Abstract
The epidermal growth factor receptor tyrosine kinase inhibitors (EGFR TKIs), gefitinib and erlotinib, are reversible competitive inhibitors of the tyrosine kinase domain of EGFR that bind to its adenosine-5′ triphosphate-binding site. Somatic activating mutations of the EGFR gene, increased gene copy number and certain clinical and pathological features have been associated with dramatic tumor responses and favorable clinical outcomes with these agents in patients with non-small-cell lung cancer (NSCLC). The specific types of activating mutations that confer sensitivity to EGFR TKIs are present in the tyrosine kinase (TK) domain of the EGFR gene. Exon 19 deletion mutations and the single-point substitution mutation L858R in exon 21 are the most frequent in NSCLC and are termed ‘classical’ mutations. The NSCLC tumors insensitive to EGFR TKIs include those driven by the KRAS and MET oncogenes. Most patients who initially respond to gefitinib and erlotinib eventually become resistant and experience progressive disease. The point mutation T790M accounts for about one half of these cases of acquired resistance. Various second-generation EGFR TKIs are currently being evaluated and may have the potential to overcome T790M-mediated resistance by virtue of their irreversible inhibition of the receptor TK domain.
Keywords: epidermal growth factor receptor, mutation, non-small-cell lung cancer, tyrosine kinase inhibitor, tyrosine kinase
Introduction
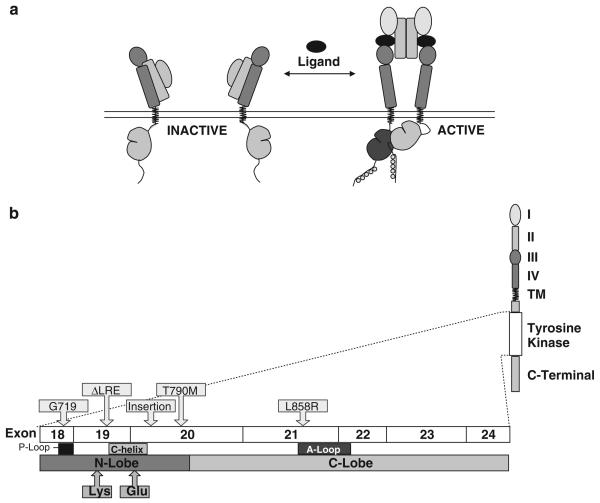
The epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases (TKs), referred to as the HER or ErbB family, consists of four members—EGFR (HER1/ErbB1), HER2 (ErbB2), HER3 (ErbB3) and HER4 (ErbB4)—that regulate many developmental, metabolic and physiological processes. The intracellular TK activity of EGFR is increased as a consequence of the binding of various cognate ligands, which include EGF, transforming growth factor-α, amphiregulin and others, leading to the homodimerization of two EGFRs or the heterodimerization of EGFR with other family members, most commonly HER2 (Bazley and Gullick, 2005). Heterodimerization with HER2, which is over-expressed in some tumors, is a more potent activator of EGFR TK than is EGFR homodimerization. The activation of receptor TK leads to the autophosphorylation of the intracellular domain of EGFR, and the phosphotyrosine residues that are formed act as a docking site for various adapter molecules, resulting in the activation of the Ras/mitogen-activated protein kinase pathway, the PI3K/Akt pathway and signal transducers and activators of transcription signaling pathways (Figure 1a) (Kumar et al., 2008).
Figure 1.
Schematic of EGFR TK activation and EGFR kinase domain mutations. (a) Upon binding of the extracellular ligand, the EGFR receptor dimerizes, leading to the activation of cytoplasmic TK activity. (b) This exon boundary map shows the location of regions within the EGFR TK domain wherein mutations activate the kinase activity by a ligand-independent mechanism. Deletions in exon 19 and the point mutation of L858R are common activating mutations and these ‘classical’ mutations are associated with sensitivity to gefitinib and erlotinib in patients with NSCLC. T790M is a secondary point mutation found in tumors that were previously responsive to these agents, but have developed acquired resistance. Adapted from Kumar et al., 2008.
