SUMMARY
Temporal changes in the prevalence of antigenic variants in Plasmodium falciparum populations have been interpreted as evidence of immune-mediated frequency-dependent selection, but evolutively neutral processes may generate similar patterns of serotype replacement. Over 4 years, we investigated the population dynamics of P. falciparum polymorphisms at the community level by using 11 putatively neutral microsatellite markers. Plasmodium falciparum populations were less diverse than sympatric P. vivax isolates, with less multiple-clone infections, lower number of alleles per locus and lower virtual heterozygosity, but both species showed significant multilocus linkage disequilibrium. Evolutively neutral P. falciparum polymorphisms showed a high turnover rate, with few lineages persisting for several months in the population. Similar results had previously been obtained, in the same community, for sympatric P. vivax isolates. In contrast, the prevalence of the 2 dimorphic types of a major antigen, MSP-2, remained remarkably stable throughout the study period. We suggest that the relatively fast turnover of parasite lineages represents the typical population dynamics of neutral polymorphisms in small populations, with clear implications for the detection of frequency-dependent selection of polymorphisms.
Keywords: Plasmodium falciparum, population dynamics, polymorphism, parasite lineages
INTRODUCTION
Understanding the forces that promote genetic diversity of human pathogens across time and space is a central question of ecology and population genetics, with clear implications for public health interventions such as drug treatment and vaccination (Gupta and Maiden, 2001; Lipsitch and O’Hagan, 2007). Although genetically diverse malaria parasite lineages are known to appear and disappear, over few hours or days, in the bloodstream of semi-immune individuals with asymptomatic infections left untreated (Farnert, 2008), the population dynamics of Plasmodium falciparum polymorphisms remains poorly understood at the community level.
The dramatic changes in serotype composition observed in P. falciparum isolates collected in a single village in Papua New Guinea over 24 months have been interpreted to result from frequency-dependent selection on a immunodominant variable protein, known as S-antigen (Forsyth et al. 1988). If evasion of naturally acquired, variant-specific immunity in the host population impacts the evolution of antigenic polymorphism in the parasite, the most common S-antigen variants found at the baseline are expected to be replaced, over the next months, by serotypes that were initially rare, against which little immunity existed.
Nevertheless, alternative mechanisms, in addition to frequency-dependent selection, could generate a similar serotype replacement pattern. Cyclic or chaotic fluctuations in the frequency of serotypes in populations, for example, may be observed for antigenic determinants that do not elicit strong variant-specific immunity (Gupta et al. 1998). Furthermore, selectively neutral processes, such as random genetic drift and migration, may also lead to a fast turnover of genetically diverse pathogen lineages (Gupta et al. 1994). Therefore, the temporal dynamics of neutrally evolving polymorphisms must be examined before a particular pattern of serotype replacement in malaria parasites and other pathogens can be attributed to immune-mediated selection. Here we use putatively neutral microsatellite markers to investigate the community-level population dynamics of P. falciparum polymorphisms, in the absence of strong frequency-dependent selection. The temporal dynamics of neutral polymorphism is compared with that of a major target of variant-specific naturally acquired immunity, the merozoite surface protein (MSP)-2 of P. falciparum.
MATERIALS AND METHODS
Study area and parasite populations
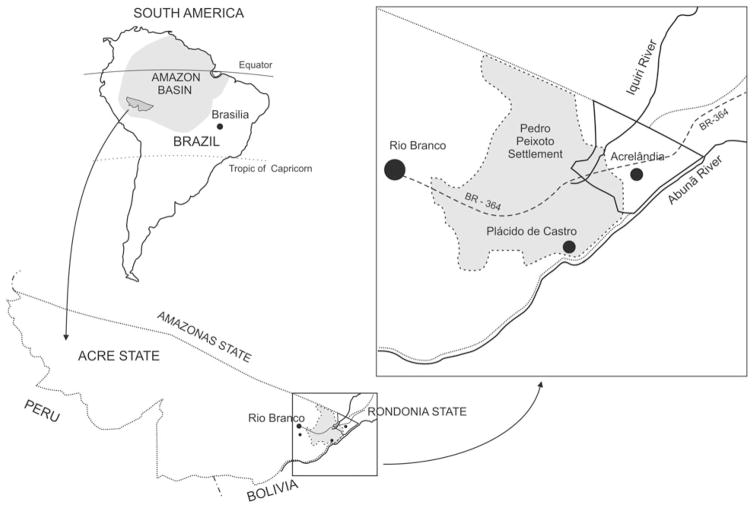
We analysed 55 isolates of P. falciparum obtained from subjects living in the eastern corner of Acre State, Western Amazon Basin of Brazil (Fig. 1). A P. falciparum isolate was defined as a sample of parasites derived from a single patient at a single occasion; one isolate may comprise one or more genetically diverse clones. Between April 2004 and October 2006, 44 isolates were collected during a population-based cohort study in the rural community of Granada (9°41′S to 94°9′S, 67°05′W to 67°07′W) (da Silva-Nunes et al. 2008). Granada is part of the frontier agricultural settlement known as Pedro Peixoto Settlement, situated 30–50 km northwest of Acrelândia (9°43′S, 66°53′W), the nearest town, and 150 km east of Rio Branco (9°58′S, 67°48′W), the capital of Acre. Both P. falciparum and P. vivax are transmitted year-round, with incidence rates between 2004 and 2006 ranging from 4·2 to 16·5 slide-confirmed infections/100 person-years at risk for P. falciparum and from 7·1 to 34·3 slide-confirmed infections/100 person-years at risk for P. vivax. The 44 isolates analysed from Granada represent 75·9% of all slide-confirmed P. falciparum infections diagnosed by active and passive case detection in this community between April 2004 and October 2006. Five pairs of P. falciparum isolates were collected from the same individuals during consecutive infections (1–4 months apart); all others were collected from unrelated subjects. Since 49 sympatric P. vivax isolates, which were collected between March 2004 and May 2005, were analysed with 14 polymorphic microsatellite markers (Ferreira et al. 2007), between-species comparisons of diversity patterns could be made for human malaria parasite populations from Granada.
