Abstract
Nuclear receptors E75, which regulates development in D. melanogaster, and Rev-erbβ, which regulates circadian rhythm in humans, bind heme within their ligand binding domains (LBD). The heme-bound ligand binding domains of E75 and Rev-erbβ were studied using electronic absorption, MCD, resonance Raman and EPR spectroscopies. Both proteins undergo redox-dependent ligand switching and CO- and NO-induced ligand displacement. In the Fe(III) oxidation state, the nuclear receptor hemes are low-spin and 6-coordinate with cysteine(thiolate) as one of the two axial heme ligands. The sixth ligand is a neutral donor, presumably histidine. When the heme is reduced to the Fe(II) oxidation state, the cysteine(thiolate) is replaced by a different neutral donor ligand, whose identity is not known. CO binds to the Fe(II) heme in both E75(LBD) and Rev-erbβ(LBD) opposite a sixth neutral ligand, plausibly the same histidine that served as the sixth ligand in the Fe(III) state. NO binds to the heme of both proteins; however, the NO-heme is 5-coordinate in E75 and 6-coordinate in Rev-erbβ. These nuclear receptors exhibit coordination characteristics that are similar to other known redox and gas sensors, suggesting that E75 and Rev-erbβ may function in heme-, redox- or gas-regulated control of cellular function.
Nuclear receptors, the largest superfamily of transcription factors, bind to specific DNA motifs in response to small molecule signaling. Errors in nuclear receptor function result in many developmental, reproductive, inflammatory and metabolic diseases (1, 2). Transcriptional regulation by nuclear receptors plays an essential role in controlling homeostasis via the circadian system, which is critical for the correct timing of physiological processes in response to external stimuli (3). The nuclear receptor superfamily is characterized by a well-conserved domain architecture: a N-terminal ‘AB’ region of variable length (activation function 1, AF-1), a ‘C’ region that encodes a highly conserved DNA binding domain (DBD) containing two zinc fingers, a flexible linker region ‘D’, a C-terminal ‘E’ region that encodes the ligand binding domain (LBD), and helix 12, which is important in transcriptional function (activation function 2, AF-2) (4). The LBD recognizes and binds ligands such as steroids, thyroid hormones, fatty acids, phospholipids, retinoids, bile acids, farnesoids, and a range of xenobiotics. In doing so, the LBD functions in regulating dimerization and association with co-regulator proteins. The structure of the LBD itself is conserved, with a fold of 11–12 helices and a β-hairpin turn; residues within the ligand binding pocket (LBP) determine the ligand specificity for the nuclear receptor (4, 5). For many nuclear receptors a ligand has not been identified, and these are referred to as orphan nuclear receptors.
In 2005, Reinking, et al. (6) published the unprecedented discoveries that heme (iron protoporphyrin IX) was the ligand for the orphan nuclear receptor E75 and that heme was required for proper folding and functioning of the E75 protein. The Drosophila nuclear receptor E75 (Eip75B, NR1D3), encoded by a target gene of the Ecdysone receptor/Ultraspiracle heterodimer (EcR/USP), was classified as an orphan nuclear receptor in the Drosophila genome (7). E75 is proposed to be part of a feedback loop that regulates a subset of the developmental processes initiated by ecdysone. The E75/DHR3 heterodimer represses expression of target genes and also functions in autoregulation, controlling ecdysone synthesis by acting upstream of the ecdysone-mediated nuclear receptor cascade (8). E75 is implicated in development of eyes and limbs as well as in coordination of juvenile hormone and ecdysone signals during larval molts and metamorphosis (9, 10). Reinking, et al. (6) demonstrated that interaction of E75 with its heterodimer partner DHR3 is modulated by the redox state of the heme as well as by the presence of CO or NO. Reduction of the heme is required for E75 to bind to DHR3 and repress transcription. Furthermore, binding of CO or NO blocked the ability of E75 to bind to a peptide derived from the AF-2 region of DHR3, suggesting that NO and CO may inhibit E75/DHR3 heterodimerization. Taken together, these findings suggest that E75 may function as an intracellular heme, redox and gas sensor.
In vertebrates, the closest homologs to Drosophila E75 are members of the NR1D/Rev-Erb family. These orphan nuclear receptors include Rev-erbα (Rev-ErbA-α, THRAL, EAR-1; NR1D1) and Rev-erbβ (Rev-ErbB-β, RVR, EAR-1R, BD-73; NR1D2), whose expression is concentrated in peripheral tissues with heavy energy demands, including skeletal muscle, brown and white adipose tissue, brain, liver and kidney (11). The Rev-erbs function as transcriptional silencers and negative regulators of RORs (retinoid-related orphan receptors), the human homologs of DHR3 (11, 12). Additionally, RORα has been found to activate Rev-erbα transcription, while Rev-erbα represses its own transcription (13, 14). Both the Rev-erbs and RORs bind as either monomers to the half-site motif AGTTCA with AAXT (X is any nucleotide) as the 5’ flanking sequence, or as homodimers to a direct repeat of this motif separated by two base pairs; furthermore, Rev-erbs recruit co-repressors (NcoR-HDAC3, SMRT, C1d, or NCOA5) (15, 16). The members of the Rev-erb (α, β) and ROR (α, β, γ) families are critical elements of the molecular circadian clock; the transcription of the clock protein BMAL1 is modulated through competition between Rev-erb repressors and ROR activators (17, 18). Rev-erbs have been linked to circadian timing in brain and liver tissue, and, upon recruitment of the nuclear receptor co-repressor histone deacetylase (NcoR-HDAC3), regulate BMAL1, and, to a lesser extent, CLOCK expression. The feedback loop observed in Drosophila for E75 is also in evidence for Rev-Erbs, which are regulated by the circadian oscillator; Rev-erb expression is activated by the BMAL1-CLOCK heterodimer, and repressed by Period (Per) and Cryptochrome (Cry) proteins. The less well-studied Rev-erbβ possesses high sequence homology, extensive overlap of tissue expression and common repression targets with Rev-erbα, suggesting that Rev-erbβ functions similarly to Rev-erbα.
Homology modeling of the putative LBD of Rev-erbα of the NR1D subgroup suggested that bulky side chains occupy the ligand-binding pocket and that the small residual cavity could not accommodate a classical ligand (19). Members of the NR1D subgroup (Rev-erbα, Rev-erbβ and E75) are the only known nuclear receptors missing helix 12 (AF-2), which is believed to be essential for transcriptional regulation upon ligand binding. Modeling revealed a hydrophobic surface, consisting of residues from helix 3, loop 3–4, helix 4, and helix 11, that is exposed by the absence of helix 12 and mediates co-repressor recruitment (16, 19). Rev-erbs function as constitutive repressors via binding of co-repressor proteins (NcoR-HDAC3, SMRT, C1d, or NCOA5) in the apparent absence of ligand. Since these proteins lack the typical regulatory features associated with ligand binding and are constitutively active as repressors, it was proposed that these orphan receptors do not bind ligands (16, 20). A recently published crystal structure of the dimeric LBD of Rev-erbβ (LBDβ, residues 386–579) revealed that residues within the ligand-binding pocket, which are replaced by smaller side chains in the heme-binding E75, may preclude the binding of heme or another ligand (21). However, the authors noted disorder in regions of the LBD; the same type of disorder is documented for other nuclear receptors that are known to accommodate ligands of varying size (21, 22). These disordered regions may contribute to a ‘flexible floor’ in the ligand-binding pocket, which may be essential to the ligand binding properties of the receptors.
Spectroscopic data is limited for the heme within these nuclear receptors, and a goal of this work is to obtain more extensive data for comparison with other heme sensor proteins. Previous spectroscopic work on the Drosophila E75 nuclear receptor reported electronic absorption and EPR studies on a LBD construct in the Fe(III) oxidation state, in wild-type and several mutant forms (6, 23). These data implicated Cys396, Cys468, His364 and His574 as potential donors to the heme iron. Electronic absorption data were also reported for the reduced and CO-and NO-bound species of the E75 LBD (6, 24). As this manuscript was in preparation, two separate reports appeared confirming that, as established for Drosophila E75, both Rev-Erbα and Rev-erbβ bind a heme prosthetic group in their LBDs (25, 26). Unlike E75, the Rev-erb nuclear receptors exhibit lowered affinity and no absolute requirement for heme for proper folding and function; however, heme appears to increase thermal stability and assist in recruitment of co-repressors. Additionally, the human receptors are reported to be insensitive to the redox state or the presence of exogenous gaseous ligands CO and NO. Herein, we present spectroscopic comparisons of LBD constructs E75(LBD) and Rev-erbβ(LBD); our data reveal that both Fe(III) species are 6-coordinate cysteine(thiolate)-bound heme proteins that lose cysteine as a heme-iron donor when reduced, and which bind CO and NO to form distinctive adducts. The implications for these observations are discussed in light of E75 and Rev-erb function and recently published reports on the Rev-erb proteins.
MATERIALS AND METHODS
Materials
Materials used in buffer preparation and glycerol were purchased from Sigma-Aldrich and used as received. Sodium dithionite was purchased from Fluka and stored under Ar(g) at −20° C until used. CO gas (99.5%) and NO gas (99%) cylinders were obtained from AGA, and 13CO gas (99%) was obtained from Sigma-Aldrich. The NO gas was passed over an anaerobic KOH column to remove higher order nitrogen oxides before use. Na15NO2 (98%) was purchased from Fischer Scientific, and NaNO2 was purchased from Sigma-Aldrich; both were used as received.
Expression and purification of E75, Rev-erbβ and Rev-erbα LBD constructs
The LBD construct for E75 (G341-S602) was subcloned into the vector pET15B (Invitrogen) as previously reported (6). The LBD constructs for Rev-erbα (T274-Q614 with an internal deletion of residues P324-M422) and Rev-erbβ (S241-P579 with an internal deletion of residues S275-R357) were subcloned into a modified pET28a vector (Novagen). All three constructs were expressed in E. coli culture supplemented with hemin (50 mg/L, Sigma). Cells were induced with IPTG at an OD600 of ~1.0 and then grown overnight at 15 °C. Expressed protein was first purified from cell lysate using Ni-NTA affinity chromatography, samples were then further purified using anion exchange and/or size exclusion chromatography as described in Reinking et al. (6). Fractions containing the desired protein constructs were identified by the 425 nm/280nm absorbance ratio and by SDS-PAGE. After isolation, E75 was dialyzed into buffer containing 10 mM Tris-HCl and 0.1 M NaCl, pH 8; Rev-erbα and Rev-erbβ were likewise stored in 10 mM Tris-HCl and 0.5 M NaCl, pH 8.2. The isolated proteins were flash frozen in liquid nitrogen. As previously reported for the E75 LBD (6), all isolated proteins appeared to contain one heme per polypeptide chain. The Soret absorption maximum to 280 nm absorption ratios were: 2.75 (A423/A280) for E75(LBD), 2.45 (A420/A280) for Rev-erbβ(LBD), and 2.51 (A413/A280) for Rev-erbα(LBD).
Electronic Absorption Spectroscopy
Electronic absorption spectra were recorded on a double-beam Varian Cary 4 Bio spectrophotometer equipped with a temperature controller and set to a spectral bandwidth of 0.5 nm. Spectra were obtained at 25 °C for samples of protein in 25 mM EPPS with 100 or 500 mM NaCl, pH 8.0, as indicated. Samples were purged of oxygen by flowing Ar(g) through the headspace of a septum sealed cuvette for ~10 minutes. Reduction of Fe(III) protein samples using sodium dithionite was performed under an atmosphere of Ar(g) using two methods: 1) the addition of several crystals of the solid reductant under Ar(g), or 2) the addition of an anaerobically prepared stock solution of sodium dithionite to a final concentration of 1–2 mM. The specific method employed for a given sample is noted in the figure legend. CO adducts were prepared by the injection of CO(g) into the headspace of a septum-sealed cuvette via gastight syringe followed by gentle agitation of the sample. The CO(g) was introduced into the Fe(III) protein before reduction and into the Fe(II) protein (pre-reduced) to observe any order of addition effects. NO adducts were prepared by the injection of NO(g) into the headspace of a septum-sealed cuvette via gastight syringe followed by gentle agitation of the sample. In general, 50–300 µL of CO or NO gas was injected as appropriate to give complete conversion to the respective adduct. The absorption spectra at room temperature were monitored until no further change was observed.
MCD Spectroscopy
Magnetic circular dichroism (MCD) spectra were recorded on a Jasco J-715 CD spectropolarimeter with the sample compartment modified to accommodate an SM-4000-8T magnetocryostat (Oxford Instruments). The buffer used for MCD samples was 50 mM EPPS, 500 mM NaCl, pH 8.0, with approximately 55% v/v glycerol present in the final sample. Addition of glycerol had no effect on the absorption spectral features of the samples at room temperature, nor was any spectral change observed at liquid helium temperatures. Sodium dithionite was introduced in excess according to the methods detailed for optical spectra. CO adducts were prepared as described above. Each sample was transferred to an Ar(g)-filled cell via gastight syringe and frozen in liquid nitrogen. MCD spectra were taken at temperatures ranging from 4.5 to 100 K. The MCD signal of each sample was recorded at positive and negative magnet polarity (7 tesla) for each temperature; negative polarity data were subtracted from the positive polarity data, and the resulting trace was divided by 2 to remove contribution of the CD.
