Abstract
For magnetic resonance spectroscopic imaging (MRSI) studies of the brain it is important to measure the distribution of metabolites in a regionally unbiased way - that is without restrictions to apriori defined regions of interest (ROI). Since MRSI provides measures of multiple metabolites simultaneously at each voxel, there is furthermore great interest in utilizing the multidimensional nature of MRSI for gains in statistical power. Voxelwise multivariate statistical mapping is expected to address both of these issues but it has not been previously employed for SI studies of brain. The aims of this study were to: 1) develop and validate multivariate voxel based statistical mapping for MRSI and 2) demonstrate that multivariate tests can be more powerful than univariate tests in identifying patterns of altered brain metabolism. Specifically, we compared multivariate to univariate tests in identifying known regional patterns in simulated data and regional patterns of metabolite alterations due to amyotrophic lateral sclerosis, a devastating brain disease of the motor neurons.
INTRODUCTION
For many magnetic resonance spectroscopic imaging (MRSI) studies of the brain it is important to measure the distribution of metabolites in a regionally unbiased way - that is without restrictions to apriori defined regions of interest (ROI). Since MRSI provides measures of multiple metabolites simultaneously at each voxel, there is furthermore great interest in utilizing the multidimensional nature of MRSI for gains in statistical power. Voxelwise multivariate statistical analysis is expected to address both issues, and the aims of this study were to: 1) develop and validate a multivariate voxel based statistical mapping for MRSI and 2) demonstrate that multivariate tests can be more powerful than univariate tests in identifying patterns of altered brain metabolism.
Statistical methods for assessing voxelwise variations in sets of co-registered brain images for a number of MR imaging modalities have been proposed and utilized (2). This includes the relatively recent introduction of a number of multivariate voxel based methods (3), for simultaneous assessment of several measures of brain anatomy and function at each voxel. Region of interest (ROI) based multivariate methods have been utilized for analysis of vector deformations to warp one brain image to another (4), diffusion tensor imaging (5) and functional MRI (3). MRSI data provide spectra of several brain metabolites simultaneously at each voxel and thus present multivariate data by design. Though interesting and useful approaches to multivariate analysis have been proposed for MRSI including: image segmentation (6), tumor classification (7), disease progression (8) and treatment outcome in mouse models of Alzheimer’s disease (9), the authors are unaware of any MRSI studies of human brain that have applied standard voxel based multivariate analysis. Given that a number of neurological pathologies may result in independent changes of multiple metabolite concentrations the availability of sensitive, regionally unbiased ways of assessing all metabolic information provided by MRSI measurements is important. In particular, a multivariate analysis may reveal and exploit meaningful relationships between the metabolites that univariate analyses would miss. This study reports on a voxel based multivariate analysis package that has been developed within the MIDAS (Metabolite Image Data Analysis System) project (1), which provides comprehensive MRI and MRSI data processing and analysis functions for proton MRSI studies of the brain.
There are a number of situations in which a multivariate voxel based analysis could significantly add to the power of MRSI studies. For example, in several neurological disorders, including Alzheimer’s disease (AD) and amyotrophic lateral sclerosis (ASL), neuronal damage can be accompanied by alterations of cell membranes and gliosis (for reviews see (10, 11). These molecular alterations could give rise to correlations between decreasing N-acetylaspartate (NAA) levels (a putative marker of neuronal integrity) and increased choline (a constituent of membrane lipids) and myo-inositol (a potential glial marker) resonances. In addition as more powerful methods are developed for the detection and quantification of low signal to noise metabolites such as glutamate, glutamine, GABA, and glutathione (12, 13) at high magnetic field, multivariate methods could significantly aid in the use of MRSI for detection of subtle changes in brain metabolism.
This report focuses on a description of the MIDAS statistical package and its use for univariate and multivariate voxel based analysis of MRSI data. Specifically, we compared multivariate to univariate tests under known conditions for simulated data as well as to those due to ALS, in which altered metabolite concentrations were observed in the motor cortex and along the known pathways of motor neuron fibers.