Fig. 1.
Map of the state of Acre, northwestern Brazil, showing the Pedro Peixoto Settlement (shaded area in the inset), 30–;50 km northwest of the town of Acrelândia. The rural community known as Granada (9°41′S to 9°49′S, 67°05′W to 67°07′W), site of the cohort study carried out between 2004 and 2006, is part of the Pedro Peixoto Settlement. The location of the town of Plácido de Castro (10°20′S, 67°11′W), site of the cross-sectional study in 2008, is also shown. Granada and Plácido de Castro are located 65–90 km apart.
Eleven additional P. falciparum isolates were collected from unrelated febrile patients, between March and May 2008, in Plácido de Castro (10°20′S, 67°11′W), a town situated 65–90 km southwest of Granada. Plasmodium falciparum infections were treated with mefloquine or quinine plus doxycycline (Ministry of Health of Brazil, 2001). Genomic DNA was isolated as described (Ferreira et al. 2007). Patients or guardians provided written informed consent, and the research protocol was approved by the ethics review board of the Institute of Biomedical Sciences, University of São Paulo (318/2002 and 538/2004).
Microsatellite typing
Eleven single-copy microsatellites with trinucleotide repeats were typed (chromosome location in brackets) : C2M3 [2], Polyα [4], TAA42 and TA81 [5], TA87 and TAA109 [6], 2490 [10], ARA2 [11], PfG377 and PfPK2 [12], and TAA60 [13]. Alleles were PCR-amplified with the oligonucleotide primers (Anderson et al. 1999; Su et al. 1999) listed in Table 1. The following amplification protocol was used: reaction mixtures (15 μl) contained 3 μl of DNA template, 0·2 units of Taq polymerase (Fermentas, Vilnius, Lithuania), 1·5 μl 10× buffer, 2 mM Mg2+, 0·2 mM of each dNTP, and 2 pM of each primer. Cycling conditions for all loci were as follows: 2 min, 94 °C; (30 sec, 94 °C; 30 sec, 42 °C; 30 sec, 40 °C; 30 sec, 65 °C) × 40 cycles; 5 min, 65 °C. After PCR amplification, products were pooled as follows (TAA60+ARA2, PfG377+TAA87, PfPK2+TAA109, TAA81+TAA42, and Polyα+ 2490+C2M3) according to their sizes and labels. Fragment size was measured on an ABI310 (Applied Biosystems, Foster City, USA) DNA sequencer. GeneScan 500-ROX (Applied Biosystems) was used as an internal size standard. The relative abundance of alleles (peak heights in electropherograms) was determined using STR and software (http://www.vgl.ucdavis.edu/informatics/strand.php). We scored 2 alleles at a locus when the minor peak was more than one-third the height of the predominant peak; infections were considered to contain multiple clones if 1 or more loci showed more than 1 allele (Anderson et al. 2000). Multilocus haplotypes, which characterize parasite lineages, were defined as unique combinations of alleles at each locus analysed; only the predominant alleles were considered for haplotype assignment in multiple-clone infections (Anderson et al. 1999).
Table 1.