EPR Spectroscopy
X-band electron paramagnetic resonance (EPR) spectra were collected on a Bruker ESP 300E equipped with an Oxford ESR 900 continuous flow liquid helium cryostat and an Oxford ITC4 temperature controller maintained at 10 K. The microwave frequency was monitored using an EIP model 625A CW microwave frequency counter. Spectra of Fe(III)E75(LBD) and Fe(III)- and Fe(II)NO Rev-erbβ(LBD) (~150 µL, 130–180 µM in heme) were recorded in 50 mM borate buffer with an additional 500 mM NaCl at pH 8.0. Reduced protein samples were prepared under Ar(g) by the addition of sodium dithionite as a solid into samples of the Fe(III) protein. Fe(II)NO samples were prepared using Na14NO2 or Na15NO2 solid reagents; NaNO2 was converted to NO gas in the aqueous protein sample by the addition of a small excess of solid sodium dithionite. Each sample was transferred to an Ar(g)-filled quartz EPR tube via gastight syringe or Ar(g)-purged small-bore tubing and frozen in liquid nitrogen. For all samples, scans of 0–10,000 G revealed no signals other than those reported. EPR data were simulated using the WEPR program written by Dr. Frank Neese (27). Parameter sets employed for the EPR simulations are given in the Supporting Information.
Resonance Raman Spectroscopy
Resonance Raman spectra were obtained with an excitation wavelength of 413.1 and 406.7 nm from a Coherent I-302C Kr+ laser in a backscattering 135° sample geometry. An Acton Research triple monochromator was used with gratings of 2400 grooves/mm. Low incident laser powers of ~50 mW, or ~7–10 mW for CO and NO adducts to minimize ligand photodissociation, were focused with a cylindrical lens onto the sample. A Princeton Instruments Spex 1877 triple spectrograph outfitted with a cooled, intensified diode array detector was operated under computer control using Spectrasense software. The frozen protein samples, prepared as described for EPR (vide infra), were placed in a quartz dewar and cooled to 77 K with liquid nitrogen to reduce local heating. Peak positions were calibrated relative to the ice peak at 228 cm−1. Windows centered at 650, 1250 and 1850 cm−1 were overlaid for a total frequency range of ~150–2250 cm−1. All spectral data were imported into and processed with IGOR Pro software (Wavemetrics, Inc.). Curve fitting utilized Gaussian peak shapes. Assignments of key vibrational modes are noted, with assignments based on comparison with those of other heme proteins and the work of Kitagawa and Spiro (28–31).
RESULTS
Characterization of the Fe(III) states of Rev-erbβ(LBD) and E75(LBD)
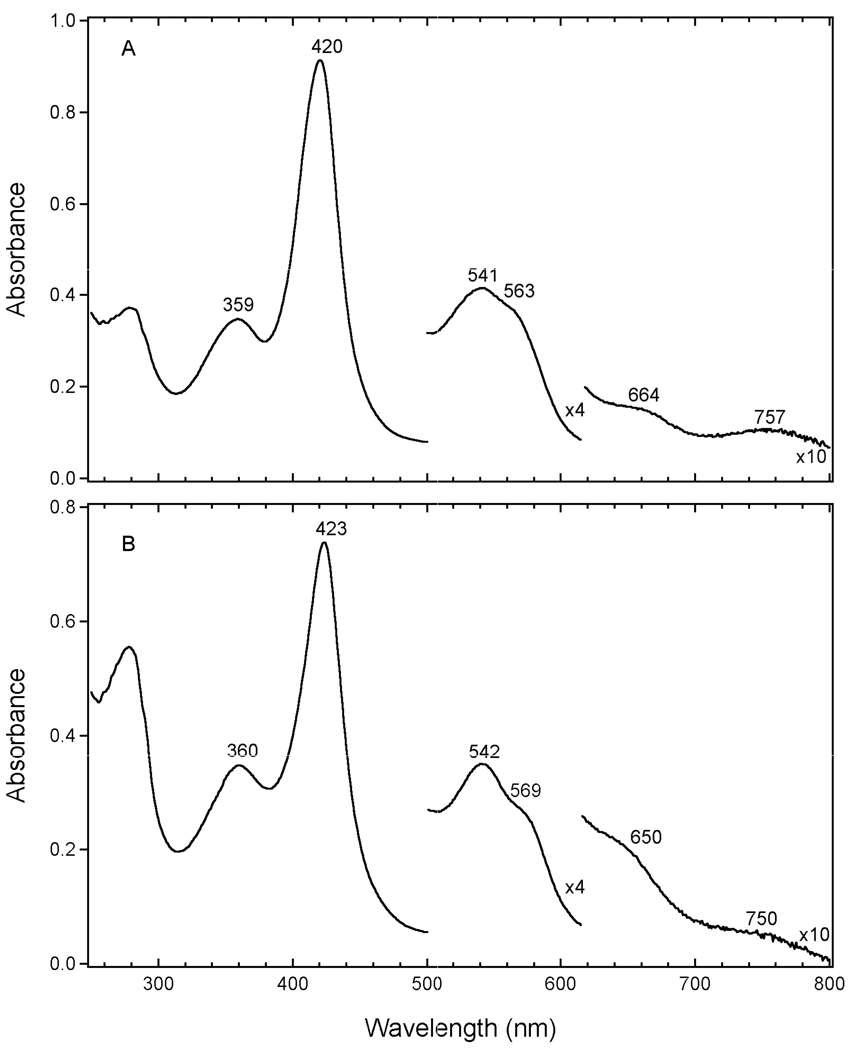
Electronic absorption spectra of the nuclear receptor LBD constructs, Fe(III)Rev-erbβ(LBD) and Fe(III)E75(LBD), reveal that these proteins each contain a low-spin, 6-coordinate Fe(III) heme in which cysteine(thiolate) is present as an axial ligand. Principal characteristics of the spectrum of Fe(III)Rev-erbβ(LBD) include a δ band at 359 nm, a Soret (γ) band at 420 nm and broad, asymmetric α–β features at 563 and 541 nm, respectively (Figure 1A). The absorption spectrum of Fe(III)E75(LBD) exhibits spectral features similar in shape and position to those of Rev-erbβ: a δ band at 360 nm, a Soret band at 423 nm, and α–β features at 569 and 542 nm, respectively (Figure 1B). In both Fe(III)Rev-erbβ(LBD) and Fe(III)E75(LBD), a pair of weak visible bands are observed at ~664 nm and ~757 nm, and ~650 nm and ~750 nm, respectively. These broad low-energy ligand-to-metal charge transfer (LMCT) bands are a distinctive feature of 6-coordinate cysteine(thiolate) coordinated hemes (32). The intense, well-defined δ (‘n’) bands present in Fe(III)Rev-erbβ(LBD) and Fe(III)E75(LBD) are further evidence for thiolate coordination. This δ band, which is typically observed between 355–365 nm, is a feature associated with mixing between a cysteine(thiolate)-associated LMCT (S(p) → Fe(dπ)) transition and the porphyrin Soret (π → π*) transition. The spectral features and peak positions for both Fe(III) Rev-erbβ(LBD) and E75(LBD) are similar to those of other 6-coordinate heme-thiolate proteins (Table 1). Absorption band positions previously reported for Fe(III)E75(LBD) and for an alternate Fe(III)E75 LBD construct vary slightly from those reported herein: the position of the α peak is blue-shifted (~6 nm) relative to that previously reported, and the asymmetry observed in the α–β features is more pronounced (6, 23).
Figure 1.
Electronic absorption spectra of purified Fe(III)Rev-erbβ(LBD) (A) and Fe(III)E75(LBD) (B) at 25 °C. Each sample was ~8–10 µM heme in 25 mM EPPS with 500 mM NaCl (Rev-erbβ(LBD)) or 100 mM NaCl (E75(LBD)), pH 8.0.
Table 1.
Comparison of electronic absorption and MCD peak positions for low-spin, 6-coordinate Fe(III) heme proteins. The experimental values for Rev-erbβ (LBD) and E75(LBD) are shaded.
| Electronic Absorption | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Ligands | δ | Soret | β | α | LMCT | Ref. | |
| Rev-erbβ (LBD) | X/Cys | 359 | 420 | 541 | 563 | 664, 757 | a | |
| E75(LBD) | X/Cys | 360 | 423 | 542 | 569 | 650, 750 | a | |
| E75(261) | N-,O-,S-?/Cysn | 359 | 424 | 542 | 575 | NRb | (23) | |
| RrCooA | Pro/Cys | 362 | 424 | 540 | 574 | 649,750 | (46) | |
| P450CAM+ImH | ImH/Cys | 358 | 425 | 542 | 574 | 638,753 | (73) | |
| hCBS | His/Cys | 365 | 428 | 550 | NR | 650,700 | (35) | |
| M80C cyt c | His/Cys | 355 | 416 | 540 | 570 | 635,734 | (73) | |
| MCD | Peak | Crossover | Trough | Peak | Crossover | Trough | Ref. | |
| Rev-erbβ(LBD) | X/Cys | 404 | 412 | 427 | 562 | 571 | 579 | a |
| E75(LBD) | X/Cys | 414 | 421 | 429 | NRb | NRb | NRb | a |
| RrCooA | Pro/Cys | 413c | 420 | 427c | 562c | 571 | 580b | (47) |
| P450CAM+ImH | ImH/Cys | 417 | 426 | 435 | 563 | 569 | 585 | (74) |
| hCBSc | His/Cys | 419 | 425 | 433 | 545 | 555 | 567 | (35) |
| M80C cyt c | His/Cys | 415c | 424c | 432 b | MSd | 560c | MSd | (74) |
This work.
Not reported.
Negligible intensity.
Peak and/or crossover positions were not reported in the original paper and are presented from inspection of the data.
Multiple max/crossover/min signals are present; only that of the central crossover position is indicated.
MCD spectra of Fe(III)Rev-erbβ(LBD) and Fe(III)E75(LBD) display nearly identical features, which are indicative of low-spin, 6-coordinate Fe(III) species. The spectra are dominated by intense temperature-dependent, derivative-shaped C-terms in the Soret region with crossovers at 412 nm for Rev-erbβ(LBD) and 421 nm for E75(LBD) (Table 1, Supporting Information Figures S1 and S2). Another low-intensity, temperature-dependent C-term is also present in the α–β region of each spectrum. Magnetization plots (Δε vs. βH/2kT) are consistent with low-spin, S = 1/2 heme systems (Supporting Information Figures S1 and S2, insets) (33). The MCD spectral characteristics of Fe(III)Rev-erb and Fe(III)E75(LBD) are similar to those of other 6-coordinate heme proteins that bear one cysteine ligand (Table 1 and references therein).
EPR spectra of the two nuclear receptor LBD constructs reveal features distinctive for low-spin, 6-coordinate Fe(III) heme species with cysteine(thiolate) coordinated opposite a neutral ligand. In our experiments Fe(III)E75(LBD) exhibited two well-resolved rhombic EPR signals spread over a narrow range of g values, as depicted in Figure 2B. Simulation of the slightly more intense features (a) is best fit with g values of 1.90, 2.26 and 2.46 for gx, gy and gz, and the slightly less intense signal (b) yields g values of 1.88, 2.26 and 2.56 for gx, gy and gz. Comparison of these EPR data with those previously reported for a similar E75 LBD construct establishes several similarities and differences. In prior EPR studies on Fe(III)E75, de Rosny, et al. (23) reported the presence of three signals for their construct E75(261); the narrowest and mid-range signals were not well-resolved and the narrowest signal was ascribed to only ~5% of the sample. The multiple features were attributed to coordination by two different cysteine residues, Cys396 and Cys468, which were identified via mutagenic analysis. The second donor ligand was His574 and an unknown neutral N-/O-donor in the two major species, respectively, and an O-/S-donor in the minor species. The two signals we observe for Fe(III)E75(LBD) are comparable to the two major signals observed in this prior study.
Figure 2.
EPR spectra of Fe(III)Rev-erbβ(LBD) (A) and Fe(III)E75(LBD) (B). A) X-band EPR spectrum of Fe(III)Rev-erbβ(LBD). The sample was ~180 µM heme in 50 mM borate with 500 mM NaCl, pH 8.0. The spectrum was recorded at 10 K, 9.3580 GHz microwave frequency, 0.319 mW microwave power, 6.3 × 105 receiver gain, 8.31 G modulation amplitude, 100 kHz modulation frequency, 163.84 ms time constant, using 8 added scans each containing 2048 data points. The background was corrected and three sets of g values (a), (b) and (c) were simulated. B) X-band EPR spectrum of Fe(III)E75(LBD). The sample was approximately 130 µM heme in 50 mM borate with 500 mM NaCl, pH 8.0. The spectrum was recorded at 10 K, 9.3588 GHz microwave frequency, 0.636 mW microwave power, 6.3 × 105 receiver gain, 8.31 G modulation amplitude, 100 kHz modulation frequency, 163.84 ms time constant, using 10 added scans each containing 2048 data points. The background was corrected and two sets of g values (a) and (b) were simulated.
