Abstract
The circumsporozoite protein (CSP) of Plasmodium vivax, a major target for malaria vaccine development, has immunodominant B-cell epitopes mapped to central nonapeptide repeat arrays. To determine whether rearrangements of repeat motifs during mitotic DNA replication of parasites create significant CSP diversity under conditions of low effective meiotic recombination rates, we examined csp alleles from sympatric P. vivax isolates systematically sampled from an area of low malaria endemicity in Brazil over a period of 14 months. Nine unique csp types, comprising six different nonapeptide repeats, were observed in 45 isolates analyzed. Identical or nearly identical repeats predominated in most arrays, consistent with their recent expansion. We found strong linkage disequilibrium at sites across the chromosome 8 segment flanking the csp locus, consistent with rare meiotic recombination in this region. We conclude that CSP repeat diversity may not be severely constrained by rare meiotic recombination in areas of low malaria endemicity. New repeat variants may be readily created by nonhomologous recombination even when meiotic recombination is rare, with potential implications for CSP-based vaccine development.
Keywords: Plasmodium vivax, malaria, circumsporozoite protein, genetic diversity, repetitive domains, recombination, mismatch distribution analysis, vaccine
1. Introduction
Plasmodium vivax is the most widespread of the four human malaria parasites, causing 132 to 391 million episodes of disease each year (Hay et al. 2004), with 2.6 billion people at risk of infection worldwide (Guerra et al. 2006). Outside of Africa, P. vivax is the main cause of malaria morbidity, with enormous public health burden. However, since P. vivax usually causes less severe symptoms than P. falciparum, it has received relatively little attention and limited funds for research and control (Price et al. 2007).
A notable feature of several malaria surface antigens (including major vaccine-candidate molecules) is the presence of tandem arrays of relatively short amino acid motifs. The circumsporozoite protein (CSP), an abundant antigen on the surface of sporozoites, has been extensively used as a vaccine development target (Nardin and Zavala, 1998). CSP has immunodominant B-cell epitopes mapped to central repeats (CR) bracketed between nonrepetitive sequences. Plasmodium vivax CSP displays two major types of nonapeptide repeats (most commonly, GDRA[D/A]GQPA and ANGAGNQPG), which define the variants known as VK210 and VK247, respectively (Rosenberg et al., 1989). Although VK210- and VK247-type sequences often occur in sympatric parasite populations, no example is known of hybrid CR array with both repeat types (Lim et al., 2005).
Insertions and deletions in the CR domain, resulting from either sexual recombination during meiosis or intrahelical strand-slippage events during mitotic DNA replication (McConkey et al., 1990), generate novel CSP variants that may be positively selected if mutant parasites evade host’s immunity. Extensive variation occurs in the CR of P. falciparum CSP, which consists of variable numbers of copies of 4-mer NANP and NVDP motifs, without breaking down the tight linkage between polymorphic sites in flanking sequences as interhelical exchanges during meiosis would do (Rich et al., 1997). This pattern suggests that CR, as other short repeats such as microsatellite- or minisatellite-type sequences (Levinson and Gutman, 1987), undergo frequent intrahelical recombination. Whether similar mechanisms create significant variation in CR arrays of P. vivax CSP, which consist of longer (9-mer) repetitive motifs, remains unknown. Frequent mitotic recombination coupled with positive selection of new variants might accelerate CR evolution even when meiotic recombination and outcrossing are relatively uncommon in malaria parasite populations (Rich et al., 2000). Whether or not length variation in CR arrays affects the recognition of B-cell epitopes is uncertain, but the conformational nature of CR epitopes in P. falciparum CSP (Monette et al., 2001) supports this hypothesis.
Here, we examine patterns of CR sequence diversity in csp alleles from sympatric P. vivax isolates from an area of low malaria endemicity. We use single-nucleotide polymorphism (SNP) typing to examine, in these same isolates, the haplotype structure of chromosome 8, where the csp gene is located. We sought to determine whether significant CR diversity at the P. vivax csp locus occurs under conditions of low malaria endemicity, which reduces effective meiotic recombination rates in local parasites.
