis factor receptor superfamily, member 10d |
NM_003840 | 1 | 1 | 1 |
| 21 | RARB | Retinoic acid receptor, β | NM_000965 | 0.95 | 1.3 | 0.58 |
| 22 | SYK | Spleen tyrosine kinase | NM_003177 | 1.43 | 1 | 1 |
| 23 | CYGB | Cytoglobin | NM_134268 | 8.83 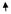
|
10.33 
|
13.08 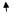
|
| 24 | GSTP1 | Glutathione S-transferase pi | NM_000852 | 0.65 | 0.93 | 0.59 |
| 25 | APAF1 | Apoptotic peptidase activating factor 1 | NM_001160 | 1.14 | 1.39 | 0.86 |
| 26 | DNMT1 | DNA (cytosine-5-)-methyltransferase 1 | NM_001379 | 0.37 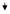
|
0.58 | 0.93 |
| 27 | RPRM | Reprimo | NM_019845 | 0.61 | 1 | 1 |
| 28 | TCF2 | Transcription factor 2 | NM_006481 | 1.28 | 0.86 | 1 |
| 29 | MGMT | O-6-methylguanine-DNA-methyltransferase | NM_002412 | 0.76 | 0.87 | 1.79 |
| 30 | IGFRB3 | Insulin-like growth factor binding protein 3 | NM_000598 | 1.2 | 1.31 | 0.73 |
| 31 | CADM1 | Cell adehesion molecule 1 | NM_014333 | 0.87 | 0.81 | 0.85 |
| 32 | MLNR | Motlin receptor | NM_001507 | 1 | 0.81 | 1 |
| 33 | ITM2B | Integral membrane protein 2B | NM_021999 | 1 | 1.13 | 1 |
| 34 | ANGPTL4 | Angiopoietin-like4 | NM_139314 | 0.86 | 1.58 | 0.94 |
| 35 | COL1A1 | Collagen, type I, α 1 | NM_000088 | 0.31 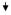
|
0.25 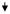
|
0.42 
|
| 36 | PRPF40 | PRP40 pre-mRNA processing factor 40 homologue A (S. cerevisiae) |
XM_371575 | 0.35 
|
0.38 
|
0.36 
|
| 37 | HDAC1 | Histone deacetylase 1 | NM_004964 | 1.25 | 0.87 | 0.85 |
| 38 | IL6 | Interleukin 6 (IFN, β 2) | NM_000600 | 1 | 1.71 | 1.46 |
| 39 | HIF1A | Hypoxia-inducible factor 1, α subunit | NM_001530 | 1.1 | 1.39 | 0.81 |
| 40 | UCP2 | Uncoupling protein 2 (mitochondrial proton carrier) |
NM_003355 | 0.3 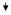
|
0.43 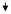
|
0.4 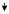
|
| 41 | VMT | Vimentin | NM_003380 | 0.67 | 0.6 | 0.62 |
| 42 | SNAIL1 | Snail homologue 1 (Drosophila) | NM_005985 | 1 | 1.42 | 1.09 |
| 43 | WNT4 | Wingless-type mouse mammary tumor virus integration site family, member 4 |
NM_030761 | 1.53 | 1 | 1 |
NOTE: The data represent fold difference, CYGB-transfected/vector-transfected sample. The data were analyzed as genes with expression up-regulated or down-regulated >2-fold or <2-fold (no change). , up-regulated;
, up-regulated;  , down-regulated.
, down-regulated.  , no change. The data are results from two independently transfected cells ( from different wells in the 6-well plate) in the same experiment, as described in Materials and Methods. The expression has to be upregulated or down-regulated>2 in both experiments to be considered significant and those data are highlighted. Arbitrarily, values with fold difference of 1 ± <0.05 are shown as 1.
, no change. The data are results from two independently transfected cells ( from different wells in the 6-well plate) in the same experiment, as described in Materials and Methods. The expression has to be upregulated or down-regulated>2 in both experiments to be considered significant and those data are highlighted. Arbitrarily, values with fold difference of 1 ± <0.05 are shown as 1.