The complex EPR spectrum of Fe(III)Rev-erbβ(LBD) is depicted in Figure 2A. Three distinct sets of g values were measured. The signal with the narrowest g anisotropy (b) is characterized by g values of 2.03, 2.29 and 2.35 for gx, gy and gz, respectively. The second, most intense signal (a) is characterized by g values of 1.99 (gx), 2.29 (gy) and 2.50 (gz). The third Fe(III)Rev-erbβ(LBD) signal with the broadest g anisotropy (c), is characterized by g values of 1.88 (gx), 2.29 (gy) and 2.60 (gz). The highest field g value, gz, was resolved from the more intense second signal at g~2.50 by increasing the pH of the Fe(III) sample to pH~9.5. Under these conditions the overall anisotropy of this third signal increased, i.e. the positions of both gz and gx changed, with gz shifted significantly to higher field and gx shifted to slightly lower field (spectrum not shown, data in Table 2). The first and second signals were also observed to shift slightly relative to their positions at pH~8.0, with near collapse of the resolved central gy signal into a single broad derivative. Addition of ~20% glycerol collapsed the resolved three signal spectrum into one dominated by a single rhombic signal (1.89 (gx), 2.30 (gy) and 2.51 (gz) with additional contributions at g~2.04 and g~2.01) (spectrum not shown). We have noted sensitivity of the EPR signals to pH and glycerol in other thiolate-ligated heme proteins, such as Fe(III)hCBS (34, 35) and Fe(III)RrCooA (36), which also exhibit dual EPR signals. We believe, based on computational studies, that the multiple signals of similar rhombicity may arise from variation in the protonation or hydrogen-bonded state of a cysteine(thiolate) ligand (37). Multiple cysteine ligands are not required, although their presence cannot be ruled out.
Table 2.
Comparison of EPR g values for selected 6-coordinate Fe(III) heme proteins. The experimental values for Rev-erbβ (LBD) and E75(LBD) are shaded.
| EPR | ||||||
|---|---|---|---|---|---|---|
| Protein | Ligands | gz | gy | gx | Ref. | |
| Rev-erbβ(LBD)(a) | X/Cys | 2.50 | 2.29 | 1.99 | pH~8 | a |
| Rev-erbβ(LBD)(b) | X/Cys | 2.35 | 2.29 | 2.03 | pH~8 | a |
| Rev-erbβ(LBD)(c) | X/Cys | 2.60 | 2.29 | 1.88 | pH~9.5b | a |
| E75(LBD) (a) | X/Cys | 2.46 | 2.26 | 1.90 | pH~8 | a |
| E75(LBD) (b) | X/Cys | 2.56 | 2.26 | 1.88 | pH~8 | a |
| E75(261)(E1) | His/CysA | 2.54 | 2.26 | 1.87 | (35–50%) | (23) |
| E75(261)(E2) | N-,O-?/CysA,CysB | 2.45 | 2.27 | 1.90 | (35–50%) | (23) |
| E75(261)(E3) | O-,S-?/CysB | 2.39 | 2.26 | 1.92 | (~5%) | (23) |
| RrCooA | Pro/Cys | 2.46 | 2.25 | 1.89 | (major) | (36) |
| RrCooA | Pro/Cys | 2.58 | 2.25 | 1.84 | (minor) | (36) |
| P450CAM+ImH | ImH/Cys | 2.56 | 2.27 | 1.87 | (75) | |
| hCBS | His/Cys | 2.49 | 2.31 | 1.87 | (major) | (35) |
| hCBS | His/Cys | 2.43 | 2.31 | 1.90 | (major) | (35) |
| M80C cyt c | His/Cys | 2.56 | 2.27 | 1.85 | (76) | |
| EcDos | His/H2O | 3.42 | - | - | (77) | |
| Cytochrome b5 | His/His | 3.03 | 2.23 | 1.43 | pH 7.0 | (78) |
| Neuroglobin | His/His | 3.14 | 2.16 | 1.34 | (79) | |
This work.
The value for gx and gz are obtained from an alternate spectrum at higher pH (pH~9.5).
The EPR signals for both Rev-erbβ(LBD) and E75(LBD) are characterized by decreased rhombicity, which is imposed by unique electronic interactions of a cysteine(thiolate) ligand with the heme iron. In contrast, low-spin, 6-coordinate Fe(III) heme proteins coordinated by two neutral donors (i.e. bis-histidine) exhibit rhombic EPR spectra characterized by a large g anisotropy, in which the position of gz (‘gmax’) is shifted to higher field, while gx and gy may be broadened and may become nearly undetectable in high symmetry ligand environment. Table 2 compares the g values of the nuclear receptors to those of low-spin heme proteins with thiolate and neutral donor ligands. Blumberg and Peisach correlated the observed g values with the nature of the axial ligand environment, constructing a ‘truth diagram’ with five distinct regions (38). When the g values for each of the EPR signals of Fe(III)Rev-erbβ(LBD) and Fe(III)E75(LBD) are used to calculate rhombicity (V/Δ) and tetragonality (Δ/λ) terms, all but the narrowest set of g values for Rev-erbβ(LBD) (signal (b)) fall within the ‘P’ region of the diagram. The narrowest set of g values observed for Rev-erbβ(LBD) do not fall within any of the assigned regions: this signal exhibits low rhombicity (V/Δ) and very high tetragonality (Δ/λ). Discounting this one anomalously placed signal, the EPR signal anisotropy for both nuclear receptors is comparable to that of other low-spin, 6-coordinate heme proteins with cysteine(thiolate) coordinated opposite a neutral sixth ligand (Table 2 and references therein), which are also within the ‘P’ region of the truth diagram.
Resonance Raman spectra of Fe(III)Rev-erbβ(LBD) and Fe(III)E75(LBD) are consistent with the assignment of a low-spin, 6-coordinate Fe(III) heme with cysteine(thiolate) as one of the axial ligands. Soret-band excitation enhances in-plane porphyrin vibrational modes, which are sensitive to the oxidation state (ν4), and the spin and coordination states (ν3, ν2, ν10) of the iron (28, 29). These high-frequency marker bands are observed at 1373 cm−1 (ν4), 1502 cm−1 (ν3), 1580 cm−1 (ν2) and 1633 cm−1 (ν10) in the spectrum of Fe(III)Rev-erbβ(LBD) (Figure 3B). Fe(III)E75(LBD) exhibits these bands at 1371 cm−1 (ν4), 1500 cm−1 (ν3), 1582 cm−1 (ν2) and 1635 cm−1 (ν10) (Supporting Information Figure S3B). The frequencies of the oxidation and spin state stretching modes of the nuclear receptors are similar to those of other low-spin, 6-coordinate hemes (28, 29). The low-frequency region (200–700 cm−1) is useful in assignment of vibrations associated with a metal-ligand bond (39). Cytochrome P450CAM, which is a high-spin, 5-coordinate thiolate-ligated Fe(III) heme in the presence of substrate, was studied by Champion, et al. (40) using 34S and 57Fe isotopic labeling. The stretching mode for the iron-sulfur bond (νFe-S) was identified at 351 cm−1. A similar study, using global 34S labeling, identified νFe-S in hCBS, a low-spin, 6-coordinate heme-thiolate protein, at 312 cm−1 (41). By analogy to this latter experiment, the νFe-S bands in Fe(III)Rev-erbβ(LBD) and Fe(III)E75(LBD) are tentatively assigned to broad bands centered at ~312 cm−1 and ~308 cm−1, respectively (Figure 3A, Supporting Information Figure S3A). The broad bandwidths observed may arise from the overlap of multiple vibrations, from different environments around the thiolate ligand, or from heterogeneity in heme coordination (e.g. multiple cysteine donors). The latter two interpretations are consistent with the multiple signals observed in the EPR.
Figure 3.
Resonance Raman for Fe(III) and Fe(II) Rev-erbβ(LBD). The samples were ~180 µM heme in 50 mM borate with 500 mM NaCl, pH 8.0. Reduction was performed using solid sodium dithionite. Spectra were acquired on the frozen samples at 77 K using the 413.1 nm Kr+ laser line with ~50 mW power at the sample. Spectral windows are presented for the A) 250–600 cm−1 and B) 1050–1650 cm−1 regions. Key vibrational modes are noted.
Characterization of the Fe(II) states of Rev-erbβ(LBD) and E75(LBD)
Electronic absorption spectra for Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) are indicative of replacement or loss of the cysteine(thiolate) ligand. Aspects of the spectra for both nuclear receptors are suggestive of heterogeneity in the Fe(II) heme axial ligand environment; the spectral features are broad and poorly resolved. When Fe(III)Rev-erbβ(LBD) is reduced to Fe(II) Rev-erbβ(LBD), the Soret band maximum is initially observed to red-shift from 420 nm to 424 nm, while the α–β peaks sharpen and blue-shift to 555 nm and 532 nm (Figure 4A). The Soret and α–β features lack the classic sharpness of low-spin Fe(II) heme. Over the course of several minutes, the Fe(II) peak positions red-shift to 428 nm (Soret), 558 (α) and 533 nm (β). The final spectrum is even more poorly resolved, with additional broadening and decreased intensity of the Soret band, and an increase in intensity of the α peak relative to the β peak. When Fe(III)E75(LBD) is reduced, the Soret band increases in intensity, and the peak maximum red-shifts slightly from 423 nm to 425 nm with sharpening of the α–β peaks, which blue-shift to 558 nm and 530 nm, respectively (Figure 5A). The positions and shape of the Fe(II)E75(LBD) spectral features do not change over time; however, the Soret band for Fe(II)E75(LBD) is broadened and the α–β features are not well-resolved. The spectrum we report for Fe(II)E75(LBD) is comparable to that reported by Reinking, et al. (6). Also notable in the Fe(II) absorption spectra of both proteins is residual intensity in the long wavelength region (600–700 nm), especially Fe(II)Rev-erbβ, which, along with the broadening of the Soret and α–β peaks, may indicate mixed coordination.
Figure 4.
Electronic absorption (A) and MCD (B) spectra of Fe(II)Rev-erbβ(LBD). A) The electronic absorption spectrum at 25 °C of Fe(II)Rev-erbβ(LBD) after the addition of solid sodium dithionite to Fe(III)Rev-erbβ(LBD) (thin gray line) is shown. The scan immediately following reduction (black dotted line) undergoes a blue shift to the final spectrum (black line) after 3–5 minutes. The sample contained ~10 µM heme in 25 mM EPPS with 500 mM NaCl, pH 8.0. B) The MCD sample was ~25 µM heme in 25 mM with 250 mM NaCl, pH 8.0, ~55% glycerol. Reduction was performed using solid sodium dithionite. The averages of three scans at 4.5, 8, 15 and 25 K at constant magnetic field (7 tesla) are shown.
Figure 5.
Electronic absorption (A) and MCD (B) spectra of Fe(II)E75(LBD). A) The electronic absorption spectrum at 25 °C of Fe(II)E75(LBD) (black line) after the addition of solid sodium dithionite to Fe(III)E75(LBD) (thin gray line) is shown. The sample contained ~8 µM heme in 25 mM EPPS with 100 mM NaCl, pH 8.0. B) The MCD sample was ~20 µM heme in 25 mM EPPS with 250 mM NaCl, pH 8.0, ~55% glycerol. Reduction was performed using solid sodium dithionite. The averages of three scans at 4.5, 8, 15 and 25 K at constant magnetic field (7 tesla) are shown.
While the possibility of heterogenous Fe(II) species cannot be disregarded, the electronic absorption peak positions for Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) are comparable to those of other low-spin, 6-coordinate heme proteins with two neutral donor ligands. Fe(II)Rev-erbβ( LBD) and Fe(II)E75(LBD) exhibit similar spectra to proteins in which two neutral axial ligands coordinate the Fe(II) heme, such as neuroglobin and cytochrome b5 (Table 3 and references therein). These peak positions, particularly that of the Soret band, are not consistent with the presence of a cysteine(thiolate) as a heme ligand. If cysteine(thiolate) were retained, the Soret band would have a distinctively red-shifted position near 450 nm, as observed in Fe(II)hCBS at pH 9 (35), the cytochromes P450 and chloroperoxidases (42). Fe(II)Rev-erbβ( LBD) or Fe(II)E75(LBD) exhibit Soret maxima between 425 nm and 428 nm; thus, the cysteine(thiolate) present in Fe(III)Rev-erbβ(LBD) and Fe(III)E75(LBD) must have been lost upon reduction. An analogous ligand replacement process occurs in RrCooA, in which Cys75 is replaced by His77 upon reduction forming a low-spin, 6-coordinate Fe(II)RrCooA species with a Soret maximum at 425 nm (43). However, in Rev-erbβ(LBD), the presence of high-spin, 5-coordinate species is also evident in the broadened Soret maximum that is shifted to longer wavelengths, more comparable to that of the globins and HRP (Table 3).
Table 3.