2. Materials and methods
2.1 Study area and parasite population
Between March 2004 and May 2005, we obtained 54 isolates of P. vivax from subjects living in the eastern corner of Acre State, Western Amazon Basin of Brazil. Study subjects participated in a population-based cohort study in the rural community of Granada (9°41′ S to 94°9′ S, 67°05′ W to 67°07′ W), which is part of the frontier agricultural settlement known as Pedro Peixoto Settlement, situated 50 km northwest of Acrelândia, the nearest town, and 150 km east of Rio Branco, the capital of Acre. Both P. falciparum and P. vivax are transmitted year-round, with average incidence rates of 30 slide-confirmed P. vivax infections/100 person-years at risk and 16.3 slide-confirmed P. falciparum infections/100 person-years at risk during the study period (da Silva-Nunes et al., 2008). Eight pairs of P. vivax isolates were collected from the same individuals during consecutive infections (40 to 210 days apart); all others were collected from unrelated subjects. The collected isolates represent 36.7% of all slide-confirmed symptomatic P. vivax infections diagnosed in Granada between March 2004 and May 2005. Previous microsatellite analysis of these isolates indicated that 49% of them comprised more than one genetically distinct clone (i. e., they contained multiple-clone infections) (Ferreira et al., 2007). Patients or guardians provided written informed consent, and the research protocol was approved by the ethics review board of the Institute of Biomedical Sciences, University of São Paulo (318/2002 and 538/2004). Plasmodium vivax infections were treated with chloroquine and primaquine (Ministry of Health of Brazil, 2001). Genomic DNA was isolated as described (Ferreira et al., 2007).
2.2 Amplification and sequencing of circumsporozoite protein (CSP) repeats of Plasmodium vivax
The CR domain of P. vivax csp was amplified by nested polymerase chain reaction (PCR) essentially as described by Imwong et al. (2005).
Briefly, the external oligonucleotide primers VCS-OF (5′-ATGTAGATCTGTCCAAGGCCATAAA-3′) and VCS-OR (5′-TAATTGAATAATGCTAGGACTAACAATATG-3′), which target conserved domains at the nonrepetitive 5′ and 3′ domains flanking the repeats, were used to amplify a fragment of ~1100 base pairs (bp), from 1 μl of purified parasite DNA, with 25 cycles and annealing temperature set at 58°C. The internal primers VCS-NF (5′-GCAGAACCAAAAAATCCACGTGAAAATAAG-3′) and VCS-NR (5′-CCAACGGTAGCTCTAACTTTATCTAGGTAT-3′) amplified a ~680-bp fragment, starting with 1 μl of the primary PCR product, with 30 cycles and annealing temperature set at 62°C. Sequencing was performed on both DNA strands directly on secondary PCR products, which were purified with HiYield Gel/PCR DNA Extraction kits (Real Biotech, Taipei, Taiwan), by using the internal primers VCS-NF and VCS-NR. BigDye v3.1 terminator chemistry (Applied Biosystems, Foster City, CA) was used in sequencing reactions and products were analyzed on a 3100 automated DNA sequencer (Applied Biosystems). Sequences of poor quality or those displaying superimposed electropherogram peaks, indicating mixed-genotype infections, were discarded, leaving a total of 45 csp sequences analyzed. Nucleotide variants that were seen in a single isolate were confirmed by independent reamplification and resequencing from purified genomic DNA of the relevant isolate. New csp sequences obtained in this study were deposited in GenBank database under the accession numbers FJ845383-FJ845391.
2.3 Single-nucleotide polymorphism (SNP) typing along chromosome 8 of Plasmodium vivax
To characterize the haplotype structure of the chromosome segment surrounding the csp locus, we typed 31 biallelic SNP markers across a 100-kilobase (kb) nontelomeric region of chromosome 8 that comprises the 1.2-kb csp locus (Feng et al., 2003; Carlton et al., 2008). We identified SNPs and characterized flanking sequences by aligning orthologous sequences from five P. vivax isolates (GenBank accession numbers AY003872, AY216936, AY216937, AY216938, AY216939). Markers were selected to meet the following criteria: (a) no SNP could be located in repetitive domains; (b) 200 bp of sequence upstream and downstream of each SNP should have no significant similarity to human sequences (as determined by BLAST search against the human genome), to prevent cross-amplification of human DNA present in the sample; and (c) both alleles of each SNP should be present in our parasite population (i. e., no monomorphic locus was selected). Thirty-one SNPs meeting these criteria (including eight synonymous and two nonsynonymous SNPs in coding sequences and 21 SNPs in intergenic regions), which were successfully typed in a large proportion of field isolates, were selected for this analysis (Supplementary Table 1 online). Supplementary Figure 1 (published online) shows the map location of these SNPs in relation to the csp locus. The physical distance between pairs of SNP markers ranged between 174 and 96315 kb. SNP typing was performed, under contract, by K-Biosciences (Cambridge, UK), with an amplifluor assay (Nazarenko et al., 1997; Newton et al., 1989). To obtain adequate DNA concentrations, parasite DNA was submitted to whole-genome amplification (WGA) prior to SNP typing (Wang et al., 2009). WGA was performed on 10 ng of genomic DNA, with high-fidelity multiple displacement technology (Dean et al., 2002), using a REPLI-g Minikit (Qiagen, Valencia, CA) according to the manufacturer’s instruction. All sequences of oligonucleotide primers (reverse-strand sequences as in Feng et al., 2003) used for SNP typing are given in Supplementary Table 2 online; the annealing temperature for all primers was 60°C. Haplotypes characterized with SNPs (unique combinations of alleles at 31 sites on chromosome 8) are referred to, throughout this article, as Chr-8 haplotypes. Thirty-one isolates had all SNPs typed, with a single allele at each site (i. e., no apparent mixed-clone infection), and had their csp gene successfully sequenced.