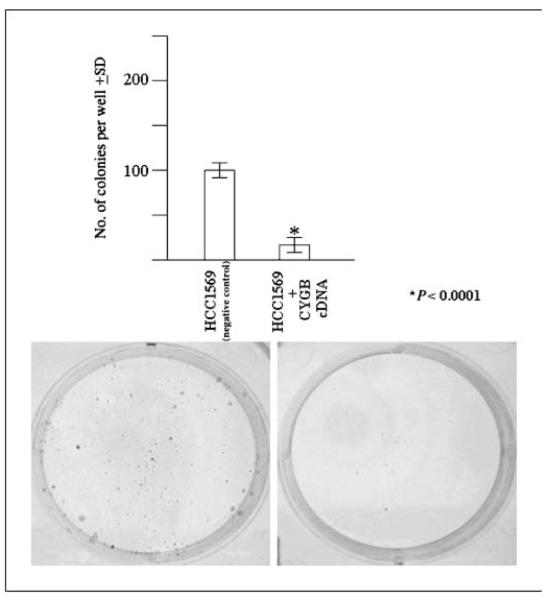
Lastly, we evaluated the effect of stable CYGB transfection on growth of NCI-H2228 (CYGB expression negative) and NCI-H2887 (CYGB expression negative) using liquid colony formation assay. We found significantly reduced number of colonies as a result of transfection and selection of the transfected cells [mean ± SD: NCI-H2228 (negative control), 137 ± 8; NCI-H2228+CYGB, 79 ± 6; P < 0.001 (Fig. 3D); mean ± SD: NCI-H2887 (negative control), 148 ± 11; NCI-H2887+CYGB, 74 ± 8; P = 0.0008]. Additionally, to further confirm tumor suppressor potential of CYGB, we also evaluated the effect of stable CYGB transfection on growth of CYGB expression–negative breast cancer cell line (NCI-HCC1569). We found significantly reduced number of colonies in liquid colony formation assay (mean ± SD: NCI-HCC1569 (negative control), 94 ± 6; NCI-HCC1569+CYGB, 14 ± 3; P > 0.0001; Fig. 4).
Figure 4.
Liquid colony formation assay of NCI-H1569 cells transfected with plasmid containing CYGB cDNA cloned within pCMVNeo vector or Neo vector alone for 48 h; the cells were replated in quadruplet in 6-well plates (1,000 cells per well), and selected in medium containing G418 for 14 d. The cells were stained with crystal violet after 14 d in selection medium and the surviving colonies were counted. Columns, mean of six replicates; bars, SD. Representative plates of the colony formation assay comparing vector alone with CYGB cDNA–transfected group are shown. −ve cont, negative control.
Gene expression analysis of H2228 clones stably transfected with CYGB (two independent transfect ions) clearly showed 6.2-fold (mean) up-regulation of CYGB and 3.6-fold (mean) down-regulation of Col1A1 (Supplementary Fig. S1) compared with clones stably transected with neovector alone. Although COL1A1, PRPF40A and UCP2, and DNMT1 were all consistently down-regulated, they did not reach criteria of >2-fold down-regulation in both of the 2 independent transfections. It is important to mention that as described at stable clone level, there were changes in gene expression of multiple genes (data not shown). However, because we were more interested in genes that are proximate down-stream target in cytoglobin signaling pathway, we focused on the limited number of genes, we earlier identified as potential down-stream targets (Table 1). In CYGB stable transfection experiments with H2887 and HCC1569, although multiple replicates were done for colony formation assay and two independent experiments at transient transfection level for gene expression studies, only one transfection experiment was done at stable transfection level for gene expression experiment. We are planning to repeat these stable transfection experiments in replicates for gene expression analysis. However, based on the single transfection experiment, significant up-regulation of CYGB (6.9-fold with H2887; 5.8-fold with HCC1569) were noted in stably transfected cells.