Set of oligonucleotide primers (F, forward; R, reverse) used to amplify 11 microsatellite loci of Plasmodium falciparum
(The fluorescent dyes used to label primers were 6-FAM (6-carboxyfluorescein, ‘blue’ label), VIC (Applied Biosystems proprietary ‘green’ fluorescent dye), and NED (Applied Biosystems proprietary ‘yellow’ fluorescent dye).)
| Name | 5′ Label | Sequence 5′–3′ | GenBank Accession number | Chromosome |
|---|---|---|---|---|
| Polyα-F | AAAATATAGACGAACAGA | G37809 | 4 | |
| Polyα-R | VIC | ATCAGATAATTGTTGGTA | ||
| TAA60-F | VIC | CTCAAAGAAAAATAATTCA | G38876 | 13 |
| TAA60-R | AAAAAGGAGGATAAATACAT | |||
| ARA2-F | NED | GTACATATGAATCACCAA | G37848 | 11 |
| ARA2-R | GCTTTGAGTATTATTAATA | |||
| PfG377-F | GATCTCAACGGAAATTAT | G37851 | 12 | |
| PfG377-R | NED | TTATGTTGGTACCGTGT | ||
| PfPK2-F | CTTTCATCGATACTACGA | G37852 | 12 | |
| PfPK2-R | NED | CCTCAGACTGAAATGCAT | ||
| TAA87-F | VIC | ATGGGTTAAATGAGGTACA | G38838 | 6 |
| TAA87-R | ACATGTTCATATTACTCAC | |||
| TAA109-F | 6-FAM | TAGGGAACATCATAAGGAT | G38842 | 6 |
| TAA109-R | CCTATACCAAACATGCTAAA | |||
| TAA81-F | 6-FAM | GAAGAAATAAGGGAAGGT | G38836 | 5 |
| TAA81-R | TTTCACACAACACAGGATT | |||
| TAA42-F | VIC | ACAAAAGGGTGGTGATTCT | G38832 | 5 |
| TAA42-R | GTATTATTACTACTACTAAAG | |||
| 2490-F | TTCTAAATAGATCCAAAG | G37790 | 10 | |
| 2490-R | 6-FAM | ATGATGTGCAGATGACGA | ||
| C2M3-F | GGTTAATATGATCACAAAATG | G37900 | 2 | |
| C2M3-R | NED | ATTGTTGATTCATGAAATGCA |
MSP-2 genotyping
To compare the dynamics of neutral and non-neutral polymorphisms in the same parasite population, we also examined variation at the MSP-2 locus of P. falciparum, which encodes a major target of variant-specific naturally acquired immunity. MSP-2 is encoded by highly divergent alleles grouped into dimorphic families or lineages known as FC27 and 3D7 (Ferreira and Hartl, 2007). Naturally acquired antibodies to MSP-2 recognize predominantly variable domains and display little cross-reactivity between dimorphic families (Taylor et al. 1995; Flück et al. 2004; Tonon et al. 2004). Nested PCR amplification and digestion with the restriction enzyme HinfI (New England Biolabs, Ipswich, USA) were used to classify the MSP-2 gene of all P. falciparum isolates into the dimorphic allelic groups. An MSP-2 fragment of ~380–740 base pairs (bp) was amplified by nested PCR with oligonucleotide primer pairs that target conserved domains on blocks 1 and 5. Primer sequences and amplification protocols are described in detail elsewhere (Felger et al. 1999). PCR products were digested with HinfI for 2 h at 37 °C and separated on 10% polyacrylamide gels. The family-specific restriction fragments resulting from HinfI digestion of FC27-type PCR products are 2 fragments of 137 bp and 115 bp, while those resulting from the digestion of 3D7-type alleles are 2 fragments of 70 bp and 108 bp. Multiple-clone infections with parasites from both families can be identified by the presence of both FC27-type and 3D7-type restriction patterns (Felger et al. 1999).
Data analysis
We used the virtual heterozygosity estimate (HE) as a measure of overall genetic diversity. It is defined as HE=[n/(n−1)][1−Σpi 2], where n is the number of isolates analysed and pi is the frequency of the i-th allele in the population. HE gives the average probability that a pair of alleles randomly obtained from the population is different. In diploid organisms, this would give the average probability of having heterozygotes at that given locus. Virtual heterozygosity ranges between 0 and 1.
The standardized index of association (ISA) was used to test for evidence of overall multilocus linkage disequilibrium in parasite populations. This test compares the variance (VD) of the number of alleles shared between all pairs of haplotypes observed in the population (D) with the variance expected under random association of alleles (VE) as follows: ISA= (VD/VE−1) (r − 1), where r is the number of loci analysed. VE is derived from 10 000 simulated data sets in which alleles were randomly reshuffled among haplotypes. Significant linkage disequilibrium is detected if VD is greater than 95% of the values derived from the reshuffled data sets. Data were analysed with LIAN 3.5 software (Haubold and Hudson, 2000) available at : http://pubmlst.org/perl/mlstanalyse/mlstanalyse.pl?site=pubmlst&page=lian&referer=pubmlst.org. Unique haplotypes were analysed separately to distinguish between clonal and ‘epidemic’ population structures of parasites (Smith et al. 1993).