2.4 Data analysis
Nucleotide sequences were aligned manually. Parasites sharing the same nucleotide sequence in the CR array were assigned to the same csp type. The different nucleotide sequences encoding the same nonapeptide motif, termed repeat allotypes (RATs; Rich et al., 1997), were recorded. To examine the mode of evolution of CR, we used the mismatch distribution analysis suggested by Hughes (2004). We aligned the nucleotide sequences encoding each nonapeptide repeat unit in each csp type and calculated the following parameters: p, the average proportion of nucleotide difference between pairs of repeat units in the same csp allele; Skewness, the skewness of the distribution of p values for all pairwise comparisons; Prop. 0, the proportion of pairwise comparisons for which p = 0; and Prop.>25, the proportion of pariwise comparisons for which p > 0.25 (Hughes, 2004).
To determine how much meiotic recombination occurs across the chromosome 8 region where csp is located, we estimated levels of linkage disequilibrium (LD) between pairs of SNP markers, measured with the coefficient of determination r2, by using the version 3.11 of the Arlequin software (available at: http://cmpg.unibe.ch/software/arlequin3/). The significance of pairwise associations between SNP sites was tested with Markov chains to explore all theoretically possible contingency tables; P values correspond to the proportion of 10,000 simulated contingency tables associated with probabilities equal or smaller to the observed table. The correlation between the physical distance between SNP markers (measured in bp) and LD was assessed using the Pearson’s correlation test.
3. Results
3.1 csp sequence diversity in sympatric Plasmodium vivax isolates from Acre, Brazil
Sequences of CR domain of csp were analyzed for 45 sympatric P. vivax isolates from an area of low malaria endemicity in the Amazon Basin of Brazil. All sequences were of the VK210 type, with no example of VK247 or hybrid VK210/VK247 allele. In fact, although both VK210 and VK247 variants circulate in Brazil, the former predominates in all areas so far studied in this country (Machado and Póvoa, 2000). Substantial diversity was observed in CR arrays; nine different csp types, consisting of variable arrangements of six different nonapeptide repeat sequences, were observed in our parasite population (Fig. 1). Table 1 shows all RATs found in the CR domains of these csp sequences. Note that csp types IVa and IVb are identical at the amino acid level, but their fourth, eighth and eleventh copies of the nonapeptide repeat are encoded by different RATs (Fig. 1). As previously observed for CR domains of P. falciparum csp (McConkey et al., 1990; Rich et al., 1997), we found two groups of csp types with a highly conserved overall structure of the nonapeptide repeat array. Within these two groups of csp types (group A comprises types I, II, VII, and VIII, found in 38 isolates; group B comprises types III, IVa, and IVb, found in 5 isolates), most CR diversity can be explained by insertions and deletions of repeat units. Few nucleotide replacements are observed when RATs, rather than nucleotide sequences, are properly aligned (Rich et al., 1997; Rich et al., 2000). Note that the RATs within these CR arrays display a remarkably similar arrangement, arguing for their relatively recent origin from a common ancestor.
Figure 1.
Types of circumsporozoite protein (CSP) central repeats in 45 sympatric isolates of Plasmodium vivax from Acre, Brazil. Different shading patterns of boxes represent each of the six variants of nonapeptide repeats found in these VK210-type sequences. The number of isolates sharing the same csp type is indicated within brackets. Numbers within the boxes represent different nucleotide sequences (termed repeat allotypes or RATs) that encode the same nonapeptide; these sequences are shown in Table 1. The csp types IVa and IVb are identical at the amino acid level (i. e., they are represented by boxes with identical shading pattern), but their fourth, eighth and eleventh copies of the nonapeptide repeat are encoded by different RATs (represented by different numbers within the boxes).
Table 1.