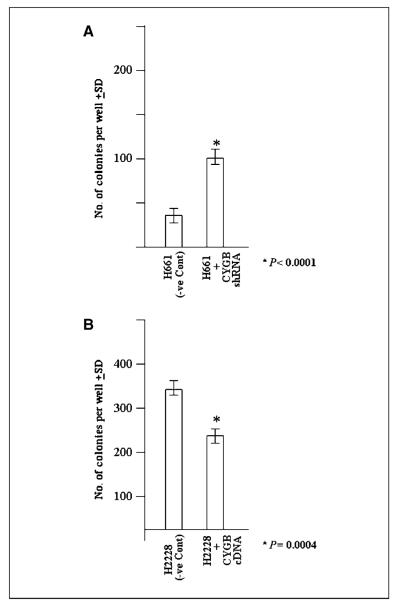
To check whether CYGB regulates the ability of cells to grow in suspension or merely their proliferation rate already established, CYGB expression–negative H661 clone and CYGB expression–positive H2228 clone established as replicates during colony formation assay (Fig. 3) and cryopreseved for future use were examined for their colony forming ability. The colony formation assay clearly showed significantly increased number of colonies in CYGB-negative H661 cells compared with respective unmodified cells [mean ± SD: NCI-H661 (negative control), 35 ± 3; H661+CGB shRNA, 93.0 ± 2; P < 0.0001; Fig. 5A] and significantly reduced number of colonies as in CYGB-positive H2228 [mean ± SD: NCI-H2228 (negative control), 325 ± 5; NCI-H2228+CGB, 226 ± 14; P = 0.0004; Fig. 5B]. The results also further confirmed that the cells that formed colonies actually were actually the cells inoculated into the liquid culture.
Figure 5.
A, liquid colony formation assay of NCI-H661 cells, stably transfected with plasmid containing CYGB shRNA cloned within pRSPuro vector (Origene) or plasmid with noneffective shRNA cassette against GFP (negative control). The cells were plated in 6-well plates (1,000 cells per well) and selected in medium containing puromycin for 14 d. The cells were stained with crystal violet after 14 d in selection medium, and the surviving colonies were counted. Columns, mean of triplicates; bars, SD. B, liquid colony formation assay of NCI-H2228 cells stably transfected with plasmid containing CYGB cDNA cloned within pCMVNeo vector or Neo vector alone. The cells were replated in quadruplet in 6-well plates (1,000 cells per well), and the cells were selected in medium containing G418 for 14 d. The cells were stained with crystal violet after 14 d in selection medium and the surviving colonies were counted. Columns, mean of triplicate; bars, SD.
Discussion
We provided evidence at various levels to support the proposed role of CYGB as a potential tumor suppressor gene inactivated by DNA methylation in multiple malignancies. Initially, our studies showed the tumor versus normal tissue specificity of CYGB methylation in multiple cancer types. We analyzed lung cancer and breast cancer cell lines, a bronchial epithelial cell line and a mammary epithelial cell line for promoter methylation and expression of CYGB. We identified a short (110 bp) CpG-rich segment in the promoter region, methylation of which is closely correlated with gene silencing. This is the first report where correlation between CYGB methylation and gene silencing was convincingly shown. We specifically targeted this short CpG-rich sequence and developed a qMSP assay, suitable for high-throughput quantitative methylation analysis. Using this assay, we showed the tumor specificity of the CYGB methylation in lung cancer and also with multiple other epithelial malignancies and acute myelogenous leukemias. Because no relation was observed between methylation levels and tumor stage in lung cancer or with invasiveness in bladder cancer, it seems that CYGB methylation might be a relatively early change in tumor pathogenesis. RNAi-mediated knock-down of CYGB in an expression-positive lung cancer cell line resulted in increased colony formation, whereas transfecting CYGB cDNA into a cancer cell lines and also a breast cancer cell line lacking CYGB expression reduced tumor cell growth. Our results involving RNAi-mediated knock-down (Supplementary Fig. S1) provide additional interesting evidence for tumor suppressor property of CYGB. A previous study (21) has described initiation of growth of cells on the top of each other as a result of P53 knockdown of HBECs. Thus, our data provide a strong evidence for tumor suppressor property of the gene.