SUBJECTS AND METHODS
Simulations
Simulation studies were performed to compare the efficacy of univariate and multivariate methods in a setting that was realistic but where the effects of varying metabolite levels could be considered independently for the purposes of the comparison. The data used for this study was slightly modified versions of MRSI data obtained from 8 normal subjects, and processed as part of the MIDAS project (1). These proton MRSI studies of the brain were carried out at 3 Tesla (Siemens Trio), and used a volumetric spin-echo acquisition with two-dimensional phase encoding and echo-planar readout. The acquisition used frequency-selective water suppression, lipid inversion nulling, TE = 70 ms, and TR = 1710 ms. After correction for oversampling in the readout spatial and spectral dimensions the resultant image was equivalent to 50×50 voxels in-plane and 18 slices, over a field-of-view (FOV) of 280×280×180 mm, with selection of a slab of 135 mm covering the cerebrum. The effective spatial resolution of MRSI was about 0.6 cc (0.31 cc nominal). Reconstructions of metabolite images for N-acetylaspartate (NAA), total creatine (Cre), and total choline (Cho) were carried out in a fully-automated manner using the MIDAS package (1), which included interpolation to 64×64×32 points, signal intensity normalization using tissue water as an internal reference, k-space extrapolation (14) to reduce ringing artifacts from subcutaneous lipids, and spatial transformation to a common spatial reference, for which the BrainWeb simulated MRI (15) was used as the target. This spatial normalization included interpolation to 4 mm isotropic voxels. The processing also included generation and spatial transformation of tissue segmentation maps for grey-matter white-matter, and CSF.
The MRSI and T1 data from the 8 normal subjects were split into 2 groups, the first considered the “control” group and a second group for which levels of NAA and Ch were varied only in posterior gray matter in successively larger amounts to generate 16 simulated sets with 4 unaffected “control” images and 4 “affected” images per set. The percentage of gray matter in the MRSI voxels that was used to determine the appropriate metabolite variations was obtained from the co-registered T1 images. After the metabolite variations were applied a Gaussian smoothing filter was applied to the altered regions in the metabolite map to simulate realistic metabolite distributions.
The control group consisted of 2 males and 2 females [age: mean ± SD of 36±8 and range of 27–43] and the second group consisted of 2 males and 2 females [age: mean ± SD of 34±13 and range of 23–51].
Between group univariate voxelwise ANOVA for the individual metabolites and between group multivariate voxelwise MANOVA for the combined metabolites were performed using the modified metabolite maps and the MIDAS statistics package described below in the Statistics section.
ALS study
To determine the gains in power provided by multivariate statistical mapping in a clinical setting, univariate and multivariate statistical mapping was performed on MRSI data acquired from 12 male subjects diagnosed with definite ALS [age: mean ± SD of 57±9 and range of 36–65; disease duration since diagnosis: range of 0–82 months; ALS type: 3 bulbar and 9 limbic] and 12 male controls [age: mean ± SD of 37±13 and range of 23–59]. The acquisition and processing of the data was identical to that described above. The level of statistical significance was fixed at a false discovery rate (FDR) threshold of 0.05 (discussed below) for all tests.
All human subjects gave written informed consent before participating in the study, which was approved by the Institutional Review Board of the University of Miami and the University of California in San Francisco.
STATISTICS
A statistical package that utilizes statistical libraries available for the R statistical language (16), including the AnalyzeFMRI package (17) and the fdrtool package (18), was developed and integrated into the MIDAS package. Among other applications, it provides for voxel based multivariate analysis of covariance (MANCOVA). For this function the observations for each voxel are first arranged into a N×q matrix Y, where N is the number of subjects and q the number of multivariate measures, for example NAA, Cho and Cr. Similarly the explanatory variables are arranged into a N×s matrix X, where s is the number of contrasts, e.g. diagnosis, age, sex, etc. Nuisance variables, such as constant terms or gray- and white-matter partial volumes of the MRSI voxel, can be arranged into a N×r matrix Z, where r is the number of nuisance variables. The objective is to relate Y to X, accounting for Z. One approach is to formalize this using a multivariate linear model according to
Here, A and B are s×q and r×q matrices, respectively of the unknown coefficients and Z is a matrix of unknown coefficients related to the nuisance variables. Ξis a N×q matrix of independent, zero mean, unit variance Gaussian errors and Σ is an unknown q×q covariance matrix of the q random variates. Formally, our objective is to test the null hypothesis that A = 0.