Amino acid and nucleotide sequences in the repeat allotypes (RATs) of the central repeats of Plasmodium vivax circumsporozoite protein (CSP) gene in Acre, Brazil
| Amino acid | RAT | Nucleotide sequence | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GDRADGQPA | 1 | G | G | A | G | A | C | A | G | A | G | C | A | G | A | T | G | G | A | C | A | G | C | C | A | G | C | A |
| 2 | G | G | T | G | A | C | A | G | A | G | C | A | G | A | T | G | G | A | C | A | A | C | C | A | G | C | A | |
| 3 | G | G | T | G | A | C | A | G | A | G | C | A | G | A | T | G | G | A | C | A | G | C | C | A | G | C | A | |
| 4 | G | G | A | G | A | T | A | G | A | G | C | A | G | A | T | G | G | A | C | A | G | C | C | A | G | C | A | |
| 5 | G | G | T | G | A | T | A | G | A | G | C | A | G | A | T | G | G | A | C | A | G | C | C | A | G | C | A | |
| 6 | G | G | C | G | A | T | A | G | A | G | C | A | G | A | T | G | G | A | C | A | G | C | C | A | G | C | A | |
| 7 | G | G | A | G | A | T | A | G | A | G | C | A | G | A | T | G | G | A | C | A | A | C | C | A | G | C | A | |
| 8 | G | G | A | G | A | C | A | G | A | G | C | A | G | A | T | G | G | A | C | A | A | C | C | A | G | C | A | |
| GDRAAGQPA | 1 | G | G | A | G | A | T | A | G | A | G | C | A | G | C | T | G | G | A | C | A | G | C | C | A | G | C | A |
| 2 | G | G | T | G | A | T | A | G | A | G | C | A | G | C | T | G | G | A | C | A | A | C | C | A | G | C | A | |
| 3 | G | G | T | G | A | T | A | G | A | G | C | A | G | C | T | G | G | A | C | A | G | C | C | A | G | C | A | |
| 4 | G | G | A | G | A | T | A | G | A | G | C | A | G | C | T | G | G | A | C | A | A | C | C | A | G | C | A | |
| 5 | G | G | A | G | A | C | A | G | A | G | C | A | G | C | T | G | G | A | C | A | G | C | C | A | G | C | A | |
| GDRAAGGAA | -a | G | G | A | G | A | T | A | G | A | G | C | A | G | C | T | G | G | A | C | A | G | G | C | A | G | C | A |
| GNGAGGQAA | -a | G | G | A | A | A | T | G | G | T | G | C | A | G | G | T | G | G | A | C | A | G | G | C | A | G | C | A |
| GNGAGGQPA | -a | G | G | A | A | A | T | G | G | T | G | C | A | G | G | T | G | G | A | C | A | G | C | C | A | G | C | A |
| GNRADGQPA | 1 | G | G | T | A | A | T | A | G | A | G | C | A | G | A | T | G | G | A | C | A | G | C | C | A | G | C | A |
| 2 | G | G | A | A | A | T | A | G | A | G | C | A | G | A | T | G | G | A | C | A | G | C | C | A | G | C | A | |
A single RAT was found for this amino acid motif. Shading indicates nucleotide differences in RATs encoding the same nonapeptide motif. The nonapeptide motifs are represented, in Figure 1, with shading patterns shown below.
 GDRADGQPA
GDRADGQPA
 GDRAAGQPA
GDRAAGQPA
 GDRAAGQAA
GDRAAGQAA
 GNGAGGQAA
GNGAGGQAA
 GNGAGGQPA
GNGAGGQPA
 GNRADGQPA
GNRADGQPA
3.2 Mismatch distribution analysis of csp repeats
Table 2 shows the parameters of the mismatch distribution analysis of aligned repeat units in the CR domain of P. vivax csp alleles. As previously shown for orthologous P. falciparum sequences, the CR domains of P. vivax csp are characterized by low p values, positive skewness in most distributions of p values, high Prop.0 and low Prop.>0.25. Therefore, CR domains have high proportions of identical or nearly identical repeats, a finding that is consistent with a recent expansion of the CR array in most csp alleles.
Table 2.
Mismatch distribution analysis of repeat units in the central domain of circumsporozoite protein (CSP) from Plasmodium vivax isolates from Acre, Brazil
| CSP typea | No. of nonapeptide repeat units | pb | Skewnessc | Prop. 0c | Prop.>25d |
|---|---|---|---|---|---|
| I | 20 | 0.0764 | −0.197 | 0.200 | 0 |
| II | 20 | 0.0636 | 0.162 | 0.295 | 0 |
| III | 18 | 0.0787 | 1.787 | 0.196 | 0.078 |
| IVa | 18 | 0.0673 | 1.967 | 0.196 | 0.085 |
| IVb | 18 | 0.0666 | 1.967 | 0.176 | 0.072 |
| V | 18 | 0.0687 | 1.554 | 0.124 | 0.026 |
| VI | 19 | 0.0713 | 1.714 | 0.181 | 0.076 |
| VII | 20 | 0.0778 | 0.077 | 0.242 | 0.089 |
| VIII | 21 | 0.0797 | −0.117 | 0.148 | 0 |
| Mean | 19.1 | 0.0722 | 0.990 | 0.195 | 0.047 |
CSP types are described in Figure 1
p = average proportion of nucleotide difference between pairs of repeat units in the same csp type