In tumor cells, the TK activity of EGFR may be dysregulated by various oncogenic mechanisms, including EGFR gene mutation, increased gene copy number and EGFR protein overexpression. (Ciardello and Tortora, 2008) Improper activation of EGFR TK results in increased malignant cell survival, proliferation, invasion and metastasis. EGFR overexpression is observed in tumors from more than 60% of patients with metastatic non-small-cell lung cancer (NSCLC) and is correlated with poor prognosis (Sharma et al., 2007). These findings have provided a rationale for the development of novel anticancer agents that target EGFR.
Treatment with the reversible EGFR TK inhibitors (TKIs), gefitinib and erlotinib, results in dramatic antitumor activity in a subset of patients with NSCLC: clinical responses have been achieved in approximately 10% of European patients and in 30% of patients from East Asia (Fukuoka et al., 2003; Kris et al., 2003; Perez-Soler et al., 2004; Shepherd et al., 2005; Thatcher et al., 2005; Sharma et al., 2007). Sequencing of the EGFR gene revealed that a majority of tumors responding to EGFR TKIs harbored mutations in the TK domain of EGFR (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004). Overall, the frequency of EGFR mutations is 5–20%, depending on the populations studied (Riely et al., 2006). For patients whose tumors exhibit EGFR mutations, the response rate to gefitinib and erlotinib is approximately 75%, suggesting that these mutations, at least in part, drive malignant transformation (Jackman et al., 2006; Riely et al., 2006).
As a result of these findings, a large amount of data on EGFR mutations occurring in patients with NSCLC have recently become available. This article reviews the types of activating and resistance EGFR mutations and the pivotal role they have in the sensitivity and resistance of NSCLC tumors to gefitinib and erlotinib. Advances in understanding EGFR mutations have led to strategies for novel EGFR TKIs that hold promise in the improvement of clinical outcomes for patients with advanced NSCLC.
Activating mutations of the EGFR gene
EGFR mutations are the most prevalent and well characterized in NSCLC, owing their relationship to clinical responses to EGFR TKIs. Because of the high frequency of EGFR mutations in NSCLC, these somatic mutations are thought to represent very early genetic events leading to the development of lung cancer (Politi et al., 2006; Gazdar and Minna, 2008). Furthermore, the susceptibility to EGFR TKIs validates the fundamental dependence of NSCLC tumors on EGFR mutations for maintaining the malignant phenotype. All of the somatic activating EGFR mutations involve the adenosine triphosphate (ATP)-binding pocket in the receptor TK domain, which is the binding site for the TKIs erlotinib and gefitinib. Kinase domain mutations in EGFR are referred to as ‘activating mutations’ because they lead to a ligand-independent activation of TK activity. In some tumors, partially activated mutant EGFRs can be rendered fully ligand independent and, therefore, constitutively active by a second mutation.
The activating mutations of the EGFR gene are found in the first four exons (18 through 21) of the TK domain (Figure 1b) (Shigematsu and Gazdar, 2006; Kumar et al., 2008). These mutations fall into three major classes, with the majority of EGFR TKI-sensitizing mutations falling into class I and II. Class I mutations are in-frame deletions in exon 19; these deletions almost always include amino-acid residues leucine-747 to glutamic acid-749 (ΔLRE), and account for about 44% of all EGFR TK mutations. Class II mutations are single-nucleotide substitutions that cause an amino-acid alteration. The predominant single-point mutation is in exon 21, which substitutes an arginine for a leucine at codon 858 (L858R). L858R has the highest prevalence of any single-point activating mutation in EGFR TK and accounts for about 41% of all EGFR TK activating mutations. Other class II activating mutations result in a glycine-719 (G719) change to serine, alanine or cysteine (4% of all EGFR TK activating mutations), and other missense mutations account for another 6% of EGFR mutations. Class III mutations are in-frame duplications and/or insertions in exon 20. These account for the remaining 5% of EGFR TK activating mutations. A variety of other activating mutations have been detected with low frequency, including V765A and T783A (<1%) in exon 20 (Sharma et al., 2007). Many of the sensitizing mutations have been detected in tumors from drug responders.