To represent the relationships between multilocus microsatellite haplotypes, we used a simple phylogenetic approach that is appropriate when little meiotic recombination occurs in the population, as indicated by the high levels of multilocus linkage disequilibrium observed in our parasite sample. We built a neighbour-joining tree with MEGA 4.0 software (http://www.megasoftware.net/) using the proportions of alleles shared between haplotypes as a measure of genetic relatedness.
To test whether microsatellite haplotypes clustered according to the date of collection of isolates, we applied the model-based clustering algorithm implemented in the Structure 2.2 software (Pritchard et al. 2000), available at : http://pritch.bsd.uchicago.edu/software/structure2_1.html. This software uses a Bayesian clustering approach to assign isolates to K populations characterized by a set of allele frequencies at each locus. We run the program 10 times at each of 6 different K values (from 1 to 6) with a burn-in period of 50 000 iterations followed by 105 iterations. We used the admixture model, in which individuals may have ancestry in 2 or more populations.
We further tested whether the pairwise genetic relatedness (proportion of alleles shared between haplotypes) decreased with increasing distance between the dates of sample collection, by using the Mantel nonparametric matrix correlation test (Mantel, 1967) implemented in Poptools 2.7.1 software (available at : http://www.cse.csriro.au/poptools/). The significance of the correlation coefficient (r) was tested with 10 000 permutations.
RESULTS
Plasmodium falciparum populations in Granada were less diverse than sympatric P. vivax isolates, with less multiple-clone infections, a lower number of alleles per locus and lower virtual heterozygosity (HE) (Table 2). Both P. falciparum and P. vivax populations in Granada showed highly significant multilocus linkage disequilibrium. We found 31 unique haplotypes in 44 isolates from Granada. The linkage disequilibrium for both parasite species remained significant when each haplotype was counted once (Table 2), indicating that recent epidemic expansions of particular lineages (Smith et al. 1993) do not account for the linkage observed. Of 5 pairs of samples from the same subject, 2 pairs (419m2 and 419m3, collected 34 days apart, and 458 and 458 m, collected 21 days apart) shared the same multilocus haplotype (Fig. 2). This pattern is consistent with parasite recrudescence after treatment.
Table 2.
Proportions of mixed-clone infections, mean number of microsatellite alleles per locus, genetic diversity (virtual heterozygosity [HE]), and linkage disequilibrium (standardized index of association [IS A]) in malaria parasite populations from rural Amazonia
| ISA by infection typed |
|||||
|---|---|---|---|---|---|
| Parasite species | Multiple-clone infections (%) | Alleles per locus, mean±S.E. (range) | HE mean±S.E. (range) | All | Those with unique haplotypes |
| P. falciparuma | 13·9 | 2·8±0·4 (2–4) | 0·43±0·06 (0·09–0·67) | 0·088 (n=44) | 0·032 (n=31) |
| P. falciparumb | 21·8 | 3·3±0·4 (2–5) | 0·46±0·04 (0·20–0·68) | 0·070 (n=55) | 0·028 (n=40) |
| P. vivaxc | 49·0 | 6·9±0·8 (3–14) | 0·71±0·04 (0·34–0·88) | 0·202 (n=49) | 0·119 (n=39) |
44 samples collected in Granada between 2004 and 2006.
44 samples collected in Granada between 2004 and 2006 and 11 samples collected in Plácido de Castro in 2008.
49 samples collected in Granada between 2004 and 2005 and genotyped with 14 microsatellites (Ferreira et al. 2007).
All IS A values were significantly larger than zero (P = 0·001), denoting significant multilocus linkage disequilibrium.
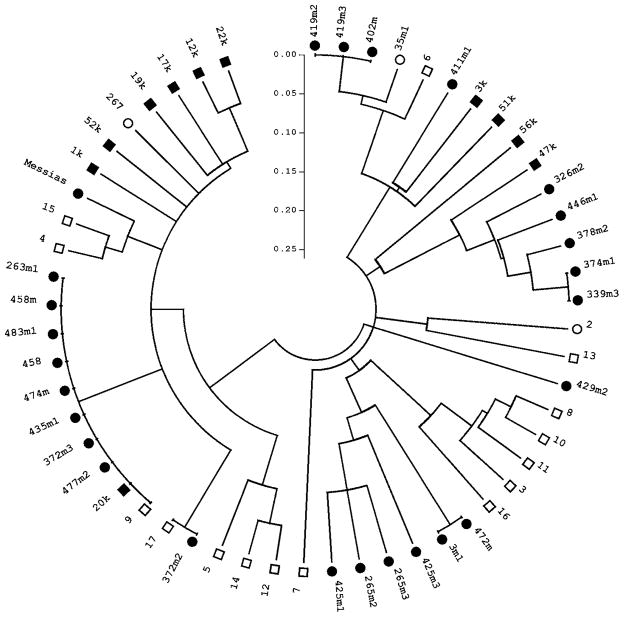
Fig. 2.