Skewness = skewness of the distribution of p values for all pairwise comparisons of nucleotide sequences
Prop. 0 = proportion of pairwise comparisons of nucleotide sequences for which p = 0
Prop.>25 = proportion of pariwise comparisons of nucleotide sequences for which p > 0.25
3.3 Temporal distribution of csp types
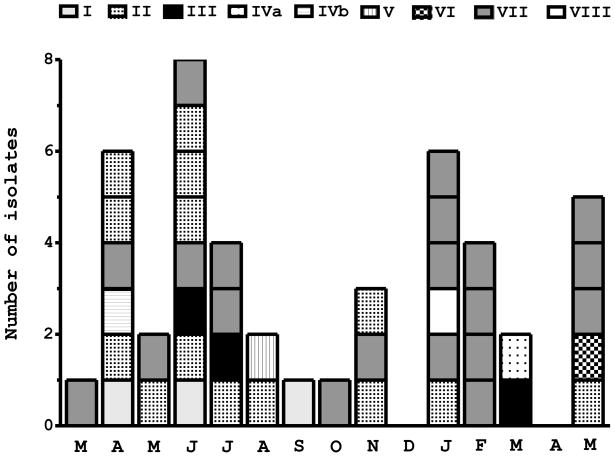
Since immune-mediated frequency-dependent selection could theoretically accelerate the turnover rate of antigenic variants in parasite populations (Forsyth et al. 1988), we next examined whether the same csp types could be sampled in parasites collected several months apart. We found no clear evidence of csp type replacement over time in our population. In fact, the most common csp types, II and VII, were found both at the beginning and the end of the follow-up, in parasites collected 13–14 months apart (Figure 2), suggesting that, within the time frame of the collection period, allele frequencies are not drastically affected by frequency-dependent selection. The temporal dynamics of relatively rare CSP variants cannot be reliably investigated given the small number of isolates analyzed, but conclusion that common CSP variants (to which the host population is predominantly exposed) are maintained over several months in the area is clear-cut and contrasts with the fast turnover of multilocus microsatellite haplotypes documented in the same parasite population (Ferreira et al., 2007; Orjuela-Sánchez et al., 2009). Whether or not functional constrains may limit the diversity of CSP repeat arrays in natural populations remains to be investigated.
Figure 2.
Monthly distribution of circumsporozoite protein (csp) gene types found in 45 sympatric isolates of Plasmodium vivax collected in Acre, Brazil, between March 2004 (denoted by the first “M” on the x-axis) and May 2005(denoted by the last “M” on the x-axis). csp types, defined in Figure 1, are represented by different shading patterns.
Of eight pairs of samples from the same subject, four pairs (collected 40–88 days apart) shared the same csp type (either type I or type VII) and four pairs (collected 154–240 days apart) had discordant csp types. If relapses account for at least some of these consecutive episodes of infection with genetically identical parasites in the same subject (Craig and Kain, 1996; Kirchgatter and del Portillo, 1998; but see also Imwong et al., 2007), this mechanism could preserve some csp types in the population over 2–3 months (the longest time interval at which the same csp type was recovered from these consecutive infections).
3.4 Haplotype structure of chromosome 8 of Plasmodium vivax
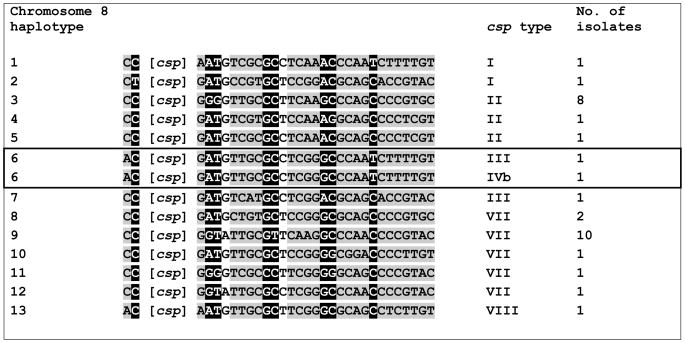
We next analyzed 31 SNP markers located both upstream and downstream from the csp locus, across a nontelomeric 100-kb segment of chromosome 8. We characterized 13 different Chr-8 haplotypes in 31 fully typed isolates (Fig. 3). Interestingly, the csp types I, II, and VII were shared by isolates with different Chr-8 haplotypes. The most striking example is provided by the pair of isolates that share the csp type I, but differ at 17 of 31 SNP sites analyzed, including those surrounding the csp locus (Fig. 3).
Figure 3.
Chromosome 8 (Chr-8) haplotypes and csp types in 31 sympatric isolates of Plasmodium vivax collected in Acre, Brazil. Each of the 13 Chr-8 haplotypes (represented with Arabic numerals) is defined as a unique combination of single-nucleotide polymorphisms (SNPs) in 31 sites typed. Grey shading indicates SNPs in intergenic (noncoding) regions; black shading represents synonymous SNPs in open reading frames, and no shading indicates nonsynonymous SNPs in open reading frames. The map position of each SNP is given in Supplementary Table 1 online. The position of csp locus is indicated by [csp]; two SNPs are located upstream of the csp locus and the remaining are located downstream of the csp locus. The csp types are represented with Roman numerals, as in Figure 1. Note that the csp types I, II, and VII were shared by isolates with different Chr-8 haplotypes, while the csp types III and IVb were found in two isolates with the same Chr-8 haplotype (boxed in the figure).