Comparison of electronic absorption and MCD peak positions for selected 6-coordinate Fe(II) heme proteins. The experimental values for Rev-erbβ (LBD) and E75(LBD) are shaded.
| Electronic Absorption | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Ligands | Soret | β | α | Ref. | |||
| Rev-erb β(LBD) | ?/? | 428 | 533 | 558 | a | |||
| E75(LBD) | ?/? | 425 | 530 | 558 | a | |||
| E75(LBD) | His/? | 426 | 531 | 559 | (6) | |||
| RrCooA | His/Pro | 425 | 529 | 559 | (55) | |||
| hCBS (pH 9.0) | His/Cys | 448 | 540 | 570 | (35) | |||
| EcDos | His/Met | 427 | 532 | 563 | (80) | |||
| neuroglobin | His/His | 426 | 530 | 560 | (81) | |||
| hemoglobin | His | 430 | 555 | 590(s)b | (82) | |||
| myoglobin | His | 434c | 520c | 560c | (82) | |||
| HRP | His | 430c | 560c | 590 | (82) | |||
| MCD | Soret | Visible | Ref. | |||||
| Protein | Ligands | Min. | Crossover | Max. | Max. | Crossover | Min. | |
| Rev-erb β(LBD) | ?/? | 427 | 434 | 441 | 555 | 558 | 561 | a |
| E75(LBD) | ?/? | 419 | 429 | 438 | 553 | 556 | 559 | a |
| RrCooA | His/Pro | 416c | 420 | 425c | 550c | 554 | 558c | (47) |
| hCBS (pH 9) | His/Cys | 445c | 448 | 452c | 563c | 566 | 569c | (35) |
| EcDos | His/Met | 430 | NRd | NRd | 558 | 561 | 565 | (83) |
| hemoglobin | His | 418c | 425c | 432c | MSe | 578c | MSe | (82) |
| myoglobin | His | 420c | 428c | 437c | MSe | 580c | MSe | (82) |
| HRP | His | 425c | 432c | 440c | MSe | 590c | MSe | (82) |
This work.
Shoulder.
Peak positions were not reported in the original paper and are presented from inspection of the data.
Not reported.
Multiple max/crossover/min signals are present; only that of a central crossover position is indicated.
The MCD spectra of Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) provide a clearer picture of the heterogeneous heme environments present in the nuclear receptor LBD constructs that result from loss of the cysteine(thiolate) ligand. Two distinct signal types are observed in each spectrum (Figure 4B, Figure 5B). The temperature-dependent C-term in the Soret region arises from a high-spin, 5-coordinate Fe(II) heme (S = 2), and the temperature-independent A-term in the visible region arises from a low-spin, 6-coordinate Fe(II) heme (S = 0). In Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD), the low-temperature MCD spectra are dominated by the C-term Soret feature with crossovers at 434 nm and 429 nm, respectively, suggesting that a substantial fraction of each protein is high-spin and 5-coordinate. The C-term peak shapes and positions of Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) are comparable to those of other high-spin, 5-coordinate heme proteins with a single histidine ligand, such as hemoglobin or myoglobin (Table 3 and references therein). The A-term peak positions are similarly inconsistent with the presence of a cysteine(thiolate). In Fe(II)Rev-erbβ, a weak temperature-independent A-term occupies the visible region with a crossover at 558 nm (Figure 4B). Fe(II)E75(LBD) exhibits a more intense A-term centered at 556 nm (Figure 5B). A crossover position of the visible region A-term between 550–560 nm is suggestive of two neutral donors, as the analogous band in low-spin, thiolate-bound Fe(II) species appears between 562–576 nm (Table 3). The relative intensities of the Soret region C-terms and the visible region A-terms in Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) indicate that both proteins contain high-spin, 5-coordinate and low-spin, 6-coordinate fractions, but the high-spin, 5-coordinate species dominates in Fe(II)Rev-erbβ(LBD). The presence of this heterogeneous coordination is responsible for the broadened peak shapes observed in the electronic absorption spectra of Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) (vide supra). Notably, the shifting and broadening of the Fe(II)Rev-erbβ(LBD) Soret peak position over time suggests instability in the heme axial ligand environment or the heme pocket, which results in loss of an axial ligand and greater conversion to a high-spin, 5-coordinate species.
Resonance Raman spectra of Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) exhibit features consistent with heterogeneous environments of high-spin, 5-coordinate and low-spin, 6-coordinate Fe(II) heme. That the heme is fully reduced is reflected in the position of the oxidation state marker bands (ν4), which occur at 1361 cm−1 in both Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) (Figure 3B, Supporting Information Figure S3B). No residual intensity is observed at the higher frequencies characteristic of Fe(III)Rev-erbβ(LBD) (1373 cm−1) and Fe(III)E75(LBD) (1371 cm−1). The complete absence of signals for Fe(III) heme confirms that the temperature-dependent C-terms in the Soret region of the Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) MCD spectra arise from high-spin Fe(II) heme (S = 2) and not residual Fe(III) (S = ½) in the samples. The frequencies of the π-sensitive band ν11, which overlap considerably with those of ν38 in the Fe(III) states, decrease to ~1540 cm−1 in Fe(II)Rev-erbβ(LBD) and to ~1541 cm−1 in Fe(II)E75(LBD), appearing as prominent shoulders in the Fe(II) states. These peak positions are consistent with those of other Fe(II) heme proteins.
The presence of multiple spin state bands in the high-frequency resonance Raman spectra of Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) provide further evidence for the presence of both 5-coordinate, high-spin and 6-coordinate low-spin heme. Pure low-spin and pure high-spin (non-thiolate) Fe(II) heme centers are differentiated by ν3 bands at ~1490–1500 cm−1 and ~1470–1475 cm−1 for low- and high-spin states, respectively. In Fe(II)Rev-erbβ, the ν3 envelope is split into a more intense band at 1469 cm−1 and a distinct smaller band at 1493 cm−1 (Figure 3B). The ν3 envelope in Fe(II)E75(LBD) is also split into two bands at 1472 cm−1 and 1494 cm−1 (Supporting Information Figure S3B). Similar frequencies are observed for the split ν3 envelopes in Fe(II)sGC (44) and Fe(II)KatG (45), which both exist as mixtures of high-spin, 5-coordinate and low-spin, 6-coordinate heme species. An admixture of coordination and spin states would be expected to give rise to ν10 absorption intensity in the ranges ~1600–1610 cm−1 and 1615–1625 cm−1. The ν10 marker bands, which appear as a shoulder in Fe(III)Rev-erbβ(LBD) (~1633 cm−1) and a distinct band in Fe(III)E75(LBD) (~1635 cm−1), shift underneath the porphyrin vibrational modes νC=C and ν37 upon reduction, giving rise to a broad band between ~1600–1625 cm−1. Thus, the positions of key marker bands in Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) further support the presence of both low-spin, 6-coordinate and high-spin, 5-coordinate Fe(II) heme in these proteins.
Characterization of the Fe(II)CO adducts of Rev-erbβ(LBD) and E75(LBD)
The electronic absorption spectra of Fe(II)CO Rev-erbβ(LBD) and Fe(II)CO E75(LBD) show features indicative of 6-coordinate CO adducts with a neutral endogenous ligand trans to CO. Fe(II)CO Rev-erbβ(LBD) exhibits a blue-shifted, very sharp and intense Soret band at 421 nm and nearly equivalent α–β peaks at 568 and 540 nm (Figure 6A). Similarly, the Fe(II)CO E75(LBD) adduct displays spectral features almost identical to those of Fe(II)CO Rev-erbβ(LBD) (Supporting Information Figure S4A). The peak positions of Fe(II)CO Rev-erbβ(LBD) and Fe(II)CO E75(LBD) are similar to those of Fe(II)CO heme proteins in which CO binds opposite a neutral donor ligand (Supporting Information Table S1 and references therein). Because we previously observed that the order of reduction and CO addition altered the spectra of the CO adduct in heme proteins with admixed high- and low-spin Fe(II) (46), we investigated whether this effect was present in the Rev-erbβ and E75 LBD constructs. The influence of order of addition was tested by introducing CO(g) to the Fe(III) proteins before reduction and comparing the absorption features to those of CO adducts formed by addition of CO(g) to pre-reduced Fe(II) proteins. When CO(g) is added to Fe(III)Rev-erbβ(LBD) before reductant, the α–β peaks exhibit nearly equivalent intensities; however, addition of CO(g) to Fe(II)Rev-erbβ(LBD) results in an ~11% decrease in the intensity of the α band relative to the β band, and the α peak maximum red-shifts to 568 nm (Figure 6A). In contrast, addition of CO(g) to Fe(II)CO E75(LBD) results in < 5% decrease in α peak intensity relative to β and no change in peak positions (Supporting Information Figure S4A).
Figure 6.
Electronic absorption (A) and MCD (B) spectra of Fe(II)CO Rev-erbβ(LBD). A) The electronic absorption spectra at 25 °C of Fe(II)CO Rev-erbβ(LBD) in 25 mM EPPS with 500 mM NaCl, pH 8.0. A sample containing ~8 µM heme with CO(g) present at time of reduction (solid line) is compared with a sample (~8 µm heme) with CO(g) injected after reduction (dotted line). The Fe(II) protein is represented by a thin gray line. Reduction was performed using a sodium dithionite solution. B) The MCD sample was ~25 µM heme in 25 mM EPPS with 250 mM NaCl, pH 8.0, ~55% glycerol. Reduction was performed after addition of CO(g) using sodium dithionite solution. The average of three scans at 7 tesla and 100 K is shown.
MCD spectral features are consistent with 6-coordinate heme in both Fe(II)CO Rev-erbβ( LBD) and Fe(II)CO E75(LBD). MCD spectra were acquired at 100 K to prevent CO photolysis, which has been observed for other 6-coordinate CO-heme adducts at low temperature (4.5–25 K) (47, 48). At 100 K, the Fe(II)CO Rev-erbβ(LBD) and Fe(II)CO E75(LBD) MCD spectra are dominated by a derivative-shaped, temperature-independent A-term in the Soret region with a crossover at 421 nm (Figure 6B, Supporting Information Figure S4B). A second, less intense A-term is observed in the α–β region with crossovers at 570 nm (Rev-erbβ) and 569 nm (E75). The shapes and peak positions of the respective A-terms in Fe(II)CO Rev-erbβ(LBD) and Fe(II)CO E75(LBD) are consistent with those of reported 6-coordinate, CO–heme adducts (Supporting Information Table S1). Fe(II)CO Rev-erbβ(LBD) shows some broadening of the visible region A-term signal, which may be a consequence of the heterogeneous heme environment in the reduced protein (Figure 6B).
Resonance Raman spectra provide evidence that the heme environments in the Fe(II)CO adducts of Rev-erbβ(LBD) and E75(LBD) are 6-coordinate with histidine trans to CO. Carbon monoxide, a π acid ligand, competes for the iron dπ electrons, giving rise to upfield shifts of π-sensitive porphyrin marker bands. In Fe(II)CO Rev-erbβ(LBD), the oxidation, spin and coordination state marker bands are observed at 1368 cm−1 (ν4), 1581 cm−1 (ν2), 1493 cm−1 (ν3) and 1631 cm−1 (ν10) (Figure 7B). In Fe(II)CO E75(LBD), the same modes are assigned as 1367 cm−1 (ν4), 1579 cm−1 (ν2), 1493 cm−1 (ν3) and 1632 cm−1 (ν10) (Supporting Information Figure S5B). These modes are shifted as expected relative to analogous modes in the Fe(II) proteins. The frequencies of these porphyrin marker bands in Fe(II)CO Rev-erbβ(LBD) and Fe(II)CO E75(LBD) are similar to those of other 6-coordinate CO-heme adducts with histidine as a trans ligand (49). In the low frequency region, isotope-sensitive modes associated with the Fe-CO unit may be observed. Two sets of stretching modes and one set of bending modes are seen in Fe(II)CO Rev-erbβ(LBD): ν(Fe-12CO), 515 cm−1; ν(Fe-13CO), 512 cm−1; ν(Fe-12CO), 500 cm−1; ν(Fe-13CO), 496 cm−1; and δ(Fe-12CO), 578 cm−1; δ(Fe-13CO), 564 cm−1 (Figure 7A). In Fe(II)CO E75(LBD), single sets of stretching modes (ν(Fe-12CO); 494 cm−1; ν (Fe-13CO), 490 cm−1) and bending modes (δ(Fe-12CO), 575 cm−1; δ(Fe-13CO), 565 cm−1) are observed (Supporting Information Figure S5A). Multiple Fe-CO stretches, such as reported here for Rev-erbβ(LBD), are correlated in other proteins with multiple conformations in the vicinity of the heme-bound CO (50, 51). In the high frequency region, C-O stretching modes were identified for both Fe(II)CO Rev-erbβ(LBD) (ν(12C-O), 1948 cm−1; ν(13C-O), 1902 cm−1) and Fe(II)CO E75(LBD) (ν(12C-O), 1958 cm−1; ν(13C-O), 1908 cm−1) (Figure 7C, Supporting Information Figure S5C). The observed ν(Fe-CO) and ν(C-O) frequencies in Fe(II)CO Rev-erbβ(LBD) and Fe(II)CO E75(LBD) compare well with those of other 6-coordinate CO-heme adducts a histidine ligand trans to the bound CO (Supporting Information Table S2 and references therein).