Overall, deletions in exon 19 and the point mutation of L858R constitute about 90% of all EGFR activating mutations, and are termed ‘classical’ activating mutations. Although the signaling events that are affected as a result of EGFR mutations are not fully understood, it is well established that the ‘on–off’ equilibrium of EGFR TK states is altered (Kumar et al., 2008). Specifically, an equilibrium shift occurs between active and inactive states of the TK that favors the activated state, resulting in a net increase in kinase activity. As a consequence, tumor cells, in which EGFR activating mutations are present, display an oncogene addiction to EGFR, with consequent selective growth and survival advantages (Gazdar and Minna, 2005; Sharma et al., 2007). Crystallographic analysis suggests that this equilibrium shift is the result of structural alterations induced by activating mutations (Kumar et al., 2008). It has been postulated that these mutations cause a constitutive activation of the kinase by destabilizing the autoinhibited conformation that is normally found in the absence of ligand binding (Zhang et al., 2006; Yun et al., 2008).
A kinetic analysis of the intracellular domains of EGFR L858R and EGFR Del (746–750) has shown that both mutants are active but show a higher KM for ATP and a lower Ki for erlotinib, relative to wild-type receptor (Carey et al., 2006). Thus, mutant kinases demonstrate a reduced affinity for ATP, which provides a molecular explanation for the increased sensitivity to erlotinib and gefitinib (Carey et al., 2006). It is notable that when expressed in a cell line that does not express EGFR or other ErbB receptors, both mutations activate downstream EGFR signaling pathways and promote cell-cycle progression.
Although common EGFR mutations have been well studied in preclinical models (in vitro and in vivo) and their effects on response to TKIs have been observed in patients, relatively little is known about rarer mutations. We now realize that not all mutations are activating, and that some activating mutations may be associated with resistance to TKIs (Kancha et al., 2009). In particular, insertion mutations in exon 20 are associated with a lack of response to TKIs.
Effect of the activating mutations on clinical response
Despite the modest response rate and overall survival benefit observed with EGFR TKIs in patients with advanced NSCLC, significant clinical benefits were achieved in a subset of 10–30% of patients. In 2004, two independent studies were published that probed the molecular basis for the dramatic responses to gefitinib observed in a series of patients with advanced NSCLC (Lynch et al., 2004; Paez et al., 2004). Somatic activating mutations in the EGFR TK domain (exons 18, 19 and 21) were found in tumor specimens from 13 of 14 patients who experienced objective responses to gefitinib. These mutations were absent in tumors from patients with progressive disease. Another study reported activating EGFR mutations in tumors from patients who responded to gefitinib or erlotinib (Pao et al., 2004). EGFR mutations were subsequently examined in several studies of unselected NSCLC tumor specimens. Activating EGFR TK mutations are significantly more common in East Asians, women, never smokers and patients with adenocarcinoma histology (Table 1) (Jänne and Johnson, 2006). Thus, the frequency of the EGFR mutation mirrors the clinically defined subgroups of patients who were most likely to achieve radiographic responses to EGFR TKIs (Miller and Kris, 2004). A germ line transmission of EGFR mutations has also been described within families that show a high incidence of lung cancer (Ikeda et al., 2008).
Table 1.
Frequency of EGFR mutations in different NSCLC patient subgroups
| Total, % | Non-east Asian, % | East Asian, % | |
|---|---|---|---|
| All subgroups | 19 | 10 | 30 |
| Smokers | 11 | 4 | 17 |
| Nonsmokers | 54 | 35 | 60 |
| Adenocarcinoma | 42 | 16 | 49 |
| Non-adenocarcinoma | 3 | 1 | 4 |
| Male | 16 | 1 | 22 |
| Female | 46 | 20 | 58 |
Adapted from Jänne and Johnson, 2006.