The exact function of CYGB is unknown, although it has been proposed to play a role in collagen synthesis (22, 23), O2 sensing and transport (24), or detoxification of reactive oxygen species (ROS; ref. 2, 24). Potential down-stream targets of CYGB may be identified through enforced expression of a gene in a cancer cell. To our knowledge, our study constitutes the first report identifying potential proximate targets down-stream of CYGB, through enforced expression of the gene into cancer cell lines. After studying gene expression patterns of 43 genes involved in various pathways frequently deregulated in cancers, we identified expressional changes in CYGB and 5 other genes after transient expression of CYGB. Of interest, three of these genes (all down-regulated) are hypoxia related (COL1A1, PRPF40A, and UCP2; refs. 18, 19, 21). COL1A1 encodes the major component of type I collagen the fibrillar collagen found in most connective tissues. COL1A is up-regulated in response to hypoxia (25). The expression of the gene is up-regulated in advanced stages of multiple tumor types (26–30). Because COL1A1 was observed to be down-regulated by CYGB in all the three cancer cell lines, it seems to be most likely one of the proximate target down-stream of the gene. This is further supported by our data, which also showed that COL1A1 was down-regulated >2-fold in H2228, stably transfected with CYGB compared with negative control.
ROS primarily originate in mitochondria during oxidative phosphorylation. Uncoupling protein-2 (UCP2) is a recently identified mitochondrial inner membrane anion carrier, which is emerging as a negative regulator of ROS production (31). In various cell types, UCP2 acts as a sensor of mitochondrial oxidative stress and may be activated by superoxide or by subsequently formed lipid peroxidation products (32). Overexpression of UCP2 in colon cancer cells inhibits ROS accumulation and apoptosis after exposure to chemotherapeutic agents. Tumor xenografts of UCP2-overexpressing human colon cancer cells retain growth in nude mice receiving chemotherapy. These findings link UCP2 with molecular mechanisms of chemoresistance. Thus, targeting UCP2 may be considered a novel treatment strategy for cancer (15). Tumor growth in general results from an imbalance between cell proliferation and cell attrition. Adaptive mechanisms in cancer cells include resistance to growth inhibition and evasion of apoptosis, cellular events appreciably affected by oxidative stress (33). Because UCP2 was observed to be down-regulated by CYGB in all the three cancer cell lines, it seems to be most likely one of the proximate target down-stream target of CYGB. PRP40 pre-mRNA (PRPF40) is a RNA-binding and pre-mRNA processing factor. PRPF40 has been reported to be up-regulated in hypoxia and during cancer progression (34, 35). Because PRPF40 was observed to be down-regulated by CYGB in all the three cancer cell lines, it seems to be most likely one of the proximate target down-stream of CYGB.
Besides, Col1A1, UCP2, and PRFP40, which were down-regulated in each of the two lung lines and one breast cancer cell line, down-regulation of DNMT1 and DAPK was observed in lung cancer cell lines, whereas down-regulation of PYCARD was observed in the breast cancer cell. Up-regulation of DNMT1 is a frequent event in many cancers and has been implicated in promoter methylation and silencing of tumor suppressor genes (20). A previous study has reported suppression of DNMT1 transcription by transient expression of APC in a human colon carcinoma cell line (36). DAP kinase and PYCARD are key genes implicated in apoptosis (37). Because these three genes were not targeted in all the three cell lines, it is possible that they could be cell-specific targets or may not be immediate down-stream targets. More comprehensive studies are needed to identify additional potential down-stream targets and role of these targets in cytoglobin signaling pathway. We report here some of the genes that are potential down-stream targets of CYGB signaling pathway and should be further explored in that context. In conclusion, overall, our data support that CYGB, the newest member of the globin family, can function such as a tumor suppressor gene methylated in several human malignancies.
Supplementary Material
Acknowledgments
Grant support: U01CA084971 from the Early Detection Research Network and the University of Texas Specialized Programs of Research Excellence in Lung Cancer P50CA70907 from National Cancer Institute, Bethesda, MD (A.F. Gazdar) and P50 CA91846-06 from National Cancer Institute, Bethesda, MD (B. Czerniak).