Linearized, circular neighbour-joining tree showing the relationships between multilocus microsatellite haplotypes derived from 55 Plasmodium falciparum isolates from Acre, Brazil. The genetic distance between pairs of haplotypes was estimated as 1 – the proportion of shared alleles. Different symbols at the branch tips show the date of collection of the parasites: 2004, black circles; 2005, white circles; 2006, white squares; and 2008, black squares. Terminal branches of zero length indicate parasites with identical multilocus haplotypes. The most common haplotype was shared by 9 isolates collected in Granada in 2004 and 2006 and by 1 isolate collected in Plácido de Castro in 2008.
Overall, 44 unique haplotypes were found in the whole P. falciparum dataset (55 isolates from Granada and Plácido de Castro), with 4 haplotypes shared by 2 isolates each, 1 shared by 3 isolates, and 1 shared by 10 isolates (Fig. 2). Significant linkage disequilibrium was also found in the whole P. falciparum dataset (Table 2).
Visual inspection of Fig. 2 does not suggest that haplotypes cluster according to the date of collection of isolates, with 2 haplotypes shared by isolates collected in different years. Fig. 3 shows the clustering patterns obtained by Bayesian analysis with 3 populations, the K value associated with the strongest statistical support (P=0·999). Each bar partitioned into 3 segments in Fig. 3 represents an isolate. The segments represent the estimated fractions of ancestry in each of the 3 populations.
Fig. 3.
Temporal structure of Plasmodium falciparum diversity, as inferred from microsatellite typing of 55 isolates from Acre, Brazil. Each individual parasite is represented by a bar, which is partitioned into 3 segments (black, white and hatched) that represent the membership (or ancestry) fraction of each individual in each of the 3 populations inferred by the analysis. Parasites are ordered according to their dates of collection. Note that identical or nearly identical isolates (as inferred by identical or similar membership fractions) were collected at rather different time-points.
We defined isolates with predominant (>70%) ancestry in 1 of the 3 populations as members of that particular population. For example, the 15 isolates with predominant ancestry in the ‘black’ population shown in Fig. 3 (including 10 isolates with identical multilocus haplotype) are considered hereafter members of the ‘black’ population; when no clear predominant ancestry could be characterized, the isolates were considered to have a ‘mixed ancestry’. Therefore, each isolate was classified as a member of the ‘black’, ‘white’, ‘hatched’ or ‘mixed ancestry’ groups. Figure 4A shows the number of isolates belonging to each population according to their dates of collection.
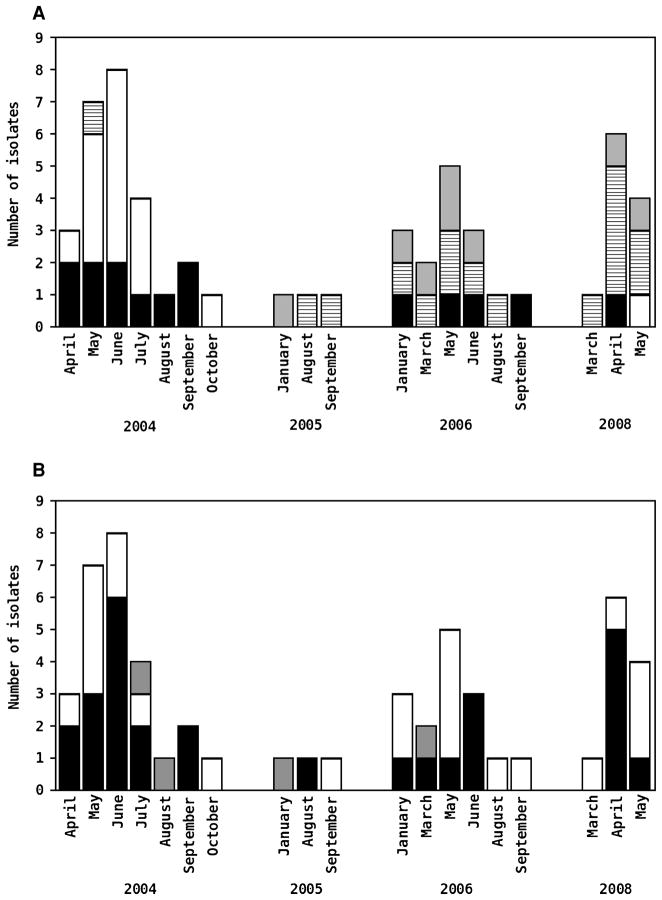
Fig. 4.
Monthly distribution of Plasmodium falciparum variants characterized in 55 isolates collected in Acre, Brazil, between April 2004 and May 2008. In the upper panel (A), isolates are grouped according to the populations in which they have predominant ancestry. They are considered as members of 1 of the 3 populations (black, white or hatched shading) if they had a membership fraction of 70% or greater in this population. When no predominant ancestry could be defined, the isolates were considered to have a ‘mixed ancestry’ and were represented with grey shading. In the lower panel (B), isolates are grouped according to the dimorphic type (either FC27 or 3D7) characterized at their MSP-2 locus. Black shading indicates FC27-type parasites, no shading (white boxes) represents 3D7-type parasites and grey shading represents mixed-clone infections with both FC27-type and 3D7-type parasites. The figure shows the number of isolates belonging to each variant according to their dates of collection.