The presence of EGFR activating mutations impacts not only on response rate but also progression-free survival and overall survival in patients with NSCLC treated with EGFR TKIs (Table 2) (Bonomi et al., 2007). In four single-arm studies of EGFR TKIs in patients with metastatic NSCLC, a significantly longer overall survival was observed in patients with EGFR mutations (Cortez-Funes et al., 2005; Han et al., 2005; Mitsudomi et al., 2005; Takano et al., 2005). In a study of NSCLC patients treated with gefitinib 250 mg/day, response and time to progression were statistically significantly correlated with EGFR mutations and there was a trend toward longer overall survival in patients harboring these mutations (Cappuzzo et al., 2005). When data from all of these studies are combined, the response rate for patients with EGFR mutations (n=110) is 60% (Bonomi et al., 2007). However, EGFR mutations were not found to be significantly associated with longer survival times in a trial comparing erlotinib with placebo, in which hazard ratios (HRs) for death were similar for patients with classical activating mutations, novel mutations and wild-type EGFR (HR, 0.65, 0.67 and 0.73, respectively) (Shepherd and Tsao, 2006). These investigators proposed that EGFR activating mutations may be a prognostic factor for NSCLC rather than being a predictive factor of EGFR TKI efficacy. This possibility is supported by a subset analysis from a phase III trial of erlotinib plus chemotherapy versus chemotherapy alone, which revealed significantly longer survival times in patients with EGFR mutations compared with those who had wild-type EGFR when treated with chemotherapy alone (Eberhard et al., 2005).
Table 2.
Wild-type EGFR vs EGFR mutations related to response rate, progression-free survival, and overall survival in patients treated with EGFR TKIs
| Investigator | Patients, n | Mutation, % |
Response rates |
PFS |
OS |
|||
|---|---|---|---|---|---|---|---|---|
| WT/mutation, % | P | WT/mutation, mo | P | WT/mutation, mo | P | |||
| Cappuzzo (2005) | 89 | 19 | 5/53 | <0.001 | 2.6/9.9 | 0.02 | 8.4/20.4 | 0.9 |
| Cortez-Funes (2005) | 83 | 12 | 9/60 | 0.001 | 3.6/12.3 | 0.002 | 4.9/13 | 0.002 |
| Han (2005) | 90 | 19 | 14/65 | <0.001 | 1.8/21.7 | <0.001 | 6.6/30.5 | <0.0001 |
| Mitsudomi (2005) | 59 | 56 | 10/84 | <0.0001 | NA | — | — | 0.0496 |
| Takano (2005) | 66 | 59 | 11/82 | 0.005 | 1.7/12.6 | <0.0001 | 6.9/20.4 | 0.0001 |
| Tsao (2005) | 100 | 37 | 7/16 | 0.37 | NA | — | — | 0.45 |
Abbreviations: NA, not available; OS, overall survival; PFS, progression-free survival. Adapted with permission from Bonomi et al., 2007.
In contrast, EGFR mutations did show a predictive value in the INTEREST study, which compared docetaxel with gefitinib in patients with NSCLC that had progressed or recurred after chemotherapy. Patients with mutations had a significantly longer progression-free survival (PFS) with gefitinib than with docetaxel (7.0 vs 4.1 months; HR, 0.16; P=0.001), whereas PFS among patients with wild-type EGFR trended in favor of docetaxel (1.7 vs 2.6 months; HR 1.24; P=0.135) (Douillard et al., 2008). A similar association was found in recently reported results from the I-PASS trial, which compared first-line gefitinib with carboplatin/paclitaxel in Asian patients with advanced NSCLC and with no history of substantial smoking. In this study, patients harboring EGFR mutations had a significantly longer PFS with gefitinib (HR, 0.48; P<0.001), whereas those with wild-type EGFR had a better PFS with chemotherapy (HR, 2.85; P<0.001) (Mok et al., 2008).
A recent prospective study in first-line gefitinib-treated patients with NSCLC reported that EGFR activating mutations were the most important independent predictors for time to treatment failure compared with other mutations, among which exon 19 deletion and L858R mutations were the best predictors for longer time to treatment failure in a multivariate analysis (Yang et al., 2008). However, additional prospective studies are needed to clarify the prognostic and predictive implications of EGFR activating mutations.
Interestingly, despite the impact of the EGFR mutation on outcomes in advanced NSCLC, the mutational status may not have a dramatic effect on the outcome of patients with early-stage NSCLC. In a study in 277 Japanese patients with early-stage lung cancer who had undergone surgical resection, a Kaplan–Meier analysis that excluded patients treated with gefitinib, as well as patients undergoing surgery for recurrent or second primary cancers, indicated that EGFR mutations did not affect the survival of these patients (P = 0.9933). However, it is noteworthy that the median follow-up period was short (788 days) (Kosaka et al., 2004).