Figure 7.
Resonance Raman spectra for Fe(II)(12CO) and Fe(II)(13CO) Rev-erbβ(LBD). The samples were ~180 µM heme in 50 mM borate with 500 mM NaCl, pH 8.0. After CO(g) was introduced into the Fe(III)Rev-erbβ(LBD) sample, reduction was performed by addition of a sodium dithionite solution. Spectra were acquired on the frozen samples at 77 K using the 413.1 nm Kr+ laser line with ~7–10 mW power at the sample. The difference spectrum (12CO–13CO) is shown for the frequency ranges containing ν(Fe-CO), δ(Fe-CO) and ν(C–O). A) The lowest frequency window, 200–600 cm−1, containing the stretching and bending modes ν(Fe-CO) and δ(Fe-CO). B) The mid-frequency window, 1050–1650 cm−1, containing the principal oxidation, spin and coordination state marker bands. Shown is a comparison of the Fe(III), Fe(II) and Fe(II)CO states. C) The high frequency window, 1750–2150 cm−1, containing the stretching mode ν(C–O). Key vibrational modes are noted.
Extensive studies of heme-CO protein adducts and model complexes have established a linear, negative correlation between ν(Fe-CO) and ν(C–O) frequencies (52, 53). The identity of the trans ligand and the environment of the heme pocket influence the observed frequencies of the two stretching modes (vide supra). The frequencies observed for 6-coordinate CO adducts with histidine trans to CO are distinct from those of either 6-coordinate CO adducts with cysteine(thiolate) trans to CO or 5-coordinate CO adducts, due to differing levels of competition for the iron dz2 orbital. Furthermore, these frequencies are sensitive to the polarity of the protein environment around the CO ligand. A positive polarity environment surrounding the CO ligand stabilizes the delocalization of π electrons from Fe(II) to CO, which strengthens the Fe-C bond, but weakens the C-O bond through increased population of the π* antibonding orbitals. The inverse effect is observed when the environment of the CO is more negative in nature. When the frequencies of the Fe-CO and C-O stretching modes for Fe(II)CO Rev-erbβ(LBD) and Fe(II)CO E75(LBD) are plotted on a ν(Fe-CO)/ ν(C-O) correlation diagram, Rev-erbβ(LBD) and E75(LBD) are placed with other 6-coordinate Fe(II)CO heme proteins and related model complexes with histidine trans to CO (Supporting Information Figure S6).
Characterization of the Fe(II)NO adducts of Rev-erbβ(LBD) and E75(LBD)
Electronic absorption spectroscopy reveals distinct differences between the two Fe(II)NO adducts: Fe(II)NO Rev-erbβ(LBD) exhibits spectral features indicative of a 6-coordinate NO-heme, while Fe(II)NO E75(LBD) exhibits features consistent with a 5-coordinate NO-heme. When NO(g) is introduced into Fe(II)Rev-erbβ(LBD), the broad Fe(II) Soret band at 428 nm is replaced by a less intense, but sharp Soret band at 420 nm accompanied by broad, undefined α–β peaks at ~571 nm and ~546 nm (Figure 8A). When anaerobic Fe(III)Rev-erbβ(LBD) is exposed to NO(g), auto-reduction and conversion to the Fe(II)NO species is also observed (data not shown). Six-coordinate Fe(II)NO hemes, in which one neutral endogenous protein ligand is retained opposite NO, typically exhibit sharp Soret bands in the 415–425 nm range (Supporting Information Table S3 and references therein). This characteristic is clearly seen for Fe(II)NO Rev-erbβ(LBD. In contrast, when NO(g) is introduced into Fe(II)E75(LBD), the NO adduct exhibits a less intense Soret band at ~390 nm, which is partially obscured by sodium dithionite absorption, and ill-defined α–β peaks at ~569 nm and ~541 nm (Figure 8B). The spectral features of Fe(II)NO E75(LBD) are consistent with those of other 5-coordinate Fe(II)NO heme proteins, such as sGC (54) and RrCooA (55), which display a broad, low intensity Soret band near 400 nm and broad α and β bands of approximately equivalent intensity (Supporting Information Table S3).
Figure 8.
Electronic absorption spectra of Fe(II)NO Rev-erbβ(LBD) (A) and Fe(II)NO E75(LBD) (B). A) The electronic absorption spectrum at 25 °C of Fe(II)NO Rev-erbβ(LBD) (thick black line) is compared to Fe(II)Rev-erbβ(LBD) (thin gray line). The sample contained ~5 µM heme in 25 mM EPPS with 500 mM NaCl, pH 8.0. B) The electronic absorption spectrum at 25 °C of Fe(II)NO E75(LBD) (thick black line) is compared to Fe(II)E75(LBD) (thin gray line). The sample contained ~5 µM heme in 25 mM EPPS with 100 mM NaCl, pH 8.0. The samples in both (A) and (B) were prepared by reduction of Fe(III) protein with solid sodium dithionite followed by addition of NO(g).
The EPR of Fe(II)NO Rev-erbβ(LBD) confirms the presence of a 6-coordinate NO-heme adduct with a nitrogenous ligand trans to NO, presumably histidine. The EPR signal observed in the Fe(II)(14NO) Rev-erbβ(LBD) spectrum exhibits a characteristic rhombic splitting (Figure 9A). Though poorly resolved, a multi-line signal due to hyperfine and superhyperfine coupling between the unpaired electron on the nitrogen of the 14NO (I = 1) ligand and a proximal nitrogenous ligand is evident. The Fe(II)(15NO) spectrum exhibits fewer lines due to the reduced nuclear spin of 15N (I = ½) (Figure 9B). The Fe(II)(14NO) and Fe(II)(15NO) Rev-erbβ(LBD) spectra exhibit consistent g values 1.974 (gx), 2.002 (gy), 2.08 (gz); the hyperfine coupling constant A1, arising from coupling of the unpaired electron to the 14N/15N nucleus, increases from approximately 22 Gauss (14NO) to approximately 33 Gauss (15NO). The spectra of Fe(II)(14NO) and Fe(II)(15NO) Rev-erbβ(LBD) each contain an additional feature at g1~2.04; a complementary feature presumed to occur near g2~1.97 was not resolved. Such additional signals have been observed in other 6-coordinate Fe(II)NO adducts and have been attributed to variation in the His-Fe(II)-NO bonding geometry (56). The g values and coupling constants measured for Fe(II)(14NO) and Fe(II)(15NO) Rev-erbβ(LBD) are consistent with those of other 6-coordinate Fe(II)NO heme proteins that possess a sixth endogenous histidine ligand (Supporting Information Table 4 and references therein). That the spectra of Fe(II)(14NO) and Fe(II)(15NO) Rev-erbβ(LBD) do not exhibit well-resolved hyperfine and superhyperfine splitting patterns may indicate that some 5-coordinate NO-heme is present. Distinct 5-coordinate NO-heme signals were observed in some sample preparations, supporting this proposal (data not shown). Alternatively, the poor resolution may arise from heterogeneity in the Fe(II)NO environment, as observed in other states of Rev-erbβ(LBD).
Figure 9.
X-band EPR spectra of (A) Fe(II)(14NO) and (B) Fe(II)(15NO) Rev-erbβ(LBD). A) Fe(II)(14NO) Rev-erbβ(LBD) was ~180 µM heme in 50 mM borate with 500 mM NaCl, pH 8.0. The spectrum was recorded at 10 K, 9.3567 GHz microwave frequency, 0.16 mW microwave power, 6.3 × 105 receiver gain, 2.09 G modulation amplitude, 100 kHz modulation frequency, 163.84 ms time constant, using 10 added scans each containing 2048 data points. B) Fe(II)(15NO) Rev-erbβ(LBD) was ~180 µM heme in 50 mM borate with 500 mM NaCl, pH 8.0. The spectrum was recorded at 10 K, 9.3567 GHz microwave frequency, 0.30 mW microwave power, 8.0 × 105 receiver gain, 1.05 G modulation amplitude, 100 kHz modulation frequency, 163.84 ms time constant, using 10 added scans each containing 2048 data points. The samples were prepared via reduction of Fe(III)Rev-erbβ(LBD) and generation of NO(g).
Resonance Raman of Fe(II)NO Rev-erbβ(LBD) reveals modes consistent with the assignment of a 6-coordinate Fe(II)NO adduct. The spectrum of Fe(II)NO Rev-erbβ(LBD) is shown in Figure 10; the modes sensitive to the oxidation, spin and coordination states are assigned as ν4 (1371 cm−1), ν2 (1577 cm−1), ν3 (1494 cm−1) and ν10 (1634 cm−1). The band positions are consistent with those of other 6-coordinate Fe(II)NO heme proteins (57). The most distinct bands associated with spin and coordination state (ν3 and ν10) are ~10–15 cm−1 higher than those expected for 5-coordinate NO-heme adducts; 5-coordinate Fe(II)NO Rev-erbβ(LBD), if present in the sample, must be at a concentration below the detection limit. In the low frequency region, isotopic substitution with 15NO resolved the weak ν(Fe-NO) stretching mode (ν(Fe-14NO), 558 cm−1; ν(Fe-15NO), 551 cm−1) (Figure 10A). The ν(Fe-14NO) frequency observed for Fe(II)NO Rev-erbβ(LBD) is comparable to those of other 6-coordinate Fe(II)NO heme proteins and distinctly higher than ν(Fe-14NO) frequencies typically observed for 5-coordinate Fe(II)NO adducts (Supporting Information Table S5 and references therein). At an excitation wavelength of 406.7 nm, the intense symmetric porphyrin modes that are enhanced at 413.1 nm were reduced, revealing a weak band at ~1640 cm−1 overlapping ν10 that is absent in the Fe(II)(15NO) spectrum (Figure 10C). This band is tentatively assigned as ν(14N-O) in Fe(II)NO Rev-erbβ(LBD). The position of this band is comparable to that of other 6-coordinate Fe(II)NO heme proteins, whose ν(N–O) bands are distinctly lower in frequency than those of 5-coordinate NO adducts. The ν(15N–O) band of Fe(II)(15NO) Rev-erbβ(LBD) cannot be assigned, as it remained buried under the porphyrin modes with excitation at 413.1, 406.7 nm and 530 nm (data not shown). Similar to heme-CO adducts, the ν(Fe-NO) and ν(N–O) frequencies for proteins and model complexes have been correlated, following a less well-behaved ‘anti-correlation’ relationship that shows variable response of the Fe-NO and N–O bonds to the surrounding environment (58). The relationship of the ν(Fe-NO) and tentative ν(N-O) frequencies for Fe(II)NO Rev-erbβ(LBD) places Rev-erbβ(LBD) near other 6-coordinate Fe(II)NO heme proteins and related model complexes with a neutral/histidine ligand trans to NO (Supporting Information Figure S7). Thus, resonance Raman and EPR data are consistent with the presence of a neutral nitrogen donor, presumably histidine, as the sixth axial ligand.
Figure 10.
Resonance Raman spectra for Fe(II)(14NO) and Fe(II)(15NO) Rev-erbβ(LBD). The samples were ~180 µM heme in 50 mM borate with 500 mM NaCl buffer, pH 8.0. The samples were prepared via reduction of Fe(III)Rev-erbβ(LBD) and generation of NO(g). Spectra were acquired on the frozen samples at 77 K using the 413.1 nm Kr+ laser line with ~10 mW power at the sample, except for (C), which was acquired using the 406.7 Kr+ laser line. The difference spectrum (14NO–15NO) is shown for the frequency ranges containing ν(Fe-NO) and ν(N-O). A) The lowest frequency window, ~250–600 cm−1, containing the stretching mode ν(Fe-NO). B) The mid-frequency window, 1100–1700 cm−1, containing the major oxidation, spin and coordination state marker bands. Shown is a comparison of the Fe(III), Fe(II) and Fe(II)NO states. C) The enlarged spectral window, ~1450–1700 cm−1, containing the stretching mode ν(N-O). Key vibrational modes are noted.