The ‘black’ population persisted in the study area, being represented in the samples collected in 2004, 2006, and 2008 (Fig. 4A). The ‘hatched’ population, however, was initially rare but became common in 2005–6 and predominated in 2008. An opposite pattern was seen for the ‘white’ population, which predominated in 2004 but became rare thereafter. We found a weak, although statistically significant, negative correlation between pairwise genetic similarity and time distance (interval between dates of collection of isolates), by applying the Mantel nonparametric test to the whole dataset (n=55) (r=−0·097, P=0·02). Nevertheless, the correlation is attenuated when the 11 samples collected in Plácido de Castro in 2008 are removed from the dataset (r=−0·043, P=0·186), suggesting that the geographical distance between Granada and Plácido de Castro, in addition to temporal distance, may have further contributed to the genetic differentiation among isolates.
Both FC27-type and 3D7-type MSP-2 alleles were found throughout the study period, with no clear temporal variation in the relative proportion of each type (Fig. 4B). Therefore, we found no evidence for replacement of MSP-2 variants over time in our population. Overall, 28 (50·9%) infections had FC27-type parasites, 23 (41·8%) had 3D7-type parasites, and 4 (7·3%) were mixed-clone infections with parasites of both dimorphic types.
DISCUSSION
Whether relative frequencies of antigenic variants vary over time has been investigated for several P. falciparum populations since the pioneering study of Forsyth et al. (1988). The patterns observed have been interpreted as evidence either for or against frequency-dependent selection (Conway, 1997). For example, clear changes in the temporal distribution of variants of MSP-1, a major vaccine candidate antigen, have been found in prospective studies in Brazil (Da Silveira et al. 1999), but not in areas with substantially higher levels of endemicity such as Gambia (Conway et al. 1992) and Vietnam (Ferreira et al. 1998). Conversely, the frequencies of variants of MSP-2 have been shown to be temporally stable in Brazil (Tonon et al. 2004 and Fig. 4B) and Gambia (Conway et al. 1992) but highly variable in Irian Jaya, Indonesia (Eisen et al. 1998). Nevertheless, diversity in T-cell epitopes of the circumsporozoite protein (CSP), the immunodominant antigen of P. falciparum sporozoites, has been shown to be remarkably stable over time in both Thailand (Kumkhaek et al. 2005) and Vietnam (Jalloh et al. 2006). These previous studies, however, differ in both temporal (months or years) and geographical scales (villages or provinces), precluding further comparisons with our community-based survey in a well-defined area of low malaria endemicity.
Differences in migration, mutation and recombination rates, random genetic drift and natural selection patterns all can affect the population dynamics of polymorphisms. Even when only antigenic determinants under diversifying selection are considered, contrasting patterns of antigenic variation over time can be observed for different human pathogens (Lipsitch and O’Hagan, 2007).
We found few microsatellite haplotypes persisting for several months in Granada, arguing for a high replacement rate. The strong linkage disequilibrium between alleles of markers on different chromosomes indicates a low rate of meiotic recombination in local malaria parasites. One particularly abundant haplotype, however, was recovered from 10 of 55 (18·2%) P. falciparum isolates collected over 4 years (April 2004 to May 2008). Similarly, the same multilocus haplotype was found in 8 of 49 (16·3%) P. vivax isolates from Granada collected between March 2004 and May 2005, despite the fast haplotype turnover (Ferreira et al. 2007). Although 4 of 11 microsatellite markers used to type P. falciparum isolates map to coding sequences, none of them map to known surface antigens putatively under strong diversifying selection.
We also investigated patterns of diversity across time for MSP-2, a P. falciparum antigen that is expected to be under frequency-dependent selection (Eisen et al. 1998). In fact, immune-mediated variant replacement has been well characterized during a clinical trial of a multivalent vaccine prototype containing the 3D7 variant of MSP-2. This vaccine partially protected Papua New Guineans from infection with parasites carrying the homologous (3D7-type) version of this antigen and, as a consequence, most infections observed during the follow-up of vaccine recipients were with parasites carrying the heterologous (FC27-type) version ofMSP-2 (Genton et al. 2002). Contrasting with the high replacement rate of microsatellite haplotypes, however, the prevalence of the 2 dimorphic types ofMSP-2 in our population remained remarkably stable throughout the study period.