Conversely, the csp types III and IVb were found in two isolates (291M2 and 474M1, respectively) with the same Chr-8 haplotype (Fig. 3). As it is unlikely that two unrelated sympatric isolates display the same Chr-8 haplotype merely by chance, we assume that isolates 291M2 and 474M1 (collected 41 days apart) have a recent common ancestor. Therefore, most nucleotide divergence between the csp types displayed by them, III and IVb, which did not change the overall Chr-8 haplotype structure, may have been created by intrahelical rearrangements in the CR array rather than independent nucleotide replacements or interhelical exchanges of homologous but divergent DNA sequences. This finding does not rule out classic meiotic recombination as a means of repeat diversification in CSP, but suggests that it plays a minor role in generating novel csp repeat types in this parasite population.
3.5 Linkage disequilibrium across chromosome 8
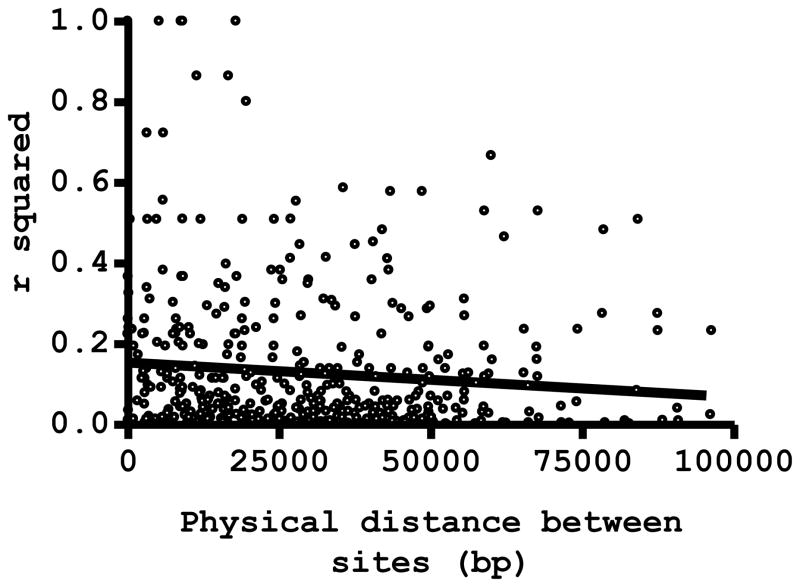
We next tested for pairwise LD between 31 SNPs across the chromosome 8 segment that comprises the csp locus. We made 465 pairwise comparisons, of which 112 (24.1%) revealed significant association between sites. We found comparable proportions of comparisons that yield significant disequilibrium for sites located <20 kb apart (58 of 193 comparisons, 30.1%), 20–50 kb apart (37 of 202 comparisons, 18.3%), and >50 kb apart (17 of 70 comparisons, 24.3%), indicating that significant LD was maintained across long segments of chromosome 8. Figure 4 shows that the strength of pairwise LD, as measured by the coefficient of determination r2, declines very slowly (but significantly, P = 0.031) with increasing physical distance (measured in bp) between SNP sites; the Pearson linear correlation coefficient r is 0.100, with a slope of −9 × 10−7 of the regression line. Results of the LD analysis are consistent with very low rates of effective meiotic recombination along the chromosome 8 segment that comprises the csp locus.
Figure 4.
Linkage disequilibrium (LD) decay with increasing physical distance between pairs of variable sites along chromosome 8 of Plasmodium vivax collected in Acre, Brazil. The LD index r2 was calculated for 465 pairwise comparisons. Circles represent individual r2 values, which are shown to decrease slowly but significantly (P = 0.031) with increasing physical distance (measured in bp) between SNP sites; the Pearson linear correlation coefficient r is 0.100, with a slope of −9 × 10−7 of the regression line shown in the figure.
4. Discussion
Sequence diversity in CSP repeat arrays, a major target of protective immunity against P. vivax, may not be severely constrained by the low effective meiotic recombination rates that are typical of malaria parasite populations in Brazil. The mode of evolution of the longer (9-mer) repeats of P. vivax csp seems to be quite similar to that hypothesized for the shorter (4-mer) repeats of P. falciparum csp (McConkey et al., 1990; Rich et al., 1997) and may involve repeated nonreciprocal, intrahelical recombination events such as strand-slippage during mitotic DNA replication. Our data suggest that these recombination events may create substantial CR diversity arrays even in a well-delimited area of relatively low endemicity, where we have carried out our systematic parasite sampling over a period of 14 months.