While there are currently a number of approaches to addressing the multiple comparison problem that arises when testing a large number of voxels (2), including Random Field Theory based and nonparametric methods (2,3), the most convenient in the context of multivariate analysis in the R statistical package for MIDAS was to use a false discovery rate (FDR) threshold via the fdrtool package (18). While the different methods constitute statistical tests with different interpretations and should each be considered in the particular context of a given study, it seemed reasonable to assume that a general study of the efficacy of multivariate approaches to analysis of MRSI data would be relatively insensitive to this choice. (19, 20).
MIDAS provides a convenient graphical user interface to the statistical package which allows researchers to conveniently design multivariate imaging studies when providing a simple comma separated data file containing clinical variables and image file names.
RESULTS
Simulations
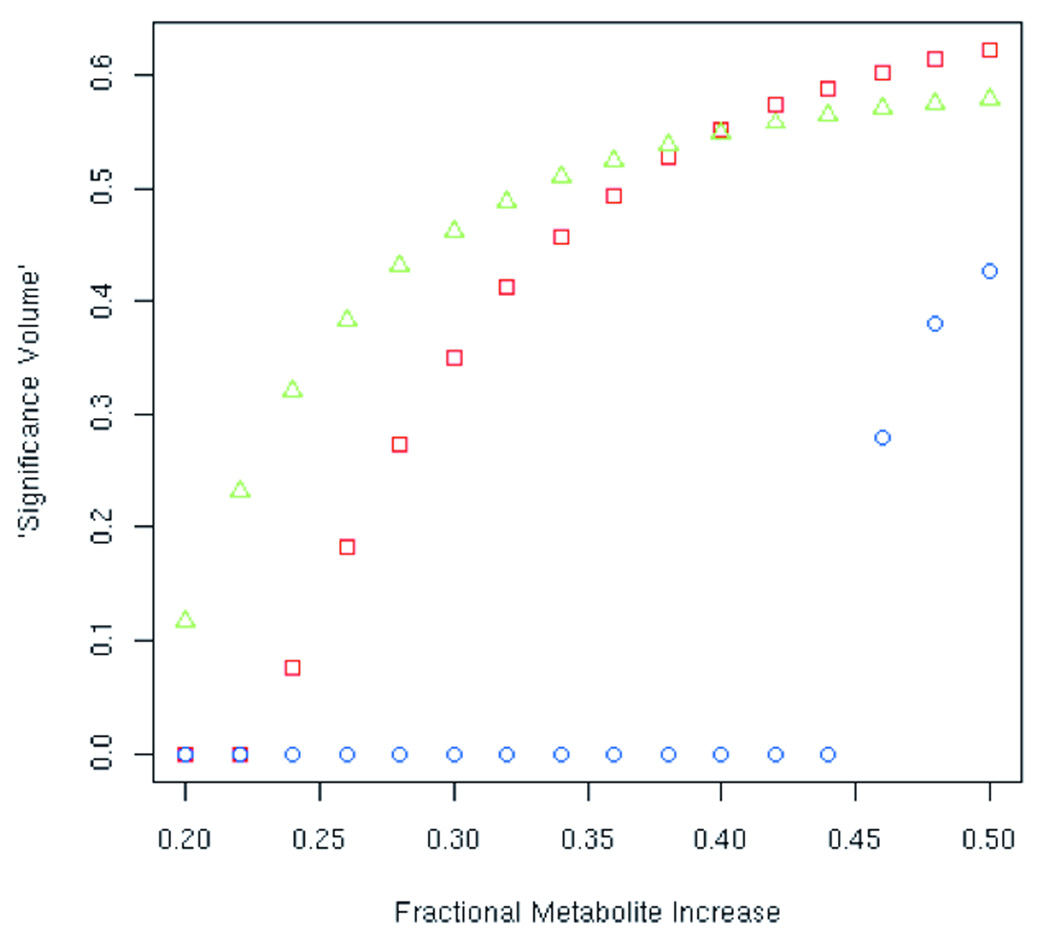
In Figure (1) is shown a comparison of the results obtained for multivariate (MANCOVA) and univariate (ANCOVA) analysis between two sets of images, i.e. “normal” images and those with increasing grey-matter metabolite levels, as previously described. For the simulations in the multivariate case the measures were NAA and Cho amplitudes in each voxel, and in the univariate cases either NAA or Cho amplitudes. For these analyses the single contrast was the binary variable indicating whether the subject was either “normal” or not. So for the simulations the analyses were really just the special cases of MANOVA and ANOVA analyses (i.e. no covariates were considered). The x-axis indicates the fractional change in the simulated posterior gray matter metabolite levels, i.e. for a given value×on the x-axis the metabolite levels in all voxels in the affected region (posterior gray matter) were increased by a factor 1+x relative to the values in the original metabolite images. The y-axis indicates an estimated “significance volume”, that is, the quantity1-p summed over all voxels in the image volume, where p is the standard significance level obtained from the MANCOVA analysis in the multivariate case and the ANVCOVA analysis in the univariate cases. For a large enough simulated increase in metabolite levels, all of the altered voxels would appear highly significant, that is, p ~ 0 and 1-p ~ 1, and the “significance volume” would equal the fraction of the total volume occupied by the altered voxels. It should be noted that though this did not appear to be the case in the simulation, voxels other than the altered voxels could by chance appear to be significant.