We therefore suggest that the fast turnover of P. falciparum haplotypes found in our population may represent the typical population dynamics of neutral polymorphisms in small populations. Understanding temporal variation in allelic frequencies of putatively neutral polymorphisms is essential to ascribe a particular pattern of serotype replacement to either evolutively neutral stochastic processes or natural selection. In other words, ‘we need to know the background genetic noise in the neutral genes of the parasite before we can understand the signals of selection ’ (Awadalla et al. 2001). Further analyses of additional neutral genetic markers, such as intergenic or synonymous single-nucleotide polymorphisms (SNP), are required to confirm whether the patterns described here are shared by other polymorphisms that differ from microsatellites in their mode of evolution. Significant differences in haplotype composition of parasites over time, detected with highly polymorphic markers, may remain undetected with more conserved markers and do not necessarily translate into biologically significant differences among populations (Hedrick, 1999). The recent availability of a high-throughput platform for genome-wide SNP typing (Neafsey et al. 2008) provides a powerful tool to examine temporal and spatial patterns of variation of these less polymorphic markers and their potential relevance across worldwide P. falciparum populations.
Acknowledgments
This study was funded by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, 470570/2006–7), Fundação de Amparo á Pesquisa do Estado de São Paulo (FAPESP, 07/51199-0), and National Institutes of Health of USA (NIH, RO1 AI 075416-01). P.O.S., N.S.dS., and M.U.F. receive scholarships from CNPq; Md.S.N., K.K.G.S., and R.M.G. receive or received scholarships from FAPESP. We thank the population of Granada for their enthusiastic participation in the study; Adamílson L. de Souza, Valdecir E. da Silva, Maria José Menezes, and Pascoal T. Muniz for help in fieldwork; and Guilherme L. S. F. Renoldi (CNPq scholarship recipient) and Carmen S. A. Takata for help with microsatellite typing.
References
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Molecular Biology and Evolution. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/S0031182099004552. [DOI] [PubMed] [Google Scholar]
- Awadalla P, Walliker D, Babiker H, Mackinnon M. The question of Plasmodium falciparum population structure. Trends in Parasitology. 2001;17:351–353. doi: 10.1016/S1471-4922(01)02034-7. [DOI] [PubMed] [Google Scholar]
- Conway DJ. Natural selection on polymorphic malaria antigens and the search for a vaccine. Parasitology Today. 1997;13:26–29. doi: 10.1016/S0169-4758(96)10077-6. [DOI] [PubMed] [Google Scholar]
- Conway DJ, Greenwood BM, McBride JS. Longitudinal study of Plasmodium falciparum polymorphic antigens in a malaria-endemic population. Infection and Immunity. 1992;60:1122–1127. doi: 10.1128/iai.60.3.1122-1127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva-Nunes M, Codeço CT, Malafronte RS, da Silva NS, Juncansen C, Muniz PT, Ferreira MU. Malaria on the Amazonian frontier : transmission dynamics, risk factors, spatial distribution, and prospects for control. American Journal of Tropical Medicine and Hygiene. 2008;79:624–635. [PubMed] [Google Scholar]
- da Silveira LA, Dorta ML, Kimura EA, Katzin AM, Kawamoto F, Tanabe K, Ferreira MU. Allelic diversity and antibody recognition of Plasmodium falciparum merozoite surface protein 1 during hypoendemic malaria transmission in the Brazilian Amazon region. Infection and Immunity. 1999;67:5906–5916. doi: 10.1128/iai.67.11.5906-5916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen D, Billman-Jacobe H, Marshall VF, Fryau D, Coppel RL. Temporal variation of the merozoite surface protein-2 gene of Plasmodium falciparum. Infection and Immunity. 1998;66:239–246. doi: 10.1128/iai.66.1.239-246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnert A. Plasmodium falciparum population dynamics: only snapshots in time? Trends in Parasitology. 2008;24:340–344. doi: 10.1016/j.pt.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Felger I, Irion A, Steiger S, Beck H-P. Epidemiology of multiple Plasmodium falciparum infections. 2. Genotypes of merozoite surface protein 2 of Plasmodium falciparum in Tanzania. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1999;93(Suppl 1):S1/3–S1/9. doi: 10.1016/s0035-9203(99)90320-6. [DOI] [PubMed] [Google Scholar]