Footnotes
Disclosure of Potential Conflicts of Interest The authors have declared that no conflict of interest exists.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Hardison RC. A brief history of hemoglobins: plant, animal, protist, and bacteria. Proc Natl Acad Sci U S A. 1996;93:5675–9. doi: 10.1073/pnas.93.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burmester T, Ebner B, Weich B, Hankeln T. Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol Biol Evol. 2002;19:416–21. doi: 10.1093/oxfordjournals.molbev.a004096. [DOI] [PubMed] [Google Scholar]
- 3.Weber RE, Fago A. Functional adaptation and its molecular basis in vertebrate hemoglobins, neuroglobins and cytoglobins. Respir Physiol Neurobiol. 2004;144:141–59. doi: 10.1016/j.resp.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Presneau N, Dewar K, Forgetta V, Provencher D, Mes-Masson AM, Tonin PN. Loss of heterozygosity and transcriptome analyses of a 1.2 Mb candidate ovarian cancer tumor suppressor locus region at 17q25.1-q25.2. Mol Carcinog. 2005;43:141–54. doi: 10.1002/mc.20096. [DOI] [PubMed] [Google Scholar]
- 5.Risk JM, Evans KE, Jones J, et al. Characterization of a 500 kb region on 17q25 and the exclusion of candidate genes as the familial Tylosis Oesophageal Cancer (TOC) locus. Oncogene. 2002;21:6395–402. doi: 10.1038/sj.onc.1205768. [DOI] [PubMed] [Google Scholar]
- 6.Langan JE, Cole CG, Huckle EJ, et al. Novel micro-satellite markers and single nucleotide polymorphisms refine the tylosis with oesophageal cancer (TOC) minimal region on 17q25 to 42.5 kb: sequencing does not identify the causative gene. Hum Genet. 2004;114:534–40. doi: 10.1007/s00439-004-1100-3. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 8.Xinarianos G, McRonald FE, Risk JM, et al. Frequent genetic and epigenetic abnormalities contribute to the deregulation of cytoglobin in non-small cell lung cancer. Hum Mol Genet. 2006;15:2038–44. doi: 10.1093/hmg/ddl128. [DOI] [PubMed] [Google Scholar]
- 9.Shivapurkar N, Stastny V, Suzuki M, et al. Application of a methylation gene panel by quantitative PCR for lung cancers. Cancer Lett. 2007;247:56–71. doi: 10.1016/j.canlet.2006.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramirez RD, Sheridan S, Girard L, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–34. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 11.Gazdar AF, Kurvari V, Virmani A, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–74. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 12.Smith LT, Lin M, Brena RM, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23–24 in lung and head and neck cancer. Proc Natl Acad Sci U S A. 2006;103:982–7. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shigematsu H, Suzuki M, Takahashi T, et al. Aberrant methylation of HIN-1 (high in normal-1) is a frequent event in many human malignancies. Int J Cancer. 2005;113:600–4. doi: 10.1002/ijc.20622. [DOI] [PubMed] [Google Scholar]
- 14.Millington-Ward S, Allers C, Tuohy G, et al. Validation in mesenchymal progenitor cells of a mutation-independent ex vivo approach to gene therapy for osteogenesis imperfecta. Hum Mol Genet. 2002;11:2201–6. doi: 10.1093/hmg/11.19.2201. [DOI] [PubMed] [Google Scholar]
- 15.Derdak Z, Mark NM, Beldi G, Robson SC, Wands JR, Baffy G. The mitochondrial uncoupling protein-2 promotes chemoresistance in cancer cells. Cancer Res. 2008;68:2813–9. doi: 10.1158/0008-5472.CAN-08-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horimoto M, Resnick MB, Konkin TA, Routhier J, Wands JR, Baffy G. Expression of uncoupling protein-2 in human colon cancer. Clin Cancer Res. 2004;10:6203–7. doi: 10.1158/1078-0432.CCR-04-0419. [DOI] [PubMed] [Google Scholar]
- 17.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–6. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 19.Toyooka KO, Toyooka S, Maitra A, et al. Establishment and validation of real-time polymerase chain reaction method for CDH1 promoter methylation. Am J Pathol. 2002;161:629–34. doi: 10.1016/S0002-9440(10)64218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. RNA interference-mediated knockdown of DNA methyltransferase 1 leads to promoter demethylation and gene re-expression in human lung and breast cancer cells. Cancer Res. 2004;64:3137–43. doi: 10.1158/0008-5472.can-03-3046. [DOI] [PubMed] [Google Scholar]