Results of the mismatch distribution analysis are consistent with a quite similar mode of evolution of the csp arrays of both major human malaria parasite species, P. vivax (Table 2) and P. falciparum (Table 1 of Hughes, 2004). In both instances, the CR domains seem to have evolved by recent duplications of short repeat units (either 4-mer or 9-mer, depending on the species considered).
Little sequence divergence between copies of repeated units has also been found in the longer (32-mer and 12-mer) repetitive motifs that characterize FC27-type alleles of another major malarial surface antigen, P. falciparum merozoite surface protein-2 (MSP-2) (Ferreira and Hartl, 2007). Since MSP-2 variants with large numbers of repeats seem to be preferentially recognized by naturally acquired antibodies (Franks et al., 2003), it has been hypothesized that duplicated repeat units are subsequently removed under immune pressure, shortening their lifespan in the array (Ferreira and Hartl, 2007). As a consequence, most duplication events are in fact recent, allowing short time for sequence divergence among paralogous copies of repeat units. Whether or not a similar mechanism could account for little sequence divergence between CSP repeat units within CR arrays remains to be determined.
In marked contrast with malaria surface antigens, mismatch analysis of repeat arrays of coding sequences of free-living organisms has revealed a pattern characterized by low or even negative skewness in the distribution of p values, low Prop.0 and high Prop.>0.25. Much more between-repeat diversity is usually observed in these repeat arrays, consistent with much older duplication events with subsequent divergence between paralogs (Hughes, 2004).
Extensive variation in repetitive domains of CSP in a well-delimited parasite population may be maintained despite relatively low levels of meiotic recombination, as inferred by the pairwise LD patterns in sites surrounding the csp locus. Strong LD has also been described in nonrepeat domains flanking the csp locus of P. falciparum (Rich et al., 1997), consistent with a major role of nonmeiotic recombination in the evolution of repeat diversity in both orthologs. The P. vivax population from our study site in Acre was also found to display strong LD among 14 putatively neutral microsatellite markers that map to different chromosomes (Ferreira et al., 2007; Orjuela-Sánchez et al., 2009), arguing for a predominantly clonal mode of reproduction (i. e., high levels of inbreeding and rare homologous meiotic recombination events) in local parasites.
Sequence variation may theoretically affect the protein structure of CSP (Monette et al., 2001) and allow parasites to evade naturally acquired, variant-specific immunity elicited in the host population by exposure to conformational B-cell epitopes. Although no clear-cut evidence is currently available for a role of variant-specific antibodies to CSP in naturally acquired or vaccine-induced immunity against either P. vivax or P. falciparum infection, it is reasonable to hypothesize that polymorphism in major parasite surface antigens tends to be preserved by balancing selection. As a consequence, a large repertoire of csp types can be observed even in areas of relatively low endemicity with small parasite populations, where random genetic drift would tend to drive rare variants to extinction. Under the hypothesis of variant-specific immunity, these findings have clear implications for naturally acquired and vaccine-induced immunity against CSP and many other major repetitive malaria antigens.
Supplementary Material
Acknowledgments
We thank the Global Health Center of Mount Sinai School of Medicine (New York, NY), for presenting this research opportunity and funding it, and Márcio M. Yamamoto (Department of Parasitology, University of São Paulo, Brazil) for help with DNA sequencing. This study was funded by the Fundação de Apoio à Pesquisa do Estado de São Paulo, Brazil (FAPESP grant 07/51199-0), the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (CNPq grant 470570/2006-7), and the National Institutes of Health of USA (NIH, RO1 grant AI 075416-01). POS and MUF are recipients of CNPq scholarships.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at: …
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang’a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AA, Kain KC. Molecular analysis of strains of Plasmodium vivax from paired primary and relapse infections. Jornal of Infectious Diseases. 1996;174:373–379. doi: 10.1093/infdis/174.2.373. [DOI] [PubMed] [Google Scholar]