- Ferreira MU, Hartl DL. Plasmodium falciparum: Worldwide sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-2 (MSP-2) Experimental Parasitology. 2007;115:32–40. doi: 10.1016/j.exppara.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Ferreira MU, Karunaweera ND, Da Silva-Nunes M, Da Silva NS, Wirth DF, Hartl DL. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. Journal of Infectious Diseases. 2007;195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- Ferreira MU, Liu Q, Zhou M, Kimura M, Kaneko O, Van Thien H, Isomura S, Tanabe K, Kawamoto F. Stable patterns of allelic diversity at the merozoite surface protein-1 locus of Plasmodium falciparum in clinical isolates from southern Vietnam. Journal of Eukaryotic Microbiology. 1998;45:131–136. doi: 10.1111/j.1550-7408.1998.tb05080.x. [DOI] [PubMed] [Google Scholar]
- Flück C, Smith T, Beck H-P, Irion A, Betuela I, Alpers MP, Anders RF, Saul A, Genton B, Felger I. Strain-specific humoral response to a polymorphic malaria vaccine. Infection and Immunity. 2004;72:6300–6305. doi: 10.1128/IAI.72.11. 6300-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth KP, Anders RF, Kemp DJ, Alpers MP. New approaches to the serotypic analysis of the epidemiology of Plasmodium falciparum. Philosophical Transactions of the Royal Society of London, B. 1988;321:485–493. doi: 10.1098/rstb.1988.0104. [DOI] [PubMed] [Google Scholar]
- Genton B, Betuela L, Felger I, Al-Yaman F, Anders RF, Saul A, Rare L, Baisor M, Lorry K, Brown GV, Pye D, Irving DO, Smith TA, Beck H-P, Alpers MP. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1–2b trial in Papua New Guinea. Journal of Infectious Diseases. 2002;185:820–827. doi: 10.1086/339342. [DOI] [PubMed] [Google Scholar]
- Gupta S, Ferguson N, Anderson R. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science. 1998;280:912–915. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- Gupta S, Maiden MC. Exploring the evolution of diversity in pathogen populations. Trends in Microbiology. 2001;9:181–185. doi: 10.1016/S0966-842X(01)01986-2. [DOI] [PubMed] [Google Scholar]
- Gupta S, Swinton J, Anderson RM. Theoretical studies of the effects of heterogeneity in the parasite population on the transmission dynamics of malaria. Proceedings of the Royal Society of London, B. 1994;256:231–238. doi: 10.1098/rspb.1994.0075. [DOI] [PubMed] [Google Scholar]
- Haubold B, Hudson RR. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics. 2000;16:847–848. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Perspective: Highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Jalloh A, Van Thien H, Ferreira MU, Ohashi J, Matsuoka H, Kanbe T, Kikuchi A, Kawamoto F. Sequence variation in the T-cell epitopes of the Plasmodium falciparum circumsporozoite protein among field isolates is temporally stable: a 5-year longitudinal study in southern Vietnam. Journal of Clinical Microbiology. 2006;44:1229–1235. doi: 10.1128/JCM.44.4.1229-1235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumkhaek C, Phra-Ek K, Renia L, Singhasivanon P, Looareesuwan S, Hirunpetcharat C, White NJ, Brockman A, Gruner AC, Lebrun N, Alloueche A, Nosten F, Khusmith S, Snounou G. Are extensive T cell epitope polymorphisms in the Plasmodium falciparum circumsporozoite antigen, a leading sporozoite vaccine candidate, selected by immune pressure? Journal of Immunology. 2005;175:3935–3939. doi: 10.4049/jimmunol.175.6.3935. [DOI] [PubMed] [Google Scholar]
- Lipsitch M, O’Hagan JJ. Patterns of antigenic diversity and the mechanisms that maintain them. Journal of the Royal Society, Interface. 2007;4:787–802. doi: 10.1098/rsif.2007.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Ministry of Health of Brazil. Manual of Antimalarial Therapeutics. Ministry of Health of Brazil; Brasilia: 2001. [Google Scholar]
- Neafsey DE, Schaffner SF, Volkman SK, Park D, Montgomery P, Milner DA, Jr, Lukens A, Rosen D, Daniels R, Houde N, Cortese JF, Tyndall E, Gates C, Stange-Thomann N, Sarr O, Ndiaye D, Ndir O, Mboup S, Ferreira MU, Moraes Sdo L, Dash AP, Chitnis CE, Wiegand RC, Hartl DL, Birren BW, Lander ES, Sabeti PC, Wirth DF. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biology. 2008;9:R171. doi: 10.1186/gb-2008-9-12-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O’Rourke M, Spratt BG. How clonal are bacteria? Proceedings of the National Academy of Sciences, USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ferdig MT, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- Taylor RR, Smith DB, Robinson VJ, McBride JS, Riley EM. Human antibody response to Plasmodium falciparum merozoite surface protein 2 is serogroup specific and predominantly of the immunoglobulin G3 subclass. Infection and Immunity. 1995;63:4382–4388. doi: 10.1128/iai.63.11.4382-4388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonon AP, Hoffmann EH, Silveira LA, Ribeiro AG, Goncalves CR, Ribolla PE, Wunderlich G, Ferreira MU. Plasmodium falciparum: sequence diversity and antibody recognition of the merozoite surface protein-2 (MSP-2) in Brazilian Amazonia. Experimental Parasitology. 2004;108:114–125. doi: 10.1016/j.exppara.2004.08.001 . Population dynamics of Plasmodium falciparum 1105. [DOI] [PubMed] [Google Scholar]