Not all activating mutations necessarily lead to a full or constitutive EGFR TK activity. Therefore, the type of EGFR mutations in NSCLC tumors seems to influence the sensitivity of the tumor to gefitinib and erlotinib. For example, NSCLC cells expressing the L858R mutant are significantly more sensitive to gefitinib than are those that express the G719S mutant (Jiang et al., 2005). Response rates to EGFR TKIs are higher in patients with NSCLC, whose tumors have exon 19 mutations (70–100%), than in patients with exon 21 mutations (20–67%) (Mitsudomi et al., 2005; Hirsch et al., 2006; Jackman et al., 2006; Riely et al., 2006). These differential response rates translated into longer survival, whereby patients with an exon 19 (ΔLRE) deletion mutation had a median overall survival ranging from 26 to 34 months and patients with exon 21 (L858R) had a median overall survival ranging from 8 to 17 months (Hirsch et al., 2006; Jackman et al., 2006; Paz-Ares et al., 2006).
EGFR mutations and resistance to EGFR TKIs
Although EGFR kinase mutations are associated with an enhanced sensitivity to gefitinib and erlotinib, not all tumors that have activating mutations are associated with an enhanced response. Tumors that fail to respond to EGFR TKIs despite the presence of an activating mutation might have an additional genetic lesion that relieves the tumor of its dependence on the EGFR signaling pathway. One mechanism that has been linked to insensitivity of NSCLC to EGFR TKIs is the occurrence of insertion point mutations in exon 20 of the EGFR gene. These include the exon 20 insertion mutants D770_N771 (ins NPG), D770_(ins SVQ) and D770_(ins G) N771T (Greulich et al., 2005; Sharma et al., 2007). In an in vitro model system, insertion mutations in exon 20 render transformed cells less responsive to EGFR TKIs compared with the sensitizing mutations of exons 19 and 21 (Greulich et al., 2005). However, exon 20 mutations are relatively rare, suggesting that other mechanisms probably contribute to EGFR TKI primary resistance in metastatic NSCLC. For many of the rare point mutations, the effect on responsiveness to EGFR TKIs remains unknown.
Acquired resistance occurs in virtually all NSCLC tumors that initially respond to EGFR TKI therapy. It is now recognized that the efficacy of gefitinib and erlotinib is of limited duration owing, in large part, to the emergence of drug resistance conferred by a second point mutation in the TK domain. The threonine-790 to methionine (T790M) point mutation is found in approximately 50% of all patients at the time of acquired resistance to EGFR TKI therapy (Kobayashi et al., 2005; Balak et al., 2006; Kosaka et al., 2006). This so-called gatekeeper mutation is believed to be acquired through selective pressure during treatment, as it is rarely detected in tumors from untreated patients (Pao et al., 2005a). Interestingly, using a highly sensitive allele-specific assay, Maheswaran et al. (2008) recently detected low levels of T790M in pretreatment NSCLC tumor samples from 10 of 26 patients. Although significant responses were achieved with EGFR TKIs in these patients, the presence of T790M before treatment was associated with a significantly shorter progression-free survival compared with that in TKI-naive patients o without detectable T790M (7.7 vs 16.5 months; P < 0.001). These results suggest that T790M may be a useful pretreatment biomarker for identifying patients who are unlikely to achieve durable responses with reversible EGFR TKIs (that is, erlotinib and gefitinib).
Preclinical studies support clinical findings implicating T790M as an underlying mechanism of resistance. It has been demonstrated in vitro that this mutation can substantially suppress the inhibitory effects of erlotinib and gefitinib, whereas TK activity is maintained (Kobayashi et al., 2005; Pao et al., 2005a). Similarly, the introduction of T790M into gefitinib-sensitive tumor cells that show activating EGFR mutations or an increased EGFR copy number also confers resistance to gefitinib treatment (Greulich et al., 2005). It is not clear how T790M imparts resistance to reversible EGFR TKIs. The T790M mutation results in an alteration of the topology of the ATP-binding pocket (Kumar et al., 2008). It has been suggested that this change in topology precludes the binding of reversible EGFR TKIs through steric hindrance, thereby resulting in resistance (Kobayashi et al., 2005; Kwak et al., 2005; Pao et al., 2005b). However, another mechanism was proposed in a recent study showing that T790M increases the affinity of the kinase domain for ATP (Yun et al., 2008). The authors suggested that this increased affinity results in reduced potency of any ATP-competitive agent.