- 21.Sato M, Vaughan MB, Girard L, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–28. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt M, Gerlach F, Avivi A, et al. Cytoglobin is a respiratory protein in connective tissue and neurons, which is up-regulated by hypoxia. J Biol Chem. 2004;279:8063–9. doi: 10.1074/jbc.M310540200. [DOI] [PubMed] [Google Scholar]
- 23.Nakatani K, Okuyama H, Shimahara Y, et al. Cytoglobin/STAP, its unique localization in splanchnic fibroblast-like cells and function in organ fibrogenesis. Lab Invest. 2004;84:91–101. doi: 10.1038/labinvest.3700013. [DOI] [PubMed] [Google Scholar]
- 24.Trent JT, III, Hargrove MS. A ubiquitously expressed human hexacoordinate hemoglobin. J Biol Chem. 2002;277:19538–45. doi: 10.1074/jbc.M201934200. [DOI] [PubMed] [Google Scholar]
- 25.Falanga V, Zhou L, Yufit T. Low oxygen tension stimulates collagen synthesis and COL1A1 transcription through the action of TGF-β1. J Cell Physiol. 2002;191:42–50. doi: 10.1002/jcp.10065. [DOI] [PubMed] [Google Scholar]
- 26.Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular-pathological prognostic factors of gastric cancer: a review. Gastric Cancer. 2005;8:86–94. doi: 10.1007/s10120-005-0320-0. [DOI] [PubMed] [Google Scholar]
- 27.Creighton CJ, Bromberg-White JL, Misek DE, et al. Analysis of tumor-host interactions by gene expression profiling of lung adenocarcinoma xenografts identifies genes involved in tumor formation. Mol Cancer Res. 2005;3:119–29. doi: 10.1158/1541-7786.MCR-04-0189. [DOI] [PubMed] [Google Scholar]
- 28.De Cecco L, Marchionni L, Gariboldi M, et al. Gene expression profiling of advanced ovarian cancer: characterization of a molecular signature involving fibroblast growth factor 2. Oncogene. 2004;23:8171–83. doi: 10.1038/sj.onc.1207979. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Miller C, Mosher R, et al. Identification of cervical cancer markers by cDNA and tissue micro-arrays. Cancer Res. 2003;63:1927–35. [PubMed] [Google Scholar]
- 30.Villar J, Arenas MI, MacCarthy CM, Blanquez MJ, Tirado OM, Notario V. PCPH/ENTPD5 expression enhances the invasiveness of human prostate cancer cells by a protein kinase C δ-dependent mechanism. Cancer Res. 2007;67:10859–68. doi: 10.1158/0008-5472.CAN-07-2041. [DOI] [PubMed] [Google Scholar]
- 31.Arsenijevic D, Onuma H, Pecqueur C, et al. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000;26:435–9. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 32.Echtay KS, Esteves TC, Pakay JL, et al. A signalling role for 4-hydroxy-2-nonenal in regulation of mitochondrial uncoupling. EMBO J. 2003;22:4103–10. doi: 10.1093/emboj/cdg412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3:420–5. doi: 10.1093/embo-reports/kvf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sluimer JC, Kisters N, Cleutjens KB, et al. Dead or alive: gene expression profiles of advanced atherosclerotic plaques from autopsy and surgery. Physiol Genomics. 2007;30:335–41. doi: 10.1152/physiolgenomics.00076.2007. [DOI] [PubMed] [Google Scholar]
- 35.Thakur A, Bollig A, Wu J, Liao DJ. Gene expression profiles in primary pancreatic tumors and metastatic lesions of Ela-c-myc transgenic mice. Mol Cancer. 2008;7:11. doi: 10.1186/1476-4598-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell PM, Szyf M. Human DNA methyltransferase gene DNMT1 is regulated by the APC pathway. Carcinogenesis. 2003;24:17–24. doi: 10.1093/carcin/24.1.17. [DOI] [PubMed] [Google Scholar]
- 37.Kato K, Iida S, Uetake H, et al. Methylated TMS1 and DAPK genes predict prognosis and response to chemotherapy in gastric cancer. Int J Cancer. 2008;122:603–8. doi: 10.1002/ijc.23143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.