Figure 1.
Comparison of multivariate and univariate analysis of images via plot of “significance volume” as a function of simulated increased metabolite levels, showing crossover point at which multivariate analysis provides more power.
This result demonstrates that the univariate NAA analysis initially shows the highest proportion of significantly altered voxels for small increases in the metabolite levels, while the univariate Cho analysis shows no significantly altered voxels despite the fact that the levels of both NAA and Cho were raised by the same fraction. This difference is due to the small sample size used for the simulations and the larger variance of Cho levels, which is known to be present in these brain metabolite images (21). The fact that Cho shows a larger variability than NAA (and a number of other brain metabolites) was studied systematically via a reproducibility study and attributed to biological effects (22). At intermediate metabolite increases, despite the fact that the univariate Cho analysis continues to exhibit no voxels with significant increases, the multivariate analysis shows increased proportions of significantly altered voxels that eventually surpass the univariate NAA analysis. This demonstrates that if a joint metabolite effect is suspected a multivariate analysis could, based on the relative strengths of the individual metabolite effects, have more power to detect between-group differences than either of the univariate tests. In fact one or more of the univariate tests could indicate no effect. Finally, at a certain level of increased metabolite there is enough of a change in the Cho amplitude for the univariate analysis to pick up, though beyond that point the multivariate analysis is clearly more sensitive than either of the univariate tests.
Analysis of Metabolic Changes in ALS
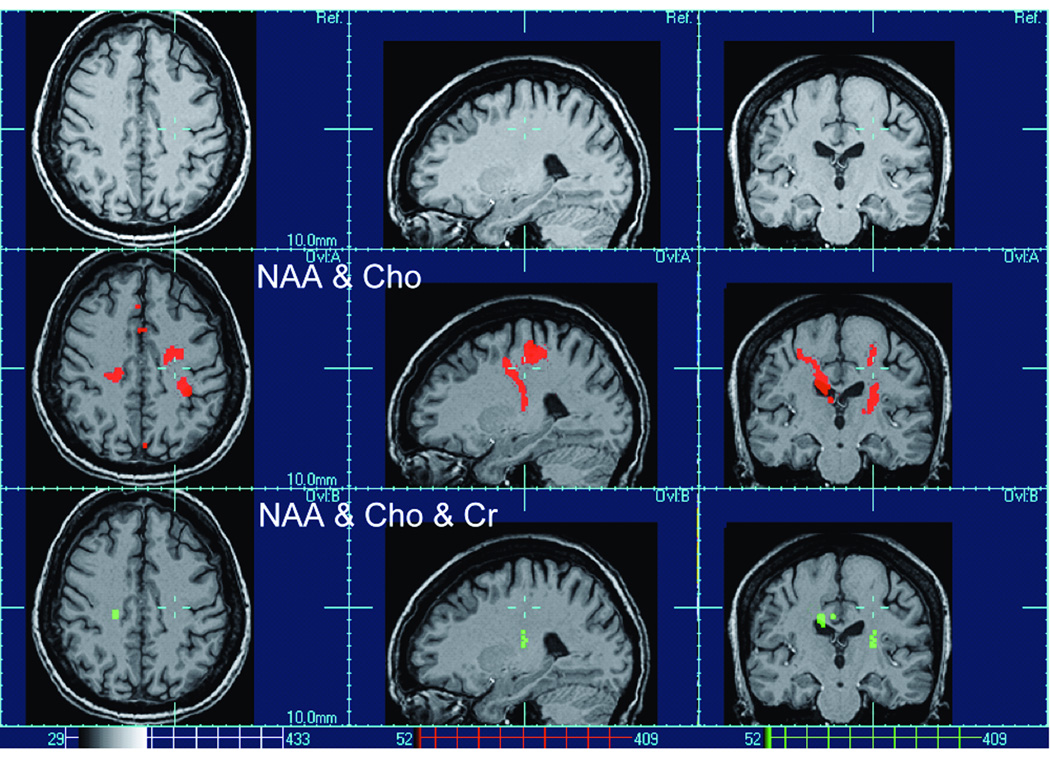
In Figure (2) are shown the results of multivariate analysis of metabolite alterations in ALS after accounting for the age-related changes. For the univariate analyses the single measures were NAA, Cho and Cr respectively, the contrast was diagnosis and the ANCOVAs were performed co-varying for age. The set of multivariate tests included a test using all three metabolites in combination as well as tests using all pair wise combinations as measures, for each the contrast was diagnosis, and the MANCOVAs were performed co-varying for age. Whereas univariate tests yielded no significant effects of ALS for any of the metabolites, multivariate tests revealed significant metabolite alterations in motor and pre-motor areas, extending along motor fibers. Moreover, multivariate tests of simultaneous NAA and Cho changes yielded on average larger clusters (in red) than multivariate tests including Cr (in green), implying that changes of these