Characterization of Rev-erba(LBD)
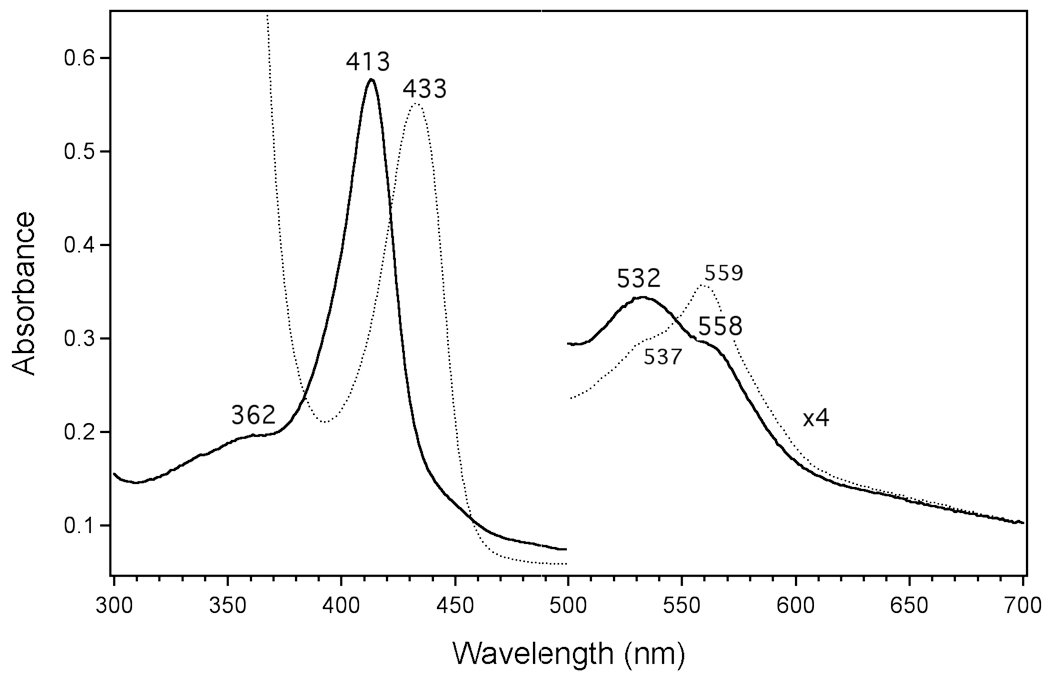
We also studied a fragmentary construct of the related human nuclear receptor Rev-erbα using electronic absorption spectroscopy. This protein construct, denoted Rev-erbα(LBD), contains 242 amino acids of the ligand binding domain, with deletion of the loop between helices 1 and 3. The precise position and size of the deletion is different from that of the Rev-erbβ(LBD). Fe(III)Rev-erbα(LBD) does not exhibit features typical of cysteine(thiolate) ligation; however, the spectral features of Fe(III)Rev-erbα(LBD), including a sharp Soret at 413 nm and broad α–β features at 558 and 532 nm, remain suggestive of a 6-coordinate, low-spin heme (Figure 11). Fe(III)Rev-erbα(LBD) may have a different coordination environment than Fe(III)E75(LBD) and Fe(III)Rev-erbβ(LBD), in which no cysteine(thiolate) serves as a ligand and two endogenous neutral donors bind to the heme, or the protein may be 6-coordinate due to coordination by an adventitious neutral protein ligand or water. Reduced Fe(II)Rev-erbα(LBD) exhibits the broadened, poorly-resolved spectral features indicative of an admixture of high-spin, 5-coordinate and low-spin, 6-coordinate heme, with a strongly red-shifted Soret at 433 nm and visible peaks at 559 nm and 537 nm (Figure 11). Furthermore, the absorption spectrum of Fe(II)CO Rev-erbα(LBD), while indicative of a 6-coordinate heme with a neutral sixth ligand, displays significant inequality in the α–β peak ratio intensity (Supporting Information Figure S8). Rev-erbα(LBD) appears to form a 6-coordinate NO adduct at room temperature, similar to that of Fe(II)NO Rev-erbβ(LBD) with a Soret at 418 nm (Supporting Information Figure S9); however, there is significant spectral intensity between ~390–410 nm, which suggests that a fraction of 5-coordinate NO-heme is present. These preliminary results for Rev-erbα(LBD) provide additional evidence that the nuclear receptor LBD constructs experience variability in heme coordination chemistry.
Figure 11.
Electronic absorption spectra of Fe(III) (solid line) and Fe(II) (dotted line) Rev-erbα( LBD) at 25 °C. The sample contained ~8 µM heme in 25 mM EPPS with 500 mM NaCl, pH 8.0. Solid sodium dithionite was used as reductant.
DISCUSSION
Heme is incorporated into a diverse array of proteins, including cytochromes, catalases, peroxidases, oxygenases, gas sensing and transport proteins, and transcriptional regulators. In these distinct environments, heme functions in an astonishing variety of biological and cellular functions, including gene expression, respiration, metabolism and stress response (59). There is mounting evidence that heme and heme proteins function as key regulators of cellular homeostasis. Heme sensor proteins are ubiquitous in regulatory responses to the bioactive gases, O2, CO and NO. Regulation of blood pressure, cell division, cell death, inflammation, metabolism, hypoxia, diurnal cycles, behavior, and memory have all been attributed to the action of NO and CO (60–62). In vertebrates, phase and period in the circadian cycle are believed to be regulated by levels of CO and NO, which are derived from light and dark environmental signals (63, 64). Furthermore, heme biosynthesis is reciprocally regulated by the circadian clock, with heme levels rising and falling in a cyclical manner (65). The human Rev-erb nuclear receptors, which are now known to bind heme, regulate circadian rhythm and energy metabolism (25, 26). Given the related regulatory roles for heme, NO and CO, the relationship between heme chemistry and the transcriptional activities of these nuclear receptors is important to elucidate.
In this work, we have characterized the heme coordination chemistry of fragmentary constructs of the heme-binding nuclear receptors E75 from Drosophila melanogaster and Rev-erbβ from Homo sapiens. The E75(LBD) construct contains 262 amino acids believed to encompass the entire 12 helix ligand binding domain; however, the full length protein, which occurs in multiple splice variants, is over 1000 amino acids. The ligand-binding domain of Rev-erbβ differs in two important ways from that of E75: there is a large loop between helices 1 and 3, and helix 12 is absent. The Rev-erbβ(LBD) construct that we have studied is modified via deletion of the loop between helices 1 and 3 to make it more similar to E75(LBD). The Rev-erbα(LBD) construct is similar, but bears a different deletion that encompassed Cys417. Because this amino acid is the analog of Cys396 in E75, which was previously shown to be a ligand to the E75 heme (23), we chose not to characterize this construct in detail. The analogous amino acid in Rev-erbβ, Cys384, is present in the Rev-erbβ(LBD) construct, and the recently published crystal structure of the heme-bound Rev-erbβ(LBD) construct confirms that Cys384 is an axial ligand to the heme (24). This structure also reveals that histidine (His568), present at the extreme C-terminus of Rev-erbβ, is the second heme ligand. This histidine is analogous to His574, implicated as a heme ligand in E75 and located the C-terminal end of the ligand binding domain of that protein (6, 23). The analogous His602 is present in the in Rev-erbα construct.
The effect of removing substantial functional regions of these proteins may explain the heterogeneity evident in our data. The Fe(II)Rev-erbβ(LBD) and Fe(II)E75(LBD) proteins contain an admixture of high-spin, 5-coordinate and low-spin, 6-coordinate heme. Importantly, Fe(II)Rev-erbβ(LBD) contains a greater proportion of high-spin heme than Fe(II)E75(LBD), suggesting that removal of the loop between helices 1 and 3 may alter the stability of the heme environment in Rev-erbβ(LBD). The observations that the Fe(II)Rev-erbβ(LBD) CO adduct exhibits broadened signals in the MCD, dual ν(Fe-CO) bands in the resonance Raman, and noticeable order of addition effects, are indicative of a heme environment that is heterogeneous. Furthermore, the 6-coordinate Fe(II)Rev-erbβ(LBD) NO adduct, which was clearly the dominant species at room temperature by absorption spectroscopy, was unstable and converted to a 5-coordinate form upon freezing for EPR or MCD. Careful sample handling was needed to produce the reported 6-coordinate EPR spectrum. Considerable spectral heterogeneity was observed for the Rev-erbα(LBD) construct, in which a different segment of the loop between helices 1 and 3 is missing. Together these observations suggest that the missing loop and functional domains may be important for the structure of Rev-erb ligand binding domains.
The behavior of the E75 and Rev-erbβ LBDs places them within a special subgroup of heme-thiolate proteins, which undergo a ligand switch upon reduction to replace the cysteine(thiolate) ligand. The spectral characteristics of Fe(III)E75(LBD) and Fe(III)Rev-erbβ(LBD) clearly show that the heme is low-spin and 6-coordinate with cysteine(thiolate) bound opposite a neutral donor ligand in both proteins. Unlike the cytochromes P450 and related enzymes (42), Rev-erbβ and E75 possess a heme environment that does not stabilize the thiolate-Fe(II) interaction. When Fe(III)E75(LBD) and Fe(III)Rev-erbβ(LBD) are reduced, the thiolate-Fe(II) bond is ruptured, and another neutral donor ligand may assume the sixth axial position. Ligand switching has been observed in heme-thiolate proteins that are CO and/or NO responsive, and which discriminate against binding O2. CooA, a CO-sensing transcription factor from the bacterium Rhodospirillum rubrum, is the best-studied example of gas-sensing heme-thiolate proteins (43). RrCooA in the Fe(III) state is coordinated by a Cys75/Pro2 ligand set; when the protein is reduced the Cys75 ligand is replaced by His77 to give a His77/Pro2 coordinated Fe(II) heme. Similarly, the CO-sensing eukaryotic transcription factor NPAS2 undergoes a Cys171 to His170 ligand switch upon reduction (66). Importantly, in E75 a change in redox state correlates with a change in repressor activity; E75 sequesters the transcriptional activator DHR3 only in the Fe(II) state (6). Thus, it appears that the ligand switch that we observe in E75 is of functional consequence to nuclear receptor function.
Intriguing similarities and differences are observed between the CO and NO adducts of E75 and Rev-erbβ. The Fe(II)CO adducts of E75(LBD) and Rev-erbβ(LBD) are 6-coordinate complexes with histidine trans to the CO ligand, with any sixth neutral donor ligand present in the Fe(II) state being replaced by CO. The CO is bound within the proteins in a relatively non-polar environment; however, the polarity of the environment is not identical in the two proteins. Whereas the Fe-CO/C-O ratio for Fe(II)CO E75(LBD) falls near proteins such as NPAS2 and EcDos on the correlation diagram, Fe(II)CO Rev-erbβ(LBD) is positioned higher on the correlation line near wild-type myglobin (Supporting Information Figure S6). This difference suggests that Rev-erbβ(LBD) contains more positive polarity residues close to the CO ligand, or, alternatively, that the CO ligand is more solvent exposed in the Rev-erbβ(LBD) adduct. In E75 functional assays, CO disrupted the ability of E75 to bind the AF-2 peptide of the activator DHR3 (6), which suggests that the change in heme coordination associated with CO binding is of functional importance for the protein. In contrast, Pardee, et al. (24) report minimal effects of CO on Rev-erb target gene expression.
The NO adducts of E75 and Rev-erbβ exhibit marked differences from one another; the NO-adduct of E75(LBD) is 5-coordinate while that of Rev-erbβ(LBD) is 6-coordinate with a histidine trans to NO. NO inhibits the interaction of E75 with the AF-2 peptide of DHR3 as well as the ability of E75 to block DHR3 transcriptional activation, thus overcoming the E75-induced repression (6). These effects plausibly may be a consequence of release of both Fe(II) axial ligands. Five-coordinate heme-NO adducts, which are also observed in sGC, eIF2α kinase/HRI and CooA, are more thermodynamically favored due to the strong negative trans influence exerted by the NO ligand, which weakens the endogenous metal-ligand bond (67). This effect is used to great advantage in the heme-based NO sensor soluble guanylyl cyclase (sGC); the 5-coordinate NO adduct activates cGMP production 200–400-fold (44, 68). In sGC, the 6-coordinate CO complex activates the enzyme to a much lesser extent. A similar case is that of eukarotic heme-regulated eIF2α kinase (HRI), which controls initiation of protein synthesis in cells (69). HRI is activated by NO binding to the heme, which displaces both endogenous ligands, but HRI is inhibited by formation of the 6-coordinate CO adduct (70). In contrast, the heme in RrCooA is specifically activated by the 6-coordinate CO-heme adduct while formation of the 5-coordinate NO-heme adduct fails to activate the protein (43). Unlike E75, whose functional behavior is equally responsive to CO and NO, Rev-erb transcriptional activity appears to be selectively responsive to NO (24). These recent data from in vivo target gene mRNA analysis and reporter assay implicate NO as a direct regulator of Rev-erb. The heme environment in Rev-erbβ may stabilize the formation of a 6-coordinate NO-heme adduct, as is observed in the thermophilic CooA homolog ChCooA (71). The limited data we collected on Rev-erbα(LBD) also implicate a largely 6-coordinate NO-heme adduct in that nuclear receptor as well. That E75 and the Rev-erbs behave differently in the presence of NO may be of biological significance, or may be a consequence of differences in the nature of the protein fragment constructs.