- Da Silva-Nunes M, Codeço CT, Malafronte RS, Da Silva NS, Juncansen C, Muniz PT, Ferreira MU. Malaria on the Amazonian frontier: transmission dynamics, risk factors, spatial distribution, and prospects for control. Am J Trop Med Hyg. 2008;79:624–635. [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Farugi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Carlton JM, Joy DA, Mu J, Furuya T, Suh BB, Wang Y, Barnwell JW, Su XZ. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc Natl Acad Sci USA. 2003:8502–8507. doi: 10.1073/pnas.1232502100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MU, Hartl DL. Plasmodium falciparum: Worldwide sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-2 (MSP-2) Exp Parasitol. 2007;115:32–40. doi: 10.1016/j.exppara.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Ferreira MU, Karunaweera ND, da Silva-Nunes M, Silva NS, Wirth DF, Hartl DL. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J Infect Dis. 2007;195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- Forsyth KP, Anders RF, Kemp DJ, Alpers MP. New approaches to the serotypic analysis of the epidemiology of Plasmodium falciparum. Philos Trans R Soc Lond, B, Biol Sci. 1988;321:485–493. doi: 10.1098/rstb.1988.0104. [DOI] [PubMed] [Google Scholar]
- Franks S, Baton L, Tetteh K, Tongren E, Dewin D, Akanmori BD, Koram KA, Ranford-Cartwright LC, Riley EM. Genetic diversity and antigenic polymorphism in Plasmodium falciparum: extensive serological cross-reactivity between allelic variants of merozoite surface protein 2. Infect Immun. 2003;71:3485–3495. doi: 10.1128/IAI.71.6.3485-3495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra CA, Snow RW, Hay SI. Mapping the global extent of malaria in 2005. Trends Parasitol. 2006;22:353–358. doi: 10.1016/j.pt.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 2004;4:327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. The evolution of amino acid repeat arrays in Plasmodium and other organisms. J Mol Evol. 2004;59:528–535. doi: 10.1007/s00239-004-2645-4. [DOI] [PubMed] [Google Scholar]
- Imwong M, Pukrittayakamee S, Grüner AC, Rénia L, Letourneur F, Looareesuwan S, White NJ, Snounou G. Practical PCR genotyping protocols for Plasmodium vivax using Pvcs and Pvmsp1. Malar J. 2005;4:20. doi: 10.1186/1475-2875-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Snounou G, Pukrittayakamee S, Tanomsing N, Kim JR, Nandy A, Guthmann JP, Nosten F, Carlton J, Looareesuwan S, Nair S, Sudimack D, Day NP, Anderson TJ, White NJ. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis. 2007;195:927–933. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- Kirchgatter K, del Portillo HA. Molecular analysis of Plasmodium vivax relapses using the MSP1 molecule as a genetic marker. J Infect Dis. 1998;177:511–515. doi: 10.1086/517389. [DOI] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanisms for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Lim CS, Tazi L, Ayala FJ. Plasmodium vivax: Recent world expansion and genetic identity to Plasmodium simium. Proc Natl Acad Sci USA. 2005;102:15523–15528. doi: 10.1073/pnas.0507413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado RLD, Póvoa MM. Distribution of Plasmodium vivax variants (VK210, VK247 and P. vivax-like) in three endemic areas of the Amazon region of Brazil and their correlation with chloroquine treatment. Trans R Soc Trop Med Hyg. 2000;94:377–381. doi: 10.1016/s0035-9203(00)90110-x. [DOI] [PubMed] [Google Scholar]
- McConkey GA, Waters AP, McCutchan TF. The generation of genetic diversity in malaria parasites. Annu Rev Microbiol. 1990;44:479–498. doi: 10.1146/annurev.mi.44.100190.002403. [DOI] [PubMed] [Google Scholar]
- Ministry of Health of Brazil. Diagnóstico e tratamento da malária. Ministry of Health of Brazil; Brasília: 2001. [Google Scholar]
- Monette M, Opella SJ, Greenwood J, Willis AE, Perham RN. Structure of a malaria parasite antigenic determinant displayed on filamentous bacteriophage determined by NMR spectroscopy: implications for the structure of continuous peptide epitopes of proteins. Protein Sci. 2001;10:1150–1159. doi: 10.1110/ps.35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardin EH, Zavala F. Acquired immunity to sporozoites. In: Sherman IW, editor. Malaria: Parasite Biology, Pathogenesis, and Protection. ASM Press; Washington, DC: 1998. pp. 495–511.pp. 495–511. [Google Scholar]
- Nazarenko IA, Bhatnagar SK, Hohman RJ. A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res. 1997;25:2516–2521. doi: 10.1093/nar/25.12.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjuela-Sánchez P, da Silva NS, da Silva-Nunes M, Ferreira MU. Parasitemia recurrences population dynamics of Plasmodium vivax polymorphisms in rural Amazonia. Am J Trop Med Hyg. 2009 doi: 10.4269/ajtmh.2009.09-0337. in press. [DOI] [PubMed] [Google Scholar]
- Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- Rich SM, Ferreira MU, Ayala FJ. The origin of antigenic diversity in Plasmodium falciparum. Parasitol Today. 2000;16:390–396. doi: 10.1016/s0169-4758(00)01741-5. [DOI] [PubMed] [Google Scholar]
- Rich SM, Hudson RR, Ayala FJ. Plasmodium falciparum antigenic diversity: evidence of clonal population structure. Proc Natl Acad Sci USA. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R, Wirtz RA, Lanar DE, Sattabongkot J, Hall T, Waters AP, Prasittisuk C. Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989;245:973–976. doi: 10.1126/science.2672336. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nair S, Nosten F, Anderson T. Multiple displacement amplification for malaria parasite DNA. J Parasitol. 2009;95:253–255. doi: 10.1645/GE-1706.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.