three metabolites in some regions are more concordant than in others. This is also consistent with previous MRSI findings in ALS demonstrating for example substantial NAA reductions without concurrent Cho changes in the motor cortex and in contrast a large increase in Cho without NAA change in the posterior internal capsule where motor fibers converge (23). It should be noted, however, that "voxel bleeding" due to the low resolution of MRSI as well as image mis-registration limit accuracy in localizing clusters. This can result in artifacts such as the NAA and Cho cluster of ALS in figure (2) which extends into the ventricles and this can happen for other CSF regions as well. Therefore, the localization and spatial extent of significant clusters should be interpreted with care.
Figure 2.
Multivariate test results of metabolite alterations in ALS after accounting for age-related changes.
DISCUSSION
There are two major findings of this study: First, the simulation study demonstrates that if a joint metabolite effect exists a multivariate analysis provides more power, based on the relative strengths of the individual metabolite effects, to detect metabolic changes than either of the univariate tests. Second, multivariate tests yielded more power to detect metabolite alterations of motor and pre-motor areas in ALS than univariate tests, suggesting that alterations of NAA and Cho are interconnected. Taken together, the results demonstrate that multivariate statistical mapping is a powerful approach for the analysis of MRSI data and should be used whenever simultaneous changes in metabolite levels are expected.
Though multivariate methods can add statistical power to the analysis of MRSI data, several issues should be considered. First, the simulations also show that multivariate tests can be inferior in power to univariate ones if the joint metabolite effect is not substantial. Although this result may seem intuitive, the decision regarding whether to use a multivariate or univariate test for MRSI analyses can be difficult. In particular, the substitution of a multivariate test by multiple univariate tests might sometimes be the most appropriate approach. However, one needs to contend with the fact that studying the significance of multiple ANOVAs does not provide any understanding of possible underlying relationships between metabolites. In principle, multivariate and univariate analysis address different research questions. The selection of which analysis to use should therefore be based on the purpose of the research. The basic question in a multivariate test that a univariate analysis cannot address is whether there are any overall interactions. In addition, a multivariate analysis may be used for the identification of underlying constructs associated with metabolite intercorrelations.