Unlike the nuclear receptor Drosophila E75, which has been implicated as both a redox and gas sensor, Rev-erbα and Rev-erbβ were initially reported to lack functional redox and gas responses. These nuclear receptors were proposed to be sensors of intracellular heme concentration via reversible binding to the ligand binding domain, which also modulate binding of co-regulator protein (25, 26). We would argue that the conservation of the heme axial ligand environment in E75 and the Rev-erb(s), which contain a distinctive Cys(thiolate)/His coordination of the Fe(III) heme, strongly suggests that the heme is intended to possess a more selective function. While cysteine(thiolate) ligation by itself is not unique, low-spin, 6-coordinate thiolate-ligated hemes have a unique instability in the Fe(II) state. This instability results in ligand switching upon reduction, which may trigger conformational changes that poise the heme to bind a regulatory gas molecule. In E75 and the Rev-erbs, the conserved histidine ligand (His574, E75; His602, Rev-erbα; His568, Rev-erbβ) is located near the C-terminus (helix 11). This ligand appears to remain bound to the heme in all redox and gas-bound states of Rev-erbβ and all but the NO-bound state of E75. In contrast, the Cys ligand present in the Fe(III) state is replaced in the Fe(II) state by an alternate neutral ligand. Based on published alignments, conserved histidine residues that may be implicated in ligand switching in E75 (Cys396 → His364/416) are located ≥ 20 residues from Cys396 (6). Alternatively, Pardee, et al. (24) note the proximity of His381 of Rev-erbβ to the Cys384 ligand, and propose that this histidine might be an Fe(II) ligand candidate. Other potential ligand amino acids occur within reasonable distance of the heme in the Rev-erbβ(LBD) structure, and since the conformational changes that occur upon ligand switching and/or gas binding are not known, amino acids outside the LBD may also be potential ligands. The NPAS2 transcription factor, which regulates circadian rhythm in conjunction with its partner BMAL1, is a heme-thiolate protein that also undergoes a redox-mediated ligand switch (Cys170 → His171) (66, 72). DNA binding by the NPAS2-BMAL1 heterodimer is dependent on the levels of heme present; furthermore, CO inhibits the heterodimerization of NPAS2-BMAL1, thus preventing DNA binding. Interestingly, NPAS2 and the Rev-erbs all function in regulation of the circadian clock, and Rev-erbs also regulate the expression of BMAL1 (17, 18). The fact that these proteins exhibit similar heme coordination characteristics, show similar gas binding properties and have complementary roles in the circadian system, seems likely to be relevant to mechanisms by which these systems are regulated.
CONCLUSION
The nuclear receptors E75 and Rev-erbβ are heme proteins whose coordination characteristics are similar to other known redox and gas sensors. The Fe(III) proteins are coordinated by a cysteine(thiolate), which is lost from the heme coordination sphere when the heme is reduced. The Fe(II) proteins bind at least one histidine residue, and are susceptible to reaction with both CO and NO. These characteristics suggest that these nuclear receptors may function in heme-, gas- or redox-mediated regulation of key processes in homeostasis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Thomas C. Brunold and his group (UW-Madison) for generous use of their instrumentation and Ms. Darci Block (NDSU) for helpful peak-fitting advice. The authors also thank Dr. Frank Neese (MPI Mülheim) for supplying free copies of the WEPR spectral fitting program.
This work was supported by N.I.H. grant HL-66147 (J.N.B.) and N.C.I.C. grant 010200 (H.M.K.)
ABBREVIATIONS
- BMAL1
brain and muscle Arnt-like protein-1, a transcription factor
- hCBS
human cystathionine β-synthase
- CO
carbon monoxide
- ChCooA
a CooA homologue from the thermophilic bacterium Carboxydothermus hydrogenoformans
- RrCooA
a CO-sensing transcription factor in Rhodospirillum rubrum
- E75(261)
a ~261 amino acid construct of the ligand binding domain of Drosophila melanogaster nuclear receptor E75 studied by de Rosny et al. (23)
- E75(LBD)
a 262 amino acid construct of the ligand binding domain of Drosophila melanogaster nuclear receptor E75 studied in this work
- EcDos
direct oxygen sensor in Escherichia coli
- EPPS
3-[4-(2-hydroxyethyl)-1-piperazinyl]propanesulfonic acid
- EPR
electron paramagnetic resonance
- FixL
an O2-sensing heme protein that regulates gene expression associated with N2 fixation
- HemAT
heme-based aerotactic transducer from Bacillus subtilis, an oxygen sensor
- HRP
horseradish peroxidase
- M80C cyt c
cytochrome c variant in which methionine is replaced by cysteine(thiolate) as an axial ligand to the Fe(III) heme
- MCD
magnetic circular dichroism
- MOPS
3-(N-morpholino)propanesulfonic acid
- NO
nitric oxide
- NPAS2
neuronal PAS domain protein 2, a CO-sensing heme protein and eukaryotic transcriptional regulator
- P450CAM
cytochrome P450 from camphor-hydroxylating Pseudomonas putida
- P450CAM+ImH
P450CAM with imidazole as an axial ligand to the Fe(III) heme
- PAS
a domain structure named for the proteins in which the motif was first identified: Period, aryl hydrocarbon receptor nuclear translocator, and Simple-minded
- Rev-erbα(LBD)
a 242 amino acid construct of the ligand binding domain of Homo sapiens nuclear receptor Rev-erbα studied in this work
- Rev-erbβ(LBD)
a 256 amino acid construct of the ligand binding domain of Homo sapiens nuclear receptor Rev-erbβ studied in this work
- sGC
soluble guanylyl cyclase an NO-sensing heme protein that catalyzes the conversion of GTP to cGMP
- Tris
tris(hydroxymethyl)aminomethane
Footnotes
SUPPORTING INFORMATION AVAILABLE
Tables supporting spectral interpretation (S1–S5). Figures showing the MCD spectra of Fe(III)Rev-erbβ(LBD) (S1) and Fe(III)E75(LBD) (S2), the rR spectra of Fe(III) and Fe(II) E75(LBD) (S3), electronic absorption and MCD spectra of Fe(II)CO E75(LBD) (S4), rR spectra of Fe(II)12/13CO E75(LBD) (S5), correlation plot of ν(Fe-CO) versus ν(C-O) (S6) and correlation plot of ν(Fe-NO) versus ν(N-O) (S7), and electronic absorption spectra of Fe(II)CO (S8) and Fe(II)NO Rev-erbα(LBD) (S9). Parameters used to fit the EPR spectrum of Figure 2. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escriva H, Safi R, Hanni C, Langlois MC, Saumitou-Laprade P, Stehelin D. Ligand binding was acquired during evolution of nuclear receptors. Proc. Natl Acad. Sci. USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowrey PL, Takahashi JS. Genetics of the mammalian circadian system: Photic entrainment, circadian pacemaker mechanisms, and posttranslational regulation. Annu. Rev. Genet. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- 4.Renaud JP, Moras D. Structural studies on nuclear receptors. Cell. Mol. Life Sci. 2000;57:1748–1769. doi: 10.1007/PL00000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weatherman RV, Fletterick RJ, Scanlan TS. Nuclear-receptor ligands and ligand-binding domains. Annu. Rev. Biochem. 1999;68:559–581. doi: 10.1146/annurev.biochem.68.1.559. [DOI] [PubMed] [Google Scholar]
- 6.Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, Krause HM. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- 8.Bialecki M, Shilton A, Fichtenberg C, Segraves WA, Thummel CS. Loss of the ecdysteroid-inducible E75A orphan nuclear receptor uncouples molting from metamorphosis in Drosophila. Dev. Cell. 2002;3:209–220. doi: 10.1016/s1534-5807(02)00204-6. [DOI] [PubMed] [Google Scholar]
- 9.D’vino PP, Thummel CS. Crooked legs encodes a family of zinc finger proteins required for leg morphogenesis and ecdysone-regulated gene expression during Drosophila metamorphosis. Development. 1998;125:1733–1745. doi: 10.1242/dev.125.9.1733. [DOI] [PubMed] [Google Scholar]
- 10.Dubrovskaya VA, Berger EM, Dubrovsky EB. Juvenile hormone regulation of the E75 nuclear receptor is conserved in Diptera and Lepidoptera. Gene. 2004;340:171–177. doi: 10.1016/j.gene.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Dumas B, Harding HP, Choi HS, Lehmann KA, Chung M, Lazar MA, Moore DD. A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol. Endocrinol. 1994;8:996–1005. doi: 10.1210/mend.8.8.7997240. [DOI] [PubMed] [Google Scholar]
- 12.Lazar MA, Hodin RA, Darling DS, Chin WW. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol. Cell. Biol. 1989;9:1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delerive P, Chin WW, Suen CS. Identification of Reverb(alpha) as a novel ROR(alpha) target gene. J. Biol. Chem. 2002;277:35013–35018. doi: 10.1074/jbc.M202979200. [DOI] [PubMed] [Google Scholar]
- 14.Adelmant G, Begue A, Stehelin D, Laudet V. A functional Rev-erb α responsive element located in the human Rev-erb α promoter mediates a repressing activity. Proc. Natl. Acad. Sci. USA. 1996;93:3553–3558. doi: 10.1073/pnas.93.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamir I, Zhang J, Lazar MA. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 16.Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol. Cell. Biol. 1995;15:4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 18.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 19.Renaud JP, Harris JM, Downes M, Burke LJ, Muscat GE. Structure-function analysis of the Rev-erbA and RVR ligand-binding domains reveals a large hydrophobic surface that mediates corepressor binding and a ligand cavity occupied by side chains. Mol. Endocrinol. 2000;14:700–717. doi: 10.1210/mend.14.5.0444. [DOI] [PubMed] [Google Scholar]
- 20.Hu X, Lazar MA. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol. Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- 21.Woo E-J, Jeong DG, Lim M-Y, Kim SJ, Kim K-J, Yoon S-M, Park B-C, Ryu SE. Structural insight into the constitutive repression function of the nuclear receptor Rev-erbβ. J. Mol. Biol. 2007;373:735–744. doi: 10.1016/j.jmb.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 22.Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, et al. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol. Endocrinol. 2005;19:1125–1134. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- 23.de Rosny E, Groot Ad, Jullian-Binard C, Gaillard J, Borel F, Pebay-Peyroula E, Fontecilla-Camps JC, Jouve HlnM. Drosophila nuclear receptor E75 is a thiolate hemoprotein. Biochemistry. 2006;45:9727–9734. doi: 10.1021/bi060537a. [DOI] [PubMed] [Google Scholar]
- 24.Pardee KI, Xu X, Reinking J, Schuetz A, Dong A, Liu S, Zhang R, Tiefenbach J, Lajoie G, Plotnikov AN, Botchkarev A, Krause HM, Edwards A. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBβ. Public Library of Science. 2009;7:0382–0398. doi: 10.1371/journal.pbio.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-Erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 26.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nature Struct. & Molec. Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neese F. Electronic structure and spectroscopy of novel copper chromophores in biology. Konztanz, Germany: University of Konztanz; 1997. [Google Scholar]
- 28.Spiro TG. Resonance Raman spectroscopic studies of heme proteins. Biochim. Biophys. Acta. 1975;416:169–189. doi: 10.1016/0304-4173(75)90006-3. [DOI] [PubMed] [Google Scholar]
- 29.Spiro TG, Strekas TC. Resonance Raman spectra of heme proteins Effects of oxidation and spin state. J. Am. Chem. Soc. 1974;96:338–345. doi: 10.1021/ja00809a004. [DOI] [PubMed] [Google Scholar]
- 30.Abe M, Kitagawa T, Kyogoku Y. Resonance Raman spectra of octaethylporphyrinato-Ni(II) and meso-deuterated and 15N substituted derivatives. II. A normal coordinate analysis. J. Chem. Phys. 1978;69:4526–4534. [Google Scholar]
- 31.Choi S, Spiro TG, Langry KC, Smith KM. Vinyl influences on protoheme resonance Raman spectra: Nickel(II) protoporphyrin IX with deuterated vinyl groups. J. Am. Chem. Soc. 1982;104:4337–4344. [Google Scholar]
- 32.Dawson JH, Andersson LA, Sono M. Spectroscopic investigations of ferric cytochrome P-450CAM ligand complexes. J. Biol. Chem. 1982;257:3606–3617. [PubMed] [Google Scholar]