Other resistance point mutations, such as aspartic acid-761 to tyrosine (D761Y), have been reported, some of which may weaken the interaction of EGFR TKI with its target (Balak et al., 2006). Clinically, the challenge remains how to best detect tumors with the T790M and other resistance point mutations on limited quantities of post-treatment tumor samples. A molecular analysis of circulating cells may provide an alternative approach for monitoring tumor mutations (Maheswaran et al., 2008). In a recent report, it was shown that the actual number of activating EGFR mutant molecules could be detected in the plasma of patients with NSCLC using a procedure called micro-fluidics digital polymerase chain reaction, which is capable of detecting single input template molecules (Yung et al., 2009). In addition, it was found that the concentration of mutant sequences from sequential measurements correlated with response to therapy (that is, decreased concentration correlated with clinical response, whereas persistence of the mutant sequence correlated with progression). These results indicate that an examination of the plasma may be a suitable surrogate test when tumor tissue is not available for determining therapy selection.
Strategies for optimizing response to EGFR TKIs
Insights gained from the treatment of patients with metastatic NSCLC with gefitinib and erlotinib are dramatically changing drug development and treatment strategies, as well as clinical outcomes. Because acquisition of the secondary resistance point mutation T790M reduces the efficacy of ATP-competitive inhibitors, one strategy for preventing or overcoming EGFR TKI resistance would be to identify novel agents that bind and inhibit EGFR by a distinct, non-ATP competitive mechanism. A second strategy may be to irreversibly inhibit the binding of ATP to the TK domain with an irreversible rather than a reversible inhibitor. As a class, the irreversible EGFR inhibitors, including BIBW 2992, HKI-272 and PF00299804, are able to inhibit EGFR phosphorylation and inhibit growth in gefitinib-resistant NSCLC or Ba/F3 cell lines that contain the EGFR T790M mutation (Kwak et al., 2005; Wong, 2007; Li et al., 2008; Engelman et al., 2008). For example, the irreversible EGFR/HER2 inhibitor, BIBW 2992, suppresses wild-type and activated EGFR and HER2 mutants, including EGFR and HER2 inhibitor-resistant isoforms (Li et al., 2008). BIBW 2992 has a higher affinity for binding to EGFR with the T790M resistance mutation than do first-generation EGFR TKIs (Table 3) (Li et al., 2008), and induces dramatic tumor regression in an L858R/T790M EGFR-driven lung cancer model. Late-stage clinical trials are evaluating BIBW 2992 and HKI-272 for the treatment of patients with NSCLC who relapse after a successful previous treatment with gefitinib or erlotinib.
Table 3.
In vitro inhibitory activities of BIBW 2992, lapatinib, canertinib and gefitinib on the receptor TK activities of wild-type and mutated EGFR
| IC50 (nm) |
||||
|---|---|---|---|---|
| BIBW 2992 | Lapatinib | Canertinib | Gefitinib | |
| EGFRWT | 0.5 | 3 | 0.3 | 3.0 |
| EGFRL858R | 0.4 | 8 | 0.4 | 0.8 |
| EGFRL858R/T790M | 10.0 | >4000 | 26.0 | 1013.0 |
Abbreviations: EGFRWT, wild-type epidermal growth factor receptor; EGFRL858R, epidermal growth factor receptor harboring the L858R resistance mutation; EGFRL858R/T790M, epidermal growth factor receptor harboring the L858R-activating mutation and the T790 resistance mutation. Adapted from Li et al., 2008.
Another approach for overcoming resistance to reversible EGFR TKIs involves targeting parallel- or convergent signaling pathways. The mammalian target of the rapamycin (mTOR) signaling pathway integrates nutrient and mitogen signals to regulate cell proliferation, survival and angiogenic pathways, and has been implicated in resistance to EGFR inhibitors. In both sensitive and resistant tumor cell lines, the mTOR inhibitor, everolimus, reduces the expression of EGFR signaling effectors and cooperates with gefitinib to overcome resistance (Bianco et al., 2008). In patients with resistance to first-generation EGFR TKIs generated by MET amplification, it is unlikely that an irreversible EGFR inhibitor alone would be effective, but the combination of an irreversible EGFR inhibitor and an mTOR inhibitor may be an effective strategy for overcoming resistance (Li et al., 2008). These hypotheses suggest novel therapeutic strategies that are yet to be validated in clinical studies.