It should be pointed out that one issue that is not addressed here (nor by any current voxel based approach) is how to asses simultaneous variations in different measured quantities in different areas of the brain, e.g. a reduction of NAA in one part of the brain, and an increase of Cho in another region. It might be the case, however, that multivariate and univariate tests such as those described in this paper could be used as important exploratory tools for forming hypothesis that could be tested with additional data. For instance various affected regions could be identified using these tests, then further studies could be used to test effects of univariate or multivariate measures in the reduced set of combined regions identified in the exploratory analysis. To explore further spatial relationships between regions of simultaneous variations, various permutations of the univariate and multivariate tests from different regions would be appropriate in the exploratory analysis. For voxelwise multivariate or univariate tests, however, even simple pair wise analysis of voxels is currently both statistically and computationally infeasible, because of the astronomical number of pair combinations; any meaningful test involving multiple voxels or regions will therefore require some reduction in the effective number of statistical tests such as that just described.
Some issues that should be addressed and will be in future studies, and that are similar to those that face any multivariate voxel based imaging study, include determining the degree to which warping of images into standard space affects metabolite relationships, and determining what degree of spatial bias is introduced by spatial smoothing (20).
Our multivariate findings in ALS patients are consistent with other MRS studies. In particular, several MRSI studies reported decreased NAA and increased Cho levels in ALS (24, 25). While decreased NAA has been interpreted to reflect neuronal loss or dysfunction, Cho increase has been thought to be associated with increased membrane syntheses and/or breakdown of the lipid membrane structures into smaller fragments. However, these earlier studies, which used univariate tests, could not address the issue of an interaction between the NAA and Cho changes in ALS. Our gain in sensitivity with multivariate tests in ALS compared directly to the results from univariate tests implies that the alterations of NAA and Cho in ALS are correlated and not independent of each other. A correlation between NAA and Cho is also consistent with new pathological findings in ALS suggesting diverse membrane proteins, which may give raise to increased Cho resonances and may contribute to motor neuron death, as reflected by decreased NAA (10).
In summary, multivariate analysis of MRSI data can add power in testing specific hypothesis regarding variations in brain metabolism.
ACKNOWLEDGEMENTS
This work was supported by NIH grant R01 EB000822 (AAM), a grant by the NIH National Center for Research Resource RR23953 (NS) and the Stanley Glaser Foundation of the University of Miami. This material is the result of work supported with resources and the use of facilities at the Veterans Affairs Medical Center in San Francisco.
REFERENCES
- 1.Maudsley AA, Darkazanli A, Alger JR, Hall LO, Schuff N, Studholme C, Yu Y, Ebel A, Frew A, Goldgof D, Gu Y, Pagare R, Rousseau F, Sivasankaran K, Soher BJ, Weber P, Young K, Zhu X X. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson PM, Miller MI, Poldrack RA, Nichols TE, Taylor JE, Worsley KJ, Ratnanather JT. Special Issue on Mathematics in Brain Imaging. NeuroImage. 2009;45:S1–S2. doi: 10.1016/j.neuroimage.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 3.Worsley KJ, Taylor JE, Tomaiuolo F, Lerch J. Unified univariate and multivariate random field theory. NeuroImage. 2004;23 Suppl 1:S189–S195. doi: 10.1016/j.neuroimage.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 4.Thompson PM, Toga AW. Detection, visualization and animation of abnormal anatomic structure with a deformable probabilistic brain atlas based on random vector field transformations. Med Image Anal. 1997;1997(4):271–294. doi: 10.1016/s1361-8415(97)85002-5. [DOI] [PubMed] [Google Scholar]