- 33.Thomson AJ, Johnson MK. Magnetization curves of haemoproteins measured by low-temperature magnetic circular dichroism spectrosopy. Biochem. J. 1980;191:411–420. doi: 10.1042/bj1910411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omura T, Hiroyuki Sadano, Tohru Hasegawa, Yuzo Yoshida, Shiro Kominami. Hemoprotein H-450 identified as a form of cytochrome P-450 having an endogenous ligand at the 6th coordination position of the heme. J. Biochem. 1996;96:1491–1500. doi: 10.1093/oxfordjournals.jbchem.a134978. [DOI] [PubMed] [Google Scholar]
- 35.Pazicni S, Lukat-Rodgers GS, Oliveriusova J, Rees KA, Parks RB, Clark RW, Rodgers KR, Kraus JP, Burstyn JN. The redox behavior of the heme is cystathionine β-synthase is sensitive to pH. Biochemistry. 2004;43:14684–14695. doi: 10.1021/bi0488496. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds MF, Shelver D, Kerby RL, Parks RB, Roberts GP, Burstyn JN. EPR and electronic absorption spectroscopies of the CO-sensing CooA protein reveal a cysteine-ligated low-spin ferric heme. J. Am. Chem. Soc. 1998;120:9080–9081. [Google Scholar]
- 37.Pazicni S. Towards understanding the role of the heme cofactor in cystathionine β-synthase. Madison, Wisconsin: University of Wisconsin-Madison; 2006. [Google Scholar]
- 38.Blumberg WE, Peisach J. Low-spin compounds of heme proteins. Adv. Chem. Ser. 1971;100:271–291. [Google Scholar]
- 39.Desbois A, Lutz M, Banerjee R. Low-frequency vibrations in resonance Raman spectra of horse heart myoglobin. Iron-ligand and iron-nitrogen vibration models. Biochemistry. 1979;18:1510–1518. doi: 10.1021/bi00575a019. [DOI] [PubMed] [Google Scholar]
- 40.Champion PM, Stallard BR, Wagner GC, Gunsalus IC. Resonance Raman detection of an Fe-S bond in cytochrome P450CAM. J. Am. Chem. Soc. 1982;104:5469–5472. [Google Scholar]
- 41.Green EL, Taoka S, Banerjee R, Loehr TM. Resonance Raman characterization of the heme cofactor in cystathionine β-synthase. Identification of the Fe-S(Cys) vibration in the six-coordinate low-spin heme. Biochemistry. 2001;40:459–463. doi: 10.1021/bi0010874. [DOI] [PubMed] [Google Scholar]
- 42.Dawson JH, Sono M. Cytochrome P-450 and chloroperoxidase: Thiolate-ligated heme enzymes. Spectroscopic determination of their active site structures and mechanistic implications of thiolate ligation. Chem. Rev. 1987;87:1255–1276. [Google Scholar]
- 43.Roberts GP, Kerby RL, Youn H, Conrad M. CooA, a paradigm for gas sensing regulatory proteins. J. Inorg. Biochem. 2005;99:280–292. doi: 10.1016/j.jinorgbio.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 44.Burstyn JN, Yu AE, Dierks EA, Hawkins BK, Dawson JH. Studies of the heme coordination and ligand binding properties of soluble guanylyl cyclase(sGC): Characterization of Fe(II)sGC and Fe(II)sGC(CO) by electronic absorption and magnetic circular dichroism spectroscopies and failure of CO to activate the enzyme. Biochemistry. 1995;34:5896–5903. doi: 10.1021/bi00017a019. [DOI] [PubMed] [Google Scholar]
- 45.Chouchane S, Girotto S, Kapetanaki S, Schelvis JPM, Yu S, Magliozzo RS. Analysis of heme structural heterogeneity in Mycobacterium tuberculosis catalase-peroxidase (KatG) J. Biol. Chem. 2003;278:8154–8162. doi: 10.1074/jbc.M208256200. [DOI] [PubMed] [Google Scholar]
- 46.Clark RW, Youn H, Parks RB, Cherney MM, Roberts GP, Burstyn JN. Investigation of the role of the N-terminal proline, the distal ligand in the CO sensor CooA. Biochemistry. 2004;43:14149–14160. doi: 10.1021/bi0487948. [DOI] [PubMed] [Google Scholar]
- 47.Dhawan IK, Shelver D, Thorsteinsson MV, Roberts GP, Johnson MK. Probing the heme axial ligation in the CO-sensing CooA protein with magnetic circular dichroism spectroscopy. Biochemistry. 1999;38:12805–12813. doi: 10.1021/bi991303c. [DOI] [PubMed] [Google Scholar]
- 48.Brittain T, Greenwood C, Springall J, Thomson AJ. The nature of ferrous haem protein complexes prepared by photolysis. Biochim. Biophys. Acta. 1982;703:117–128. [Google Scholar]
- 49.Tsubaki M, Srivastava RB, Yu N-T. Resonance Raman investigation of carbon monoxide bonding in (carbonmonoxy)hemoglobin and -myoglobin: Detection of Fe-CO stretching and Fe-C-O bending vibrations and influence of quaternary structure change. Biochemistry. 1982;21:1132–1140. doi: 10.1021/bi00535a004. [DOI] [PubMed] [Google Scholar]
- 50.Das TK, Friedman JM, Kloek AP, Goldberg DE, Rousseau DL. Origin of the anomalous Fe-CO stretching mode in the CO complex of Ascaris hemoglobin. Biochemistry. 2000;39:837–842. doi: 10.1021/bi9922087. [DOI] [PubMed] [Google Scholar]
- 51.Das TK, Lee HC, Duff SMG, Peisach RDH, Rousseau DL, Wittenberg BA, Wittenberg JB. The heme environment in barley hemoglobin. J. Biol.Chem. 1999;274:4207–4212. doi: 10.1074/jbc.274.7.4207. [DOI] [PubMed] [Google Scholar]
- 52.Ray GB, Li X-Y, Ibers JA, Sessler JL, Spiro TG. How far can proteins bend the FeCO unit? Distal polar and steric effects in heme proteins and models. J. Am. Chem. Soc. 1994;116:162–176. [Google Scholar]
- 53.Vogel KM, Kozlowski PM, Zgierski MZ, Spiro TG. Determinants of the FeXO [X=C,N,O] vibrational frequencies in heme adducts from experiment and density functional theory. J. Am. Chem. Soc. 1999;121:9915–9921. [Google Scholar]
- 54.Stone JR, Sands RH, Dunham WR, Marletta MA. Electron paramagnetic resonance evidence for the formation of a pentacoordinate nitrosyl-heme complex in soluble guanylate cyclase. Biochem. Biophys. Res. Commun. 1995;207:572–577. doi: 10.1006/bbrc.1995.1226. [DOI] [PubMed] [Google Scholar]
- 55.Reynolds MF, Parks RB, Burstyn JN, Shelver D, Thorsteinsson MV, Kerby RL, Roberts GP, Vogel KM, Spiro TG. Electronic absorption, EPR and resonance Raman spectroscopies of CooA, a CO-sensing transcription factor from R. rubrum, reveals a five-coordinate NO-heme. Biochemistry. 2000;39:388–396. doi: 10.1021/bi991378g. [DOI] [PubMed] [Google Scholar]
- 56.Morse RH, Chan SI. Electron paramagnetic resonance studies of nitrosyl ferrous heme complexes: Determination of an equilibrium between two conformations. J. Biol. Chem. 1980;255:7876–7882. [PubMed] [Google Scholar]
- 57.Mackin HC, Benko B, Yu NT, Gersonde K. Resonance Raman study on pentacoordinated and hexacoordinated ferrous nitrosyl myoglobin. The influence of pH. FEBS Lett. 1983;158:199–202. [Google Scholar]
- 58.Ibrahim M, Xu C, Spiro TG. Differential sensing of protein influences by NO and CO vibrations in heme adducts. J. Am. Chem. Soc. 2006;128:16834–16845. doi: 10.1021/ja064859d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsiftsoglou AS, Tsamadoua AI, Papadopoulou LC. Heme as key regulator of major mammalian cellular functions: Molecular, cellular, and pharmacological aspects. Pharmacol. Ther. 2006;111:327–345. doi: 10.1016/j.pharmthera.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 60.Bredt DS, Snyder SH. Nitric oxide: A physiological messenger molecule. Annu. Rev. of Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- 61.Ryter SW, Otterbein LE. Carbon monoxide in biology and medicine. Bioessays. 2004;26:270–280. doi: 10.1002/bies.20005. [DOI] [PubMed] [Google Scholar]
- 62.Jaffrey SR, Snyder SH. Nitric oxide: A neural messenger. Annu. Rev.Cell Dev. Biol. 1995;11:417–440. doi: 10.1146/annurev.cb.11.110195.002221. [DOI] [PubMed] [Google Scholar]
- 63.Artinian LR, Ding JM, Gillette MU. Carbon monoxide and nitric oxide: interacting messengers in muscarinic signaling to the brain’s circadian clock. Exp. Neurol. 2001;171:293–300. doi: 10.1006/exnr.2001.7781. [DOI] [PubMed] [Google Scholar]
- 64.Ding JM, Chen D, Weber ET, Faiman LE, Rea MA, Gillette MU. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science. 1994;266:1713–1717. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- 65.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 66.Uchida T, Sato E, Sato A, Sagami I, Shimizu T, Kitagawa T. CO-dependent activity-controlling mechanism of heme-containing CO-sensor protein, neuronal PAS domain protein 2. J. Biol. Chem. 2005;280:21358–21368. doi: 10.1074/jbc.M412350200. [DOI] [PubMed] [Google Scholar]
- 67.Traylor TG, Sharma VS. Why NO? Biochemistry. 1992;31:2847–2849. doi: 10.1021/bi00126a001. [DOI] [PubMed] [Google Scholar]
- 68.Ignarro LJ, Degnan JN, Baricos WH, Kadowitz PJ, Wolin MS. Activation of purified guanylate cyclase by nitric oxide requires heme: Comparison of the heme-deficient, heme-reconstituted and heme-containing forms of soluble enzyme from bovine lung. Biochim. Biophys. Acta. 1982;718:49–59. doi: 10.1016/0304-4165(82)90008-3. [DOI] [PubMed] [Google Scholar]
- 69.Chen J-J, London IM. Regulation of protein synthesis by heme-regulated eIF-2α kinase. Trends Biochem. Sci. 1995;20:105–108. doi: 10.1016/s0968-0004(00)88975-6. [DOI] [PubMed] [Google Scholar]
- 70.Uma S, Yun BG, Matts RL. The heme-regulated eukaryotic initiation factor 2 alpha kinase - A potential regulatory target for control of protein synthesis by diffusible gases. J. Biol. Chem. 2001;276:14875–14883. doi: 10.1074/jbc.M011476200. [DOI] [PubMed] [Google Scholar]
- 71.Clark RW, Lanz ND, Lee AJ, Kerby RL, Roberts GP, Burstyn JN. Unexpected NO-dependent DNA binding by the CooA homolog from Carboxydothermus hydrogenoformans. Proc. Natl. Acad. Sci. USA. 2006;103:891–896. doi: 10.1073/pnas.0505919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 73.Lu Y, Casimiro DR, Bren KL, Richards JH, Gray HB. Structurally engineered cytochromes with unusual ligand-binding properties: expression of Saccharomyces cerevisiae Met-80-Ala iso-1-cytochrome c. Proc. Natl. Acad. Sci. USA. 1993;90:11456–11459. doi: 10.1073/pnas.90.24.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rux JJ, Dawson JH. Magnetic circular dichroism spectroscopy as a probe of axial heme ligand replacement in semisynthetic mutants of cytochrome c. FEBS Lett. 1991;290:49–51. doi: 10.1016/0014-5793(91)81222-t. [DOI] [PubMed] [Google Scholar]
- 75.Lipscomb JD. Electron paramagnetic resonance detectable states of cytochrome P-450CAM. Biochemistry. 1980;19:3590–3599. doi: 10.1021/bi00556a027. [DOI] [PubMed] [Google Scholar]
- 76.Wuttke DS. Preparation, characterization and intramolecular electron transfer studies of ruthenium-modified cytochromes c. Pasadena, California: California Institute of Technology; 1993. pp. 408–413. [Google Scholar]
- 77.Gonzalez G, Dioum EM, Bertolucci CM, Tomita T, Ikeda-Saito M, Cheesman MR, Watmough NJ, Gillez-Gonzalez M-A. Nature of the displaceable heme-axial residue in the EcDos protein, a heme-based sensor from Escherichia coli. Biochemistry. 2002;41:8414–8421. doi: 10.1021/bi025845x. [DOI] [PubMed] [Google Scholar]
- 78.Boiss-Poltoratzsky R, Ehrenberg A. Magnetic and spectrophotometric investigations of cytochrome b5. Eur. J. Biochem. 1967;2:361–365. doi: 10.1111/j.1432-1033.1967.tb00146.x. [DOI] [PubMed] [Google Scholar]
- 79.Nistor SV, Goovaerts E, Van Doorslaer S, Dewilde S, Moens L. His2013Fe III2013His EPR-spectroscopic evidence of a dominant His–Fe III–His coordination in ferric neuroglobin Chem. Phys. Lett 2002361355–361. [Google Scholar]
- 80.Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez M-A. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 2000;39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- 81.Dewilde S, Kiger L, Burmester T, Hankeln T, Baudin-Creuza V, Aerts T, Marden MC, Caubergs R, Moens L. Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J. Biol. Chem. 2001;276:38949–38955. doi: 10.1074/jbc.M106438200. [DOI] [PubMed] [Google Scholar]
- 82.Sharonov YA, Mineyev AP, Livshitz MA, Sharonova NA, Zhurkin VB, Lysov YP. Magnetic circular dichroism studies of myoglobin, hemoglobin and peroxidase at room and low temperatures. Ferrous high spin derivatives. Biophys. Struct. Mechanism. 1978;4:139–158. doi: 10.1007/BF00539228. [DOI] [PubMed] [Google Scholar]
- 83.Hirata S, Matsui T, Sasakura Y, Sugiyama S, Yoshimura T, Sagami I, Shimizu T. Characterization of Met95 mutants of a heme-regulated phosphodiesterase from Escherichia coli. Optical absorption, magnetic circular dichroism, circular dichroism, and redox potentials. Eur. J. Biochem. 2003;270:4771–4779. doi: 10.1046/j.1432-1033.2003.03879.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.