Conclusion
Various clinical characteristics and molecular factors have been associated with sensitivity and resistance to EGFR TKIs. The discovery and characterization of EGFR activating mutations and their relationship to sensitivity to gefitinib and erlotinib have provided a basis for transforming NSCLC from a disease treated with conventional combination chemotherapy to one in which subsets of patients with specific EGFR mutations can be effectively treated with targeted therapy. It is reasonable to suggest that personalized therapy for NSCLC patients should include a genetic assessment of the EGFR mutational status for individual patients. Current research is directed at optimizing the accuracy and sensitivity of EGFR mutational testing so that it might be introduced into routine clinical practice. The appropriate role of an EGFR mutation analysis in the treatment of patients with NSCLC continues to evolve, awaiting prospective clinical studies with an adequate documentation of the EGFR mutational status.
Several novel targeted therapies are currently in clinical development for patients with advanced NSCLC. The irreversible EGFR TKIs are one class of agents that may have the potential to prevent and overcome resistance that emerges during treatment with gefitinib and erlotinib. Results of ongoing phase III studies on this class of compounds in erlotinib-resistant NSCLC populations are eagerly awaited.
Acknowledgements
I thank Johnathan C Maher, PhD of BlueSpark Healthcare Communications for medical and editorial assistance with this article. Financial support for medical and editorial assistance was provided by Boehringer Ingelheim Pharmaceuticals Inc. Supported by grant P50CA70907 from the Specialized Programme for Research Excellence in Lung Cancer from the National Cancer Institute, Bethesda, MD, USA.
Footnotes
Conflict of interest
AF Gazdar has received consulting fees/paid advisory board fees from AstraZeneca, Boehringer and Genentech. AF Gazdar has also received lecture fees from AstraZeneca.
References
- Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor–mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- Bazley LA, Gullick WJ. The epidermal growth factor receptor family. Endocr Relat Cancer. 2005;12(Suppl 1):S17–S27. doi: 10.1677/erc.1.01032. [DOI] [PubMed] [Google Scholar]
- Bianco R, Gaofalo S, Rosa R, Damiano V, Gelardi T, Daniele G, et al. Inhibition of mTOR pathway by everolimus cooperates with EGFR inhibitors in human tumours sensitive and resistant to anti-EGFR drugs. Br J Cancer. 2008;98:923–930. doi: 10.1038/sj.bjc.6604269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomi PD, Buckingham L, Coon J. Selecting patients for treatment with epidermal growth factor tyrosine kinase inhibitors. Clin Cancer Res. 2007;13(15 Suppl):4606s–4612s. doi: 10.1158/1078-0432.CCR-07-0332. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- Carey KJ, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- Ciardello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- Cortez-Funes H, Gomez C, Rosell R, Valero P, Garcia-Giron C, Velasco A, et al. Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small-cell lung cancer patients. Ann Oncol. 2005;16:1081–1086. doi: 10.1093/annonc/mdi221. [DOI] [PubMed] [Google Scholar]
- Douillard JY, Hirsch V, Mok TS, Socinski MA, Watkins C, Lowe E, et al. Molecular and clinical subgroup analyses from a phase III trial comparing gefitinib with docetaxel in previously treated non-small cell lung cancer (INTEREST) (abstract) J Clin Oncol. 2008;26(Suppl) abstract no. 8001. [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, et al. PF00299804, an irreversible pan ErbB inhibitor, is effective in lung cancer models with EGFR and HER2 mutations that are resistant to gefitinib. Cancer Res. 2008;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Duillard JY, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small cell lung cancer. J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Minna JD. Inhibition of EGFR signaling: all mutations are not created equal. PLoS Med. 2005;2:e377. doi: 10.1371/journal.pmed.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Minna JD. Deregulated EGFR signaling during lung cancer progression: mutations, amplicons, and autocrine loops. Cancer Prev Res. 2008;1:156–160. doi: 10.1158/1940-6207.CAPR-08-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich H, Chen TH, Feng W, Jänne PA, Alvarez JV, Zappaterra M, et al. Oncogenic transformation by inhibitor-sensitive and-resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SE, Kim TY, Hwang PG, Jeong S, Kim J, Choi IS, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Franklin WA, McCoy J. Predicting clinical benefit from EGFR TKIs: not all EGFR mutations are equal (abstract) J Clin Oncol. 2006;24:382s. [Google Scholar]