- 5.Avants B, Duda JT, Kim J, Zhang H, Pluta J, Gee JC, Whyte J. Multivariate analysis of structural and diffusion imaging in traumatic brain injury. Acad Radiol. 2008;15(11):1360–1375. doi: 10.1016/j.acra.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luts J, Laudadio T, Idema AJ, Simonetti AW, Heerschap A, Vandermeulen D, Suykens JA, Van Huffel S. Nosologic imaging of the brain: segmentation and classification using MRI and MRSI. NMR Biomed. 2008 Dec. 23 doi: 10.1002/nbm.1347. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 7.Marjanska M, Curran GL, Wengenack TM, Henry PG, Bliss RL, Poduslo JF, Jack CR, Jr, Ugurbil K, Garwood M. Monitoring disease progression in transgenic mouse models of Alzheimer's disease with proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 2005;102(33):11906–11910. doi: 10.1073/pnas.0505513102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westman E, Spenger C, Oberg J, Reyer H, Pahnke J, Wahlund LO. In vivo 1H-magnetic resonance spectroscopy can detect metabolic changes in APP/PS1 mice after donepezil treatment. BMC Neurosci. 2009;10:33. doi: 10.1186/1471-2202-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devos A, Simonetti AW, van der Graaf M, Lukas L, Suykens JA, Vanhamme L, Buydens LM, Heerschap A, Van Huffel S. The use of multivariate MR imaging intensities versus metabolic data from MR spectroscopic imaging for brain tumour classification. J Magn Reson. 2005;173(2):218–228. doi: 10.1016/j.jmr.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Masters CL, Cappai R, Barnham KJ, Villemagne VL. Molecular mechanisms for Alzheimer's disease: implications for neuroimaging and therapeutics. J Neurochem. 2006;97(6):1700–1725. doi: 10.1111/j.1471-4159.2006.03989.x. [DOI] [PubMed] [Google Scholar]
- 11.Neusch C, Bähr M, Schneider-Gold C. Glia cells in amyotrophic lateral sclerosis: new clues to understanding an old disease? Muscle Nerve. 2007;35(6):712–724. doi: 10.1002/mus.20768. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser LG, Young K, Meyerhoff DJ, Mueller SG, Matson GB. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4 T. NMR Biomed. 2008;21(1):22–32. doi: 10.1002/nbm.1150. [DOI] [PubMed] [Google Scholar]
- 13.Terpstra M, Marjanska M, Henry PG, Tkác I, Gruetter R. Detection of an antioxidant profile in the human brain in vivo via double editing with MEGA-PRESS. Magn Reson Med. 2006;56(6):1192–1199. doi: 10.1002/mrm.21086. [DOI] [PubMed] [Google Scholar]
- 14.Haupt CI, Kiefer AP, Maudsley AA. In-plane motion correction for MR spectroscopic imaging. Magn Reson Med. 1998;39(5):749–753. doi: 10.1002/mrm.1910390512. [DOI] [PubMed] [Google Scholar]
- 15.Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging. 1998;17(3):463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 16.Ihaka R, Gentleman. R R. A Language for Data Analysis and Graphics. Journal of Computational and Graphical Statistics. 1996;5(3):299–314. [Google Scholar]
- 17.Marchini JL, Lafaye de Micheaux P. AnalyzeFMRI: Functions for analysis of fMRI datasets stored in the ANALYZE or NIFTI format. R package version 1.1–10. 2008 [Google Scholar]
- 18.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57(1):289–300. [Google Scholar]
- 20.Genovese CR, Lazar NA, Nichols T. Thresholding of Statistical Maps in Functional Neuroimaging Using False Discovery Rate. NeuroImage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 21.Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI) Magn Reson Med. 2009;61(3):548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chard DT, McLean MA, Parker GJ, MacManus DG, Miller DH. Reproducibility of in vivo metabolite quantification with proton magnetic resonance spectroscopic imaging. J Magn Reson Imaging. 2002;15(2):219–225. doi: 10.1002/jmri.10043. [DOI] [PubMed] [Google Scholar]
- 23.Schuff N, Rooney WD, Miller R, Gelinas DF, Amend DL, Maudsley AA, Weiner MW. Reanalysis of multislice (1)H MRSI in amyotrophic lateral sclerosis. Magn Reson Imaging. 2001;45(3):513–516. doi: 10.1002/1522-2594(200103)45:3<513::aid-mrm1067>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 24.Block W, Karitzky J, Träber F, Pohl C, Keller E, Mundegar RR, Lamerichs R, Rink H, Ries F, Schild HH, Jerusalem F. Proton magnetic resonance spectroscopy of the primary motor cortex in patients with motor neuron disease: subgroup analysis and follow-up measurements. Arch Neurol. 1998;55(7):931–936. doi: 10.1001/archneur.55.7.931. [DOI] [PubMed] [Google Scholar]
- 25.Sarac H, Zagar M, Vranjes D, Henigsberg N, Bilić E, Pavlisa G. Magnetic resonance imaging and magnetic resonance spectroscopy in a patient with amyotrophic lateral sclerosis and frontotemporal dementia. Coll Antropol. 2008;32 Suppl 1:205–210. [PubMed] [Google Scholar]