- Ikeda K, Nomori H, Takeshi M, Sasaki K, Kobayashi T. Novel germline mutation: EGFR V843I in patient with multiple lung adenocarcinomas and family members with lung cancer. Ann Thorac Surg. 2008;85:1430–1432. doi: 10.1016/j.athoracsur.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- Jänne PA, Johnson BE. Effect of epidermal growth factor receptor tyrosine kinase domain mutations on the outcome of patients with non-small cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors. Clin Cancer Res. 2006;12(14 Suppl):4416s–4420s. doi: 10.1158/1078-0432.CCR-06-0555. [DOI] [PubMed] [Google Scholar]
- Jiang J, Greulich H, Jänne PA, Sellers WR, Meyerson M, Griffin JD. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- Kancha RK, von Bubnoff N, Peschel C, Duyster J. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin Cancer Res. 2009;15:460–467. doi: 10.1158/1078-0432.CCR-08-1757. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- Kris MG, Natale RB, Herbst RS, Lynch TJ, Jr, Prager D, Belani CP, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- Kumar A, Petri ET, Halmos B, Boggon TJ. Structure and clinical relevance of the epidermal growth factor receptor in human cancer. J Clin Oncol. 2008;26:1742–1751. doi: 10.1200/JCO.2007.12.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, et al. Irreversible inhibitors of the EGFR receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L. Detection of mutations in EGFR in circulating lung cancer cells. N Engl J Med. 2008;359:1–12. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VA, Kris MG. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- Mok T, Wu Y-L, Thongprasert S, Yang C-H, Chu D, Saijo N, et al. Phase III, randomised, open-label, first-line study of gefitinib (G) vs carboplatin/paclitaxel (C/P) in clinically selected patients (pts) with advanced non-small-cell lung cancer (NSCLC) (IPASS) (abstract) Ann Oncol. 2008;19(Suppl 8):LBA2. [Google Scholar]
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib and erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005a;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005b;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares L, Sanchez JM, Garcia-Velasco B. A prospective phase II trial of erlotinib in advanced non-small cell lung cancer (NSCLC) patients (p) with mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR) (abstract) J Clin Oncol. 2006;24(Suppl):369s. [Google Scholar]
- Perez-Soler R, Chachoua A, Hammond LA, Rowinski EK, Huberman M, Karp D, et al. Determinants of tumor response and survival and erlotinib in patients with non-small cell lung cancer. J Clin Oncol. 2004;22:3238–3247. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- Politi K, Zakowski MF, Fan PD, Schoenfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancer respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Shepherd F, Tsao MS. Unraveling the mystery of prognostic and predictive factors in epidermal growth factor receptor therapy. J Clin Oncol. 2006;24:1219–1220. doi: 10.1200/JCO.2005.04.4420. [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Pereria J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in previously treated non-small cell lung cancer. N Eng J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor-signaling pathway in lung cancers. Int J Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- Takano T, Ohe Y, Sakamotto H, Tsuta K, Matsuno Y, Tateishi U, et al. Epidermal growth factor receptor gene mutations and increased copy number predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- Thatcher N, Chang A, Parikh P, Pereira JR, Ciuleanu T, von Pawel J, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomized, placebo-controlled, multi-centre study (Iressa survival evaluation in lung cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- Wong KK. HKI-272 in non-small cell lung cancer. Clin Cancer Res. 2007;13(15 Part 2):S4593–S4596. doi: 10.1158/1078-0432.CCR-07-0369. [DOI] [PubMed] [Google Scholar]
- Yang C-H, Yu C-J, Shih J-Y, Cheng Y-C. Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib therapy. J Clin Oncol. 2008;26:2745–2753. doi: 10.1200/JCO.2007.15.6695. [DOI] [PubMed] [Google Scholar]
- Yun C-H, